Introduction
The tumor microenvironment consists of extracellular
matrix, stromal cells (for example, endothelial cells, fibroblasts,
myofibroblasts and leukocytes), intratumoral hypoxia and nutrient
starvation, and cytokines, chemokines and proteins secreted by
epithelial, cancer or stromal cells (1,2).
Cytokines, chemokines and growth factors, including interleukin
(IL) superfamily, form complex immune signaling networks and have
important roles in various aspectes of cancer initiation and
progression (2–4). Several cytokines including IL-2,
IL-12, TNF-α, type I IFNs and GM-CSF possess anticancer potential
via intratumoral delivery (2,4,5).
IL-2, which has antitumor functions that include the activation of
natural killer (NK) and cytotoxic T cells, can induce durable
remissions in 5–10% of patients with metastatic melanoma and renal
cell carcinoma, malignancies with poor prognoses (6). During an immune response, cancer
cells encounter extrinsic and intrinsic factors, including
oxidative stress, nutrient availability, and inflammation, that can
modulate their capacity to activate, proliferate, and survive.
Autophagy occurs at a constitutive basal level, but
it can be enhanced in response to various types of stress, mainly
including oxygen tension, nutrient deprivation and chemicals such
as rapamycin, all of which can inhibit mechanistic target of
rapamycin (mTOR) pathway and thus initiate autophagy (7–9).
Steps in the autophagy pathway involve nucleation of targeted
macromolecules on the ER membrane, trafficking of autophagosomes to
lysosomes and, finally, fusion of the autophagosome-lysosome,
resulting in targeted protein degradation (10,11).
This process is controlled by the products of numerous
autophagy-specific genes (ATGs). A key regulator of autophagy is
microtubule-associated protein 1 light chain 3 (LC3, a mammalian
homolog of yeast Atg8), which controls major steps in the
autophagic pathway including the growth of autophagic membranes,
recognition of autophagic cargoes, and the fusion of autophagosomes
with lysosomes (12). Recent
studies demonstrate that the phosphorylation of LC3 at threonine 50
(Thr50) plays a critical role in mediating fusion of autophagosomes
with lysosomes (13).
Increasing evidence shows that the posttranslational
modifications (PTMs), including phosphorylation, acetylation,
methylation, and ubiquitination, are important for regulating the
autophagy process by providing structural and functional diversity
among proteins (12,14–16).
The majority of research is dedicated to the PTMs of key
autophagy-related molecules containing autophagy receptors,
providing a layer of regulation for the specificity and efficiency
of selective autophagy. Selective autophagy refers to the selective
degradation of, for instance, organelles (mitophagy and pexophagy),
bacteria (xenophagy), ribosomes, macromolecular structures,
specific proteins and protein aggregates (aggrephagy) by autophagy
(17). Optineurin, an autophagy
receptor, is phosphorylated by the protein kinase TBK1 (TANK
binding kinase 1) at serine 177, which enhances the LC3 binding
affinity and autophagic clearance of cytosolic Salmonella
enterica (18). However,
little is known about the PTMs of substrates determining their
specific recognition by autophagy receptors. NDP52, which is an
autophagy adaptor that contains an LC3-interacting region (LIR)
motif (17), is induced by Nrf2
and specifically directs phosphorylated tau to the autophagic
degradative pathway (19). Mad1, a
member of the Myc/Max/Mad family, can also be phosphorylated at
serine 145 and introduced into the autophagic degradation (20). Interestingly, EPG-11/PRMT-1
directly methylates arginines in the RGG domains of PGL-1 and PGL-3
and promotes their autophagic removal in C. elegans during
embryogenesis (21). However, the
link between arginine methylation and selective autophagy has not
been clearly demonstrated in cancer.
Arginine methylation is one common PTM mainly of
nuclear proteins in eukaryotic cells, and is catalyzed by a family
of enzymes termed protein arginine methyltransferases (PRMTs)
(22,23). Three main forms of methylarginine
have been identified in eukaryotes:
NG-monomethylarginine (MMA), NGNG
(asymmetric) dimethylarginine (aDMA), and
NGNG (symmetric) dimethylarginine (sDMA)
(24). In humans, PRMTs are
classified into type I (PRMT1, PRMT2, PRMT3, PRMT4 and PRMT6), type
II (PRMT5 and PRMT7) and type III (PRMT7) methyltransferases, based
on their corresponding aDMA, sDMA and MMA activities, respectively.
PRMT1 and PRMT5 are the major asymmetric and symmetric arginine
methyltransferases, respectively (25). Arginine methylation has received
increasing attention over the last years as several recent reports
have illustrated a novel role for this posttranslational
modification in regulating protein-protein interaction and
transcriptional induction (26,27),
and is often deregulated in cancer (25). However, how arginine methylation
could be regulated by interleukins or autophagy in the context of
the tumor microenvironment has not yet been investigated.
In this study, we identified for the first time that
p90aDMA, which is a 90-kDa protein of aDMA in the nucleus and
accumulated by IL-2, IL-4 and IL-6, can serve as a unique substrate
for selective autophagy. Conversely, p90sDMA was a 90-kDa protein
of sDMA and accumulated in a dose-dependent manner in response to
rapamycin treatment. Taken together, our study provides evidence
for immunity regulation through crosstalk between arginine
methylation and selective autophagy in the tumor microenvironment
by using in vitro models.
Materials and methods
Chemicals and reagents
Recombinant human IL-2, IL-4, IL-6 and IL-8 were
purchased from Peprotech (NJ, USA). They were dissolved in water to
a concentration of 5 μg/ml. 3-Methyladenine (3-MA) was purchased
from Sigma (MO, USA) using as an inhibitor of autophagy. 3-MA (100
mg) was dissolved in phosphate-buffered saline (PBS) to make a
100-mM stock solution. Adenosine-2,3 dialdehyde (AdOx) was
purchased from Sigma and used as an inhibitor of methyltransferase.
AdOx (5 mg) was dissolved in 0.2 M HCl to make a 10-mM stock
solution. Rapamycin was also purchased from Sigma.
Cell culture
Human cancer cell lines including MDA-MB-231,
MDA-MB-468, MCF-7, SKBR3, T47D and HeLa cells were obtained from
American Type Tissue Culture Collection (ATCC, USA). The cells were
grown in Dulbecco's modified Eagle's medium (DMEM, Gibco, CA, USA)
containing 10% fetal bovine serum (FBS, CA, USA) and 100 U/ml
penicillin-streptomycin (Gibco) in a humidified incubator of 5%
CO2 at 37°C. The standard hypoxic conditions were 1%
O2 and 5% CO2. Hypoxia was done in a
multi-gas incubator chamber with a compact gas oxygen controller
(MCO-5M, Sanyo, Osaka-SHI, Japan) to maintain oxygen concentration
at 1% by injecting a gas mixture of 95% N2 and 5%
CO2.
Small interfering RNA (siRNA)
transfection
siRNA targeting ATG5 (5′-CCAUCAAUCGGAAACUCAUTT-3′)
and negative control siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′), which was
used for normalisation, were synthesised by Genepharma (Shang Hai,
China). For transfection, MCF-7 cells were seeded in 6-well plates
at a density of 2×105 cells/well. After 24 h, cells were
transfected with siRNA (50 pmol) using Lipofectamine®
2000 Reagent (Invitrogen, CA, USA) according to the instructions of
the manufacturer. After 24 h of transfection, cells were treated
with 100 nM rapamycin for another 24 h. Then the cells were
harvested and the knockdown of ATG5 was confirmed by western blot
analysis.
Western blot analysis
Monolayer cultures of respective cell lines at an
80–90% confluence were prepared with pre-cold RIPA lysis and
extraction buffer (Thermo Scientific, CA, USA) containing protease
inhibitor cocktail (Roche, Basel, Switzerland) on ice. The total
cell lysate was centrifuged and the supernatant was denatured by
boiling. Protein concentrations of supernatants were analyzed by
bicinchoninic acid (BCA) assay kit (Beyotime, Nan Tong, China).
Equivalent amounts of total proteins (20 μg) were subjected to
8–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and then transferred to a 0.45-μM PVDF membrane
(Millipore, MD, USA). The membranes were blocked for 2 h in 5%
bovine serum albumin (BSA) at room temperature and incubated with
specific primary antibodies at 4°C overnight. A list of the primary
antibodies used for western blot analysis are characterized in
Table I. Further incubation was
performed with the corresponding horseradish peroxidase-coupled
secondary antibodies (1:10,000, cat. nos. sc-2004 and sc-2005;
Santa Cruz, CA, USA) at room temperature for 2 h. Then the bands
were detected using Super Signal® West Pico
Chemiluminescent Substrate kit (Thermo Scientific), and the results
were recorded using the ChemiDox™ XRS+ system. Relative protein
expression was normalized with β-actin.
 | Table IPrimary antibodies used in western
blot analysis. |
Table I
Primary antibodies used in western
blot analysis.
| Target | Source | Host | Dilution | Catalog |
|---|
| SYM11 | Merck Millipore,
MA, USA | Rabbit | 1:1,000 | 07-413 |
| ASYM24 | Merck Millipore,
MA, USA | Rabbit | 1:500 | 07-414 |
| LC3B | Cell Signaling
Technology, Inc. (CST), MA, USA | Rabbit | 1:1,000 | #3868 |
| ATG5 | Cell Signaling
Technology, Inc. (CST), MA, USA | Rabbit | 1:1,000 | #12994 |
| TBP | Proteintech, Wu
Han, China | Rabbit | 1:500 | 22006-1-AP |
| β-actin | Zen Bioscience,
Cheng Du, China | Mouse | 1:20,000 | 70068 |
Nuclear cytoplasmic fractionation
Nuclear and cytoplasmic extracts were prepared using
NE-PER Nuclear and Cytoplasmic Extraction reagents (Pierce, Thermo
Fisher Scientific, CA, USA). The quality of nuclear and cytoplasmic
extracts was verified by immunoblotting with protein differentially
enriched in the nucleus (TBP) or the cytoplasm (β-actin).
Results
Comparison of basal sDMA or aDMA levels
in breast cancer cell lines
Breast cancer is a heterogeneous group of diseases
with different histological prognostic and clinical aspects
(28). To investigate the
preference for the substrate methylation state, we collected the
lysates of five human breast cancer cell lines and evaluated the
basal aDMA and sDMA using immublotting. Endogenous proteins of aDMA
and sDMA can be specifically recognized by antibodies ASYM24 and
SYM11, respectively. Both SYM11 and ASYM24 antibodies recognized
unknown endogenous proteins containing methylated arginine in
breast cancer cells with molecular mass of 90, 70, 55 and 34 kDa.
Proteins of 90 and 70 kDa recognized by SYM11 were termed p90sDMA
and p70sDMA. Proteins of 90, 70 and 34 kDa recognized by ASYM24
were termed p90aDMA, p70aDMA and p34aDMA. p90sDMA, p90aDMA and
p70aDMA were widely and dominantly expressed in breast cancer cells
but at relatively higher levels in MCF-7 cells and lower levels in
the MDA-MB-231 cells (Fig. 1).
Interestingly, the p90sDMA and p70aDMA proteins were predominantly
expressed in breast cancer cells at relatively higher levels in the
MCF-7, MDA-MB-468, T47D and SKBR3 cells but lower levels in the
MDA-MB-231 cells (Fig. 1). In
contrast, p90aDMA and p70sDMA proteins were constitutively
expressed at a lower level in the breast cancer cell lines
(Fig. 1). As a result, the ratio
of p90sDMA to p70sDMA was higher than the ratio of p90aDMA to
p70aDMA in each line (Fig. 1).
Expression of p90aDMA is specifically
enhanced by IL-2, IL-4 or IL-6 but not by IL-8 in cancer cells
To determine whether the cancer immune
microenvironment enriched with cytokines could mobilize arginine
methylation, we treated MDA-MB-231 cells with IL-2, IL-4, IL-6 or
IL-8 for 24 h because of their action on cancer cell proliferation
(29). As a result, IL-2, IL-4,
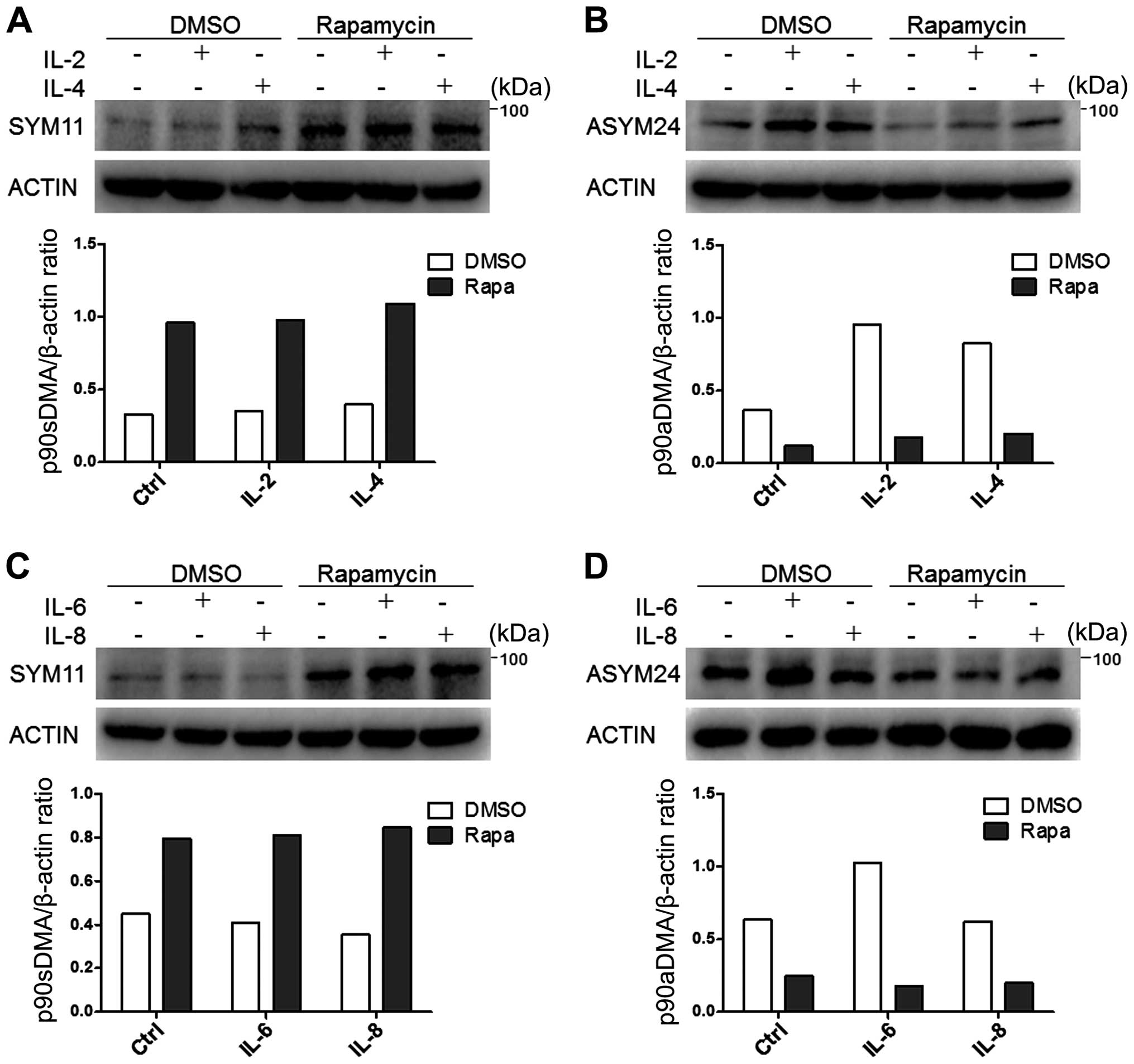
IL-6 but not IL-8 specifically enhanced expression of p90aDMA in
the MDA-MB-231 cells (Fig. 2C and
D). In accordance with Fig. 2B and
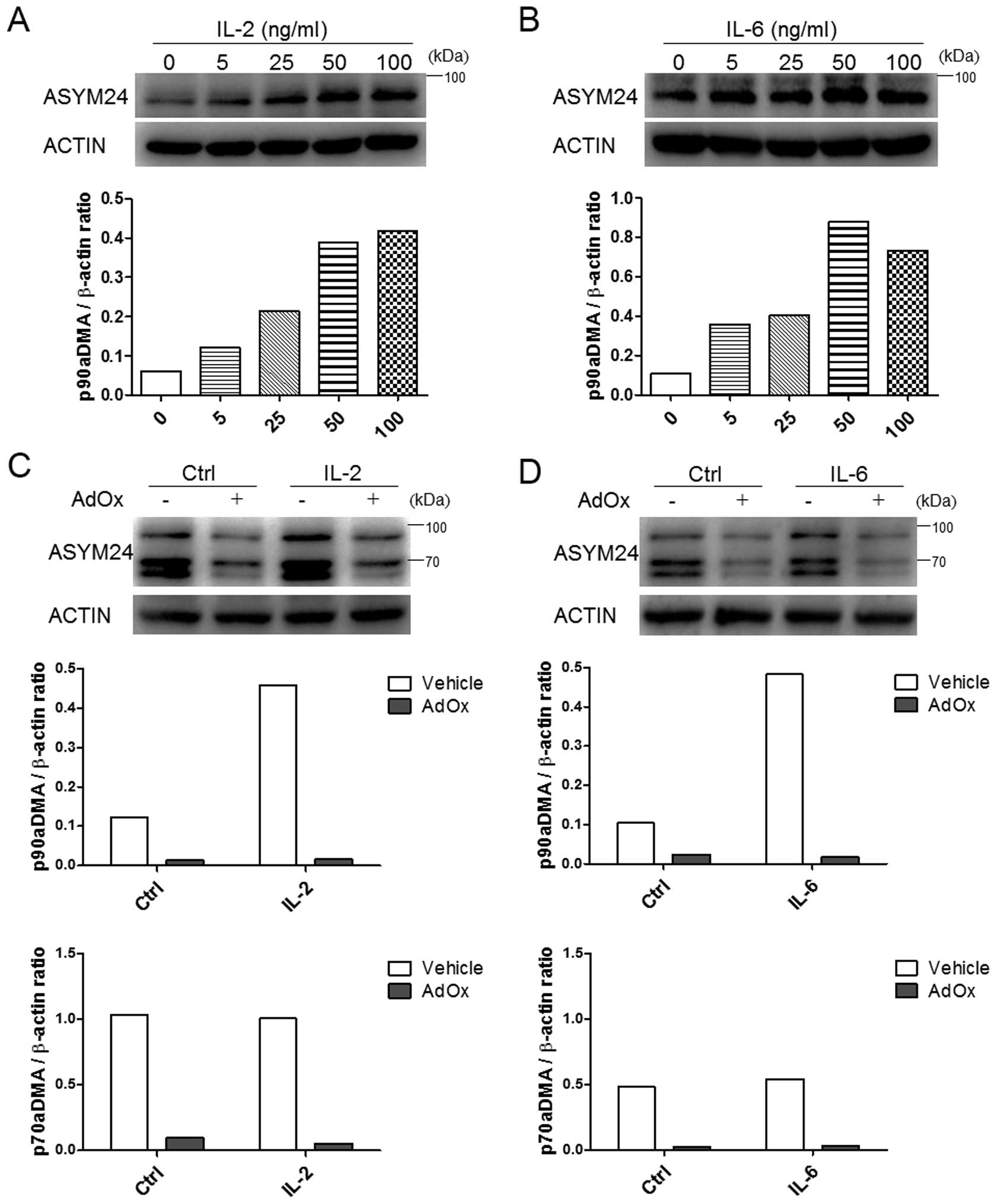
D, we observed that 90aDMA was increased in a dose-dependent
manner upon simulation with IL-2/IL-6 in MDA-MB-231 cells (Fig. 3A and B). However, these cytokines
had no effect on the expressions of p90sDMA, p70sDMA and p70aDMA
(Fig. 2).
To confirm that the methylation signals detected by
the ASYM24 antibody were specific, similar experiments were
performed in IL-2 or IL-6 treated MDA-MB-231 cells incubated with
or without adenosine-2′, 3′-dialdehyde (AdOx), a methyltransferase
inhibitor. It was found that basal or IL-2/6 induced aDMA was
almost blocked in the presence of AdOx (Fig. 3C and D). Of note, IL-2 or IL-6 did
not alter the expression of p70aDMA (Fig. 3C and D). Therefore, it could be
considered that IL-2 and IL-6 specifically induced the accumulation
of p90aDMA in MDA-MB-231 cells. The reason for the specific
increase in p90aDMA is unclear.
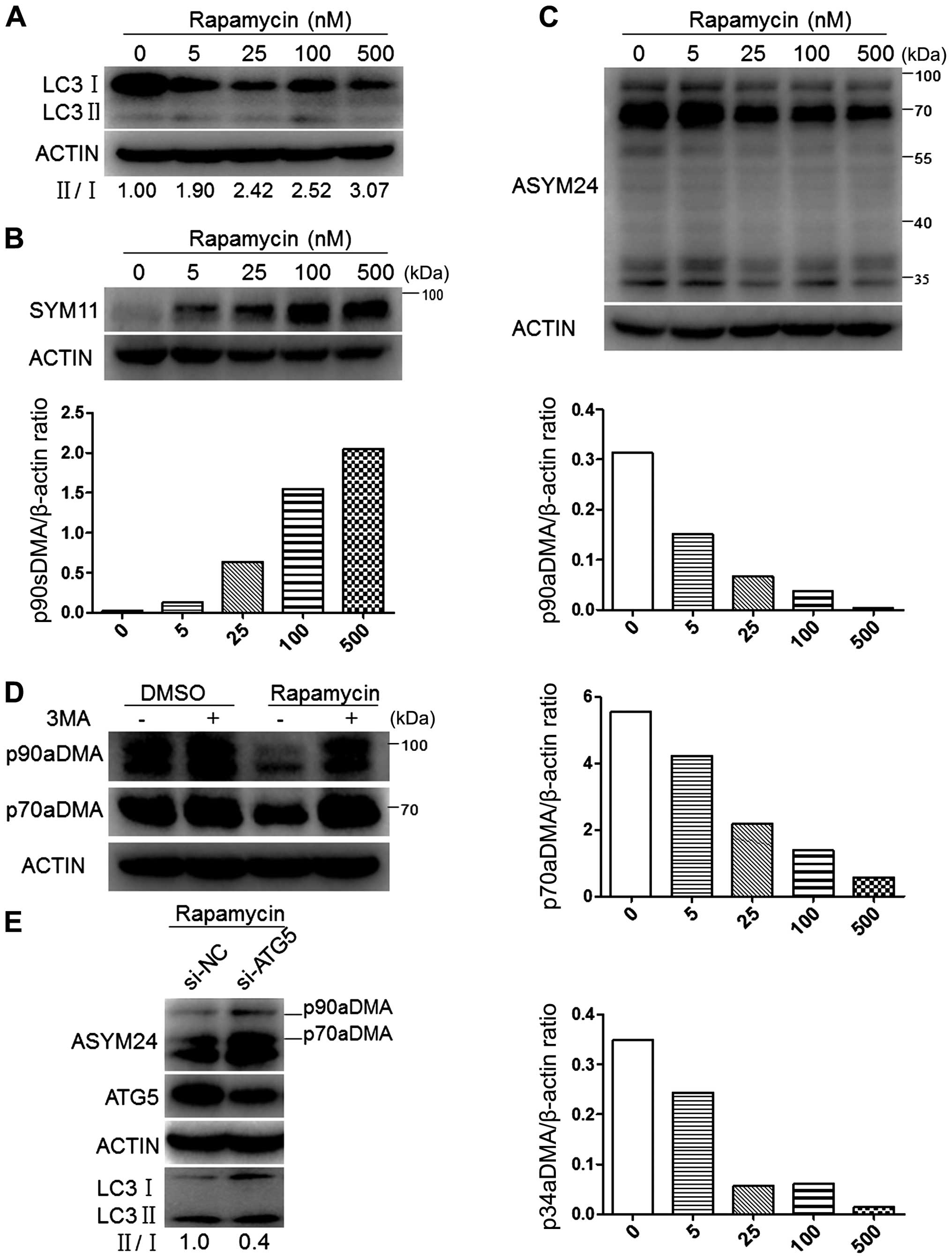
Rapamycin specifically inhibits p90aDMA
expression in cancer cells independent of cytokine stimulation
Arginine methylation has been considered to be an
irreversible post-translational modification until recently.
However, a recent study suggested that autophagy may regulate
arginine methylation (30). Then,
rapamycin, an autophagy enhancer, was administrated in control or
IL-2/4/6/8 treated MDA-MB-231 cells. It was the aDMA levels rather
than the sDMA levels that were significantly reduced regardless of
the interleukin stimulation (Fig.
2). On the contrary, p90sDMA was increased in rapamycin treated
MDA-MB-231 cells (Fig. 2A and C).
These results drew our attention to the possibility that aDMA
serves as a novel and specific signaling molecule that provokes
selective autophagic degradation.
To determine if the increased nuclear p90aDMA
resulting from IL-2, IL-4 or IL-6 exposure could be due to
autophagy inhibition, we monitored whether it affects the ratio of
LC3-II/I, a canonical hallmark of autophagy. MDA-MB-231 cells were
exposed to IL-2, IL-4, IL-6 or IL-8 for 24 h, lysed and subjected
to western blotting for detecting the abundance of autophagic
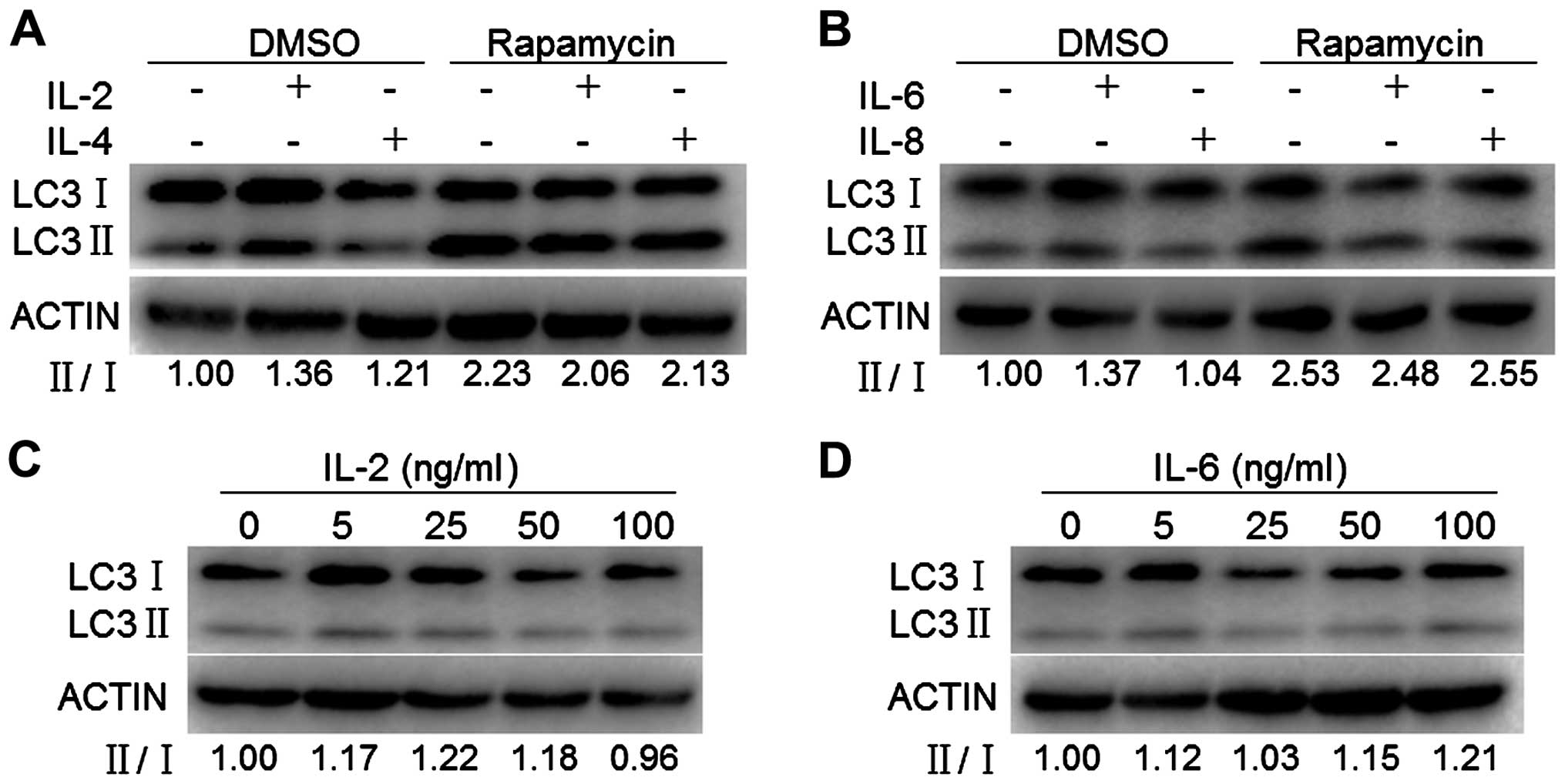
markers. As shown in Fig. 4A and
B, IL-2, IL-4, IL-6 but not IL-8 slightly increased the ratio
of LC3-II/I. To determine the dose curve of IL-2/IL-6 exposure for
the expression of autophagic markers, MDA-MB-231 cells were exposed
to varying doses of IL-2 or IL-6 (5, 25, 100 and 500 μM) for 24 h
and assessed for expression of autophagic markers by western
blotting. The ratio of LC3-II/I exhibited a similar dose-dependent
trend in response to IL-2 or IL-6 with a slight increase of 20% at
25 ng/ml IL-2 and 100 ng/ml IL-6 exposure, respectively (Fig. 4C and D). We next determined whether
IL-2, IL-4, IL-6 or IL-8 affects autophagy induction. We monitored
rapamycin-induced autophagy by assessing conversion of LC3-I to
LC3-II. IL-2, IL-4, IL-6 but not IL-8 slightly decreased conversion
of LC3-I to LC3-II induced by rapamycin (Fig. 4A and B). Taken together, these
findings demonstrated that IL-2, IL-4, IL-6 but not IL-8 slightly
increased the basal autophagy but slightly decreased the
rapamycin-induced autophagy in MDA-MB-231 cells, indicating that
IL-2/IL-4/IL-6 accumulated p90aDMA in an autophagy-independent
pathway.
The aDMA proteins are localized in the
nucleus of cancer cells
Extensive studies have focused on autophagic
turnover of cytoplasmic materials, little is known about the role
of autophagy in degrading nuclear components (31). To determine the cellular
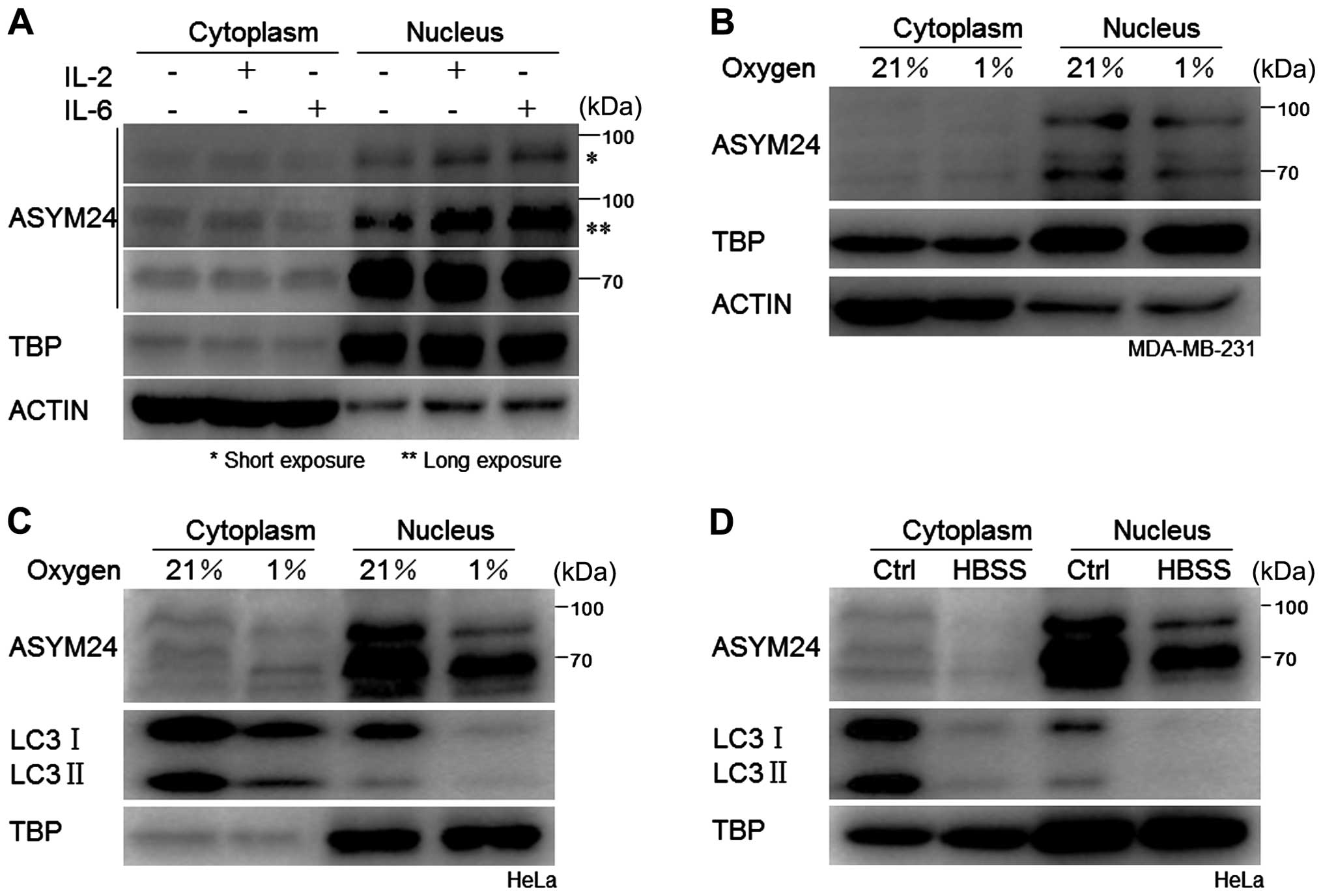
localization of proteins containing aDMA, we examined p90aDMA and
p70aDMA in the cytoplasm and nucleus through cellular fractionation
experiments followed by western blot analysis in the IL-2 or IL-6
treated MDA-MB-231 cells, and found that both p90aDMA and p70aDMA
distributed and accumulated predominantly in the nucleus regardless
of cytokine stimulation, suggesting that the aDMA proteins were
constitutively localized in the nucleus and might play a critical
role (Fig. 5A). To monitor
cytoplasmic contamination of the nuclear extracts, immunoblotting
was performed using the anti-β-actin antibody and TBP (TATA box
binding protein) antibody as the internal controls. The dramatic
reduction of β-actin or the presence of TBP only in the nuclear
fractions confirmed that cytoplasmic contamination did not occur
(Fig. 5A).
Hypoxia and nutrient starvation
disablizes nuclear p90aDMA and p70aDMA in cancer cells
Tumor cells are continually subjected to diverse
stress conditions of the tumor microenvironment, including hypoxia,
nutrient deprivation, oxidative or genotoxic stress (1). Among the stresses, hypoxia or
starvation is a classical autophagy inducing stimulus (32,33).
As shown in Fig. 5, two WB bands
representing 70 kDa (p70aDMA) and 90 kDa (p90aDMA)
nuclear-localized proteins, respectively, were detected in breast
cancer MDA-MB-231 cells and cervical cancer HeLa cells. In
accordance with the deacetylated nuclear LC3 being transported into
the cytoplasm to carry out PE conjugation to pre-autophagic
membranes (34), we observed that
LC3 in the nucleus vanished under hypoxia or starvation (Fig. 5C and D) and aDMA proteins
dominantly expressed in the nucleus were diminished synchronously
(Fig. 5B, C and D).
As is well known, LC3 proteins play a key role in
the selective recruitment of autophagic cargoes into
autophagosomes, and serve as docking sites for adaptor proteins
(12,31). Therefore, it is conceivable that
proteins of aDMA including p90aDMA and p70aDMA as LC3 cargo
substrates were translocated from the nucleus to the cytoplasm and
tethered to the site of engulfing autophagosomes. The hypothesis
needs further investigation under various stress conditions in our
future work.
Autophagy inhibition reverses the
degradation of p90aDMA and p70aDMA proteins in cancer cells
We first examined the effects of rapamycin treatment
on LC3 protein levels in MCF-7 cells. Immunoblot analyses showed a
concentration-dependent increase in the ratio of LC3-II/I,
representing mounting activation of autophagy (Fig. 6A). Concomitantly with the
activation of autophagy, a 90-kDa protein (p90sDMA) showed a marked
dose-dependent increase in sDMA levels (Fig. 6B). Whereas the effects of rapamycin
treatment on cellular aDMA in MCF-7 cells were tested by immunoblot
using ASYM24 antibody, and exhibited to a concentration-dependent
decrease in the intensity of multiple bands, including p90aDMA,
p70aDMA and p34aDMA (Fig. 6C).
To determine if p90aDMA and p70aDMA degradation in
rapamycin-treated MCF-7 cells was mediated by autophagy, we blocked
the early stage of autophagy with 3-MA, a class III PI3K inhibitor
(35), and examined its effect on
p90aDMA and p70aDMA expression levels. One-hour pretreatment with
3-MA effectively blocked the rapamycin-induced decrease in the
expression of p90aDMA and p70aDMA (Fig. 6D). Similarly, siRNA-mediated
knockdown of ATG5 prevented the rapamycin mediated decrease in
p90aDMA and p70aDMA expression levels, whereas the non-targeting
control siRNA had no effect (Fig.
6E). Therefore, we conclude that rapamycin triggers p90aDMA and
p70aDMA degradation through the autophagy pathway and aDMA serves
as a specific degradation signal for autophagy.
Discussion
We demonstrate for the first time that IL-2, IL-4
and IL-6 specifically promote nuclear accumulation of p90aDMA,
which is abrogated by AdOx, confirming that IL-2/4/6 treatment can
effectively increase asymmetric dimethylarginine proteins (Figs. 2, 3 and 5A). In addition, the interleukins
slightly increased basal autophagy but had an opposite effect on
rapamycin-induced autophagy, which may decrease p90aDMA expression
(Figs. 3 and 4). It can be concluded that IL-2/4/6
stimulated p90aDMA expression is not caused by inhibiting
autophagy. Moreover, indirect evidence suggests that p90aDMA,
p70aDMA and even p34aDMA could be degraded by LC3-mediated
selective autophagy in response to hypoxia, starvation and
rapamycin (Figs. 2B and D, 5B–D
and 6C), whereas rapamycin can accumulate p90sDMA in a
dose-dependent manner (Fig. 6B).
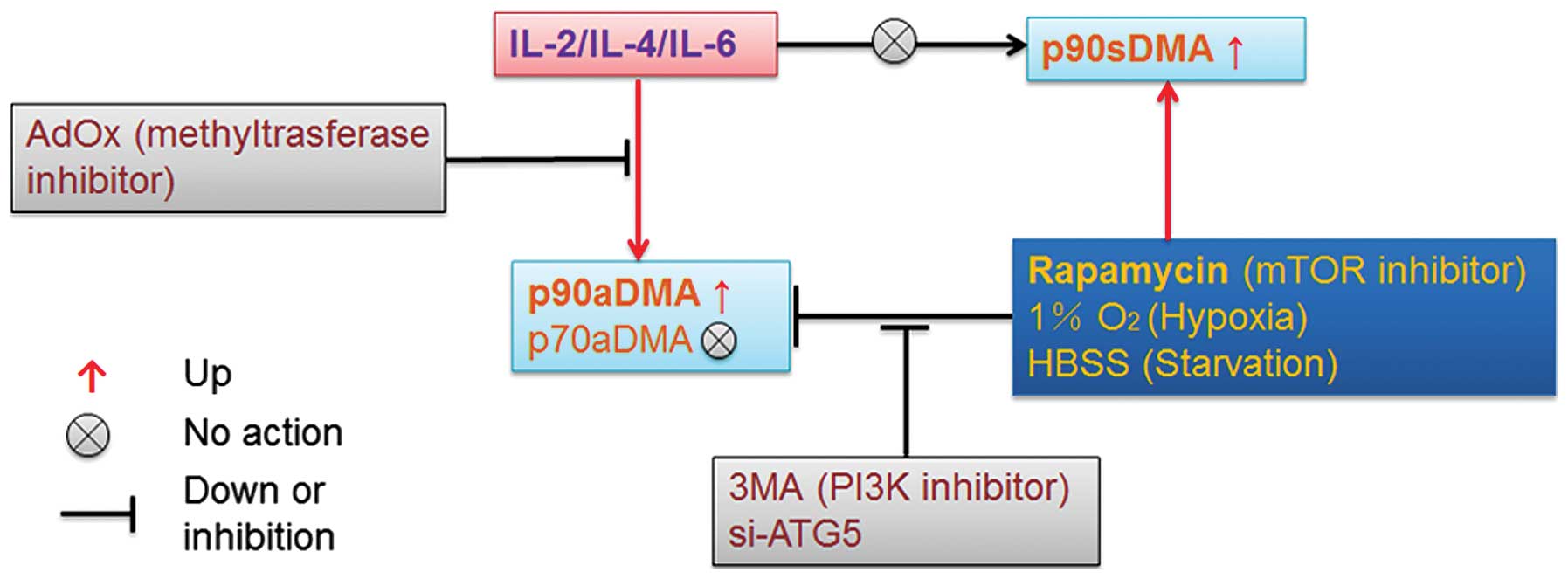
It is obvious that the aDMA induced by IL-2, IL-4 or IL-6
determines a highly processive initial reaction on the substrate
for specific autophagic degradation. Our findings reveal a novel
role for selective autophagy in the regulation of immunologic
responses and highlight the role of the posttranslational
modification of aDMA in controlling nuclear protein aggregation
induced by interleukins and selective autophagy of ubiquitinated
proteins (Fig. 7). One fundamental
question not addressed here is which endogenous proteins are
methylated at arginine sites and translocated into the nucleus in
response to IL-2/4/6 treatment.
Our results raise the fundamental questions how
IL-2/IL-4/IL-6 regulates the biogenesis of p90aDMA and whether aDMA
takes place in the cytoplasm or the nucleus of cancer cells
(Figs. 2B and D and 3A and B). Protein arginine methylation is
a common posttranslational modification in higher eukaryotes, but
its precise role in providing structural and functional diversity
among proteins is not well understood. Arginine methylation has
been shown to affect several cellular processes, including
intracellular localization, protein-protein interactions as well as
transcription (36,37). Signal transducers and activators of
transcription (STAT) proteins, for example, are a family of latent
cytoplasmic transcription factors which mediate interferons (IFNs),
interleukins, and some growth factors and peptide hormone signaling
in cells (38). Once tyrosine
phosphorylated, STAT proteins form homo- or heterodimers, which are
actively imported into the nucleus and bind to DNA consensus motifs
to elicit specific transcriptional responses (38–40).
However, arginine 31 methylation of STAT1 enhances its DNA binding
by reducing association with the specific inhibitor PIAS1, thus
intensifying the growth-restraining activities of the interferons
(41). In addition, arginine
methylation of STAT1 controls the rate of STAT1 dephosphorylation
by modulating its interaction with PIAS1 and the nuclear tyrosine
phosphatase TcPTP (42). STAT1 and
STAT2 have a structural arginine/lysine-rich element involved in
IFN-induced nuclear import (43).
Considering: i) interleukins mainly activate STAT signaling
pathways (39,40), and ii) p90aDMA derived from
endogenous proteins has a similar molecular mass to the 90 kDa and
nuclear location of activated STAT proteins (39,40),
it can be deduced that p90aDMA may be one or several proteins of
the STATs family that could be dimethylated by IL-2/4/6 commonly or
differentially and promote its or their translocation into the
nucleus. Further research involves the use of ASYM24 antibody to
proteins of 90 kDa containing aDMA for immunoprecipitation (IP)
experiments. The purified proteins are then digested and subjected
to immublotting or mass spectrometric analysis to confirm our
hypothesis.
Selective autophagy is a degradative pathway that
controls the quality and abundance of proteins and cellular
organelles and is mediated by autophagy receptors that
simultaneously bind the designated target and LC3/GABARAP proteins
on autophagosomal membranes (11).
The autophagic/signaling adaptor LC3 is known to exert its
functions through multiple domains containing a ubiquitin core with
two α helices, α1 and α2 attached at its N-terminus, which is
utilized as the interaction site with its target proteins (44). Physical linkages between autophagy
adaptor proteins via polyubiquitin chains are required for
autophagy flux. A recent research demonstrates that the
deacetylated nuclear LC3 is transported into the cytoplasm to carry
out PE conjugation to pre-autophagic membranes by sequential
interaction with Atg7 and Atg3 (34). Especially, nuclear lamina protein
lamin B1 degradation is achieved by nucleus to cytoplasm transport
degradation that delivers lamin B1 to the lysosome via LC3-lamin B1
interaction in the nucleus (31).
The reduction in p90aDMA, p70aDMA and p34aDMA levels implicates the
specificity and apparent affinity of aDMA for autophagy receptors
(Figs. 5B–D and 6C–E). Based on the above reports and
results that p90aDMA and p70aDMA are dominantly expressed in the
nucleus, we may deduce that selective autophagy via LC3 is required
for the translocation to the autophagosomes and degradation of
p90aDMA and p70aDMA. Once autophagy is stimulated in response to
stress such as hypoxia, starvation or rapamycin, proteins
undergoing aDMA can be transported to autophagosomes by
deacetylated LC3 and suffer from autophagic degradation
specifically. Based on our findings that p90aDMA and p70aDMA,
dominantly expressed in the nucleus, are recognized and bound by
LC3 translocating to the cytoplasm (Figs. 2, 3, 5 and
6A and C), we conclude that aDMA
as a signal is required for LC3-mediatad selective autophagy
traffic.
As a stress integrator pathway, autophagy is a major
mechanism that mediates protein and organelle degradation in
response to external and internal signals. Additionally, autophagy
has been verified in different contexts to regulate immune
responses to various stimuli (45–50).
In addition to the role of p62, NDP52 and optineurin as adaptors in
selective autophagy, these proteins have also recently been shown
to regulate innate immunity signaling pathways and, thus, were
suggested to represent a new class of pattern recognition
receptors, the sequestosome-1-like receptors (SLRs) (51,52).
When IL-2 and IL-6 accumulate p90aDMA, which may be a STAT protein,
to activate some signaling pathways or transcription, external
stresses such as hypoxia, starvation or rapamycin antagonize the
signaling transduction via autophagic degradation of p90aDMA
(Fig. 2B and D). A growing body of
evidence indicates that similar elimination of signaling molecules
play key roles in autophagy-regulated immune responses (53–57).
For instance, microglial autophagy plays an important role in the
clearance of extracellular Aβ (β-amyloid) fibrils and the
regulation of Aβ-induced inflammation, thereby affecting neuronal
viability (53). One the other
hand, emerging data have suggested that additional mechanisms
involved in cancer-related inflammation (CRI) are induction of
angiogenesis, metastasis, invasion of surrounding tissues and
genetic instability by inflammatory mediators, leading to
accumulation of random genetic alterations in cancer cells
(29,58,59).
Based on the above, the possibility is raised that a negative
feedback via the targeted regulation of p90aDMA is established in
our study bridging immune microenvironment and selective autophagy
that may have a potentially pivotal role in shaping the
oncogenesis, immunogenic cell death and even heterogeneity in
response to dynamic changes in a cancer cell metabolic,
environmental, or developmental status.
In conclusion, our data support that a model for the
link between arginine methylation and selective autophagy in the
immune microenvironment could be proposed for further
investigation. Additionally, it remains unclear how aDMA (or sDMA)
is regulated by interleukins (or rapamycin) and their function need
to be further explored. Collectively, our study expands what is
known about the tumor microenvironment and supports the idea of the
regulation of arginine methylation as a new immune-therapeutic
method in the future.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (J.X.G., nos. 81171940 and 81372188;
L.F.L., no. 81402287); Science and Technology Support Program,
Science and Technology Commission of Shanghai Municipality
(12431900704 to J.X.G.); the Special Fund for Innovation and
Development of Science and Technology and Cultivation Fund for
Major Projects and Innovative Team (J.X.G., 2014), Shanghai Jiao
Tong University, China; the State Key Laboratory of Oncogenes and
Related Genes in China (J.X.G., no. 90-14-06); the University
Doctorate Research Fund for Freshly Recruited Teachers (L.F.L., no.
20130073120010), Ministry of National Education, China; and Startup
Funds (J.X.G.) from Renji Hospital and School of Medicine, Shanghai
Jiao Tong University, China; the Fund for Key Disciplines and
Specialties, Shanghai Health and Family Planning Committee, China
(J.X.G.), and Shandong Outstanding Young and Middle-aged Scientists
Research Award Fund (2014BSE27021 to Shao-Hua Zhao).
Abbreviations:
|
IL
|
interleukin
|
|
ATG
|
autophagy-related gene
|
|
LC3
|
microtubule-associated protein 1 light
chain 3
|
|
PTMs
|
posttranslational modifications
|
|
sDMA
|
symmetric dimethylarginine
|
|
aDMA
|
asymmetric dimethylarginine
|
|
AdOx
|
adenosine-2′,3′-dialdehyde
|
|
HBSS
|
Hank′s balanced salt solution
|
References
|
1
|
Semenza GL: The hypoxic tumor
microenvironment: A driving force for breast cancer progression.
Biochim Biophys Acta. S0167-4889(15)00192-5. 2015.PubMed/NCBI
|
|
2
|
Johansson A, Hamzah J and Ganss R: More
than a scaffold: Stromal modulation of tumor immunity. Biochim
Biophys Acta. S0304-419X(15)00044-X. 2015.PubMed/NCBI
|
|
3
|
Pitt LA, Tikhonova AN, Hu H, Trimarchi T,
King B, Gong Y, Sanchez-Martin M, Tsirigos A, Littman DR, Ferrando
AA, et al: CXCL12-producing vascular endothelial niches control
acute T cell leukemia maintenance. Cancer Cell. 27:755–768. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dranoff G: Cytokines in cancer
pathogenesis and cancer therapy. Nat Rev Cancer. 4:11–22. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van der Jeught K, Bialkowski L,
Daszkiewicz L, Broos K, Goyvaerts C, Renmans D, Van Lint S, Heirman
C, Thielemans K and Breckpot K: Targeting the tumor
microenvironment to enhance antitumor immune responses. Oncotarget.
6:1359–1381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosenberg SA: IL-2: The first effective
immunotherapy for human cancer. J Immunol. 192:5451–5458. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blagosklonny MV: Hypoxia, MTOR and
autophagy: Converging on senescence or quiescence. Autophagy.
9:260–262. 2013. View Article : Google Scholar :
|
|
8
|
Kim YC and Guan K-L: mTOR: A pharmacologic
target for autophagy regulation. J Clin Invest. 125:25–32. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun L, Li T, Wei Q, Zhang Y, Jia X, Wan Z
and Han L: Upregulation of BNIP3 mediated by ERK/HIF-1α pathway
induces autophagy and contributes to anoikis resistance of
hepatocellular carcinoma cells. Future Oncol. 10:1387–1398. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mochida K, Oikawa Y, Kimura Y, Kirisako H,
Hirano H, Ohsumi Y and Nakatogawa H: Receptor-mediated selective
autophagy degrades the endoplasmic reticulum and the nucleus.
Nature. 522:359–362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stolz A, Ernst A and Dikic I: Cargo
recognition and trafficking in selective autophagy. Nat Cell Biol.
16:495–501. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wild P, McEwan DG and Dikic I: The LC3
interactome at a glance. J Cell Sci. 127:3–9. 2014. View Article : Google Scholar
|
|
13
|
Wilkinson DS, Jariwala JS, Anderson E,
Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V,
Dillin A, et al: Phosphorylation of LC3 by the Hippo kinases
STK3/STK4 is essential for autophagy. Mol Cell. 57:55–68. 2015.
View Article : Google Scholar :
|
|
14
|
Xie Y, Kang R, Sun X, Zhong M, Huang J,
Klionsky DJ and Tang D: Posttranslational modification of
autophagy-related proteins in macroautophagy. Autophagy. 11:28–45.
2015. View Article : Google Scholar :
|
|
15
|
Hunter T: The age of crosstalk:
Phosphorylation, ubiquitination, and beyond. Mol Cell. 28:730–738.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lazarou M, Sliter DA, Kane LA, Sarraf SA,
Wang C, Burman JL, Sideris DP, Fogel AI and Youle RJ: The ubiquitin
kinase PINK1 recruits autophagy receptors to induce mitophagy.
Nature. 524:309–314. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Birgisdottir AB, Lamark T and Johansen T:
The LIR motif - crucial for selective autophagy. J Cell Sci.
126:3237–3247. 2013.PubMed/NCBI
|
|
18
|
Wild P, Farhan H, McEwan DG, Wagner S,
Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et
al: Phosphorylation of the autophagy receptor optineurin restricts
Salmonella growth. Science. 333:228–233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jo C, Gundemir S, Pritchard S, Jin YN,
Rahman I and Johnson GV: Nrf2 reduces levels of phosphorylated tau
protein by inducing autophagy adaptor protein NDP52. Nat Commun.
5:34962014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu J, Blenis J and Yuan J: Activation of
PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by
phosphorylating and promoting the degradation of Mad1. Proc Natl
Acad Sci USA. 105:6584–6589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li S, Yang P, Tian E and Zhang H: Arginine
methylation modulates autophagic degradation of PGL granules in C.
elegans. Mol Cell. 52:421–433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pahlich S, Zakaryan RP and Gehring H:
Protein arginine methylation: Cellular functions and methods of
analysis. Biochim Biophys Acta. 1764:1890–1903. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yagoub D, Hart-Smith G, Moecking J, Erce
MA and Wilkins MR: Yeast proteins Gar1p, Nop1p, Npl3p, Nsr1p, and
Rps2p are natively methylated and are substrates of the arginine
methyltransferase Hmt1p. Proteomics. 15:3209–3218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McBride AE and Silver PA: State of the
arg: Protein methylation at arginine comes of age. Cell. 106:5–8.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Y and Bedford MT: Protein arginine
methyltransferases and cancer. Nat Rev Cancer. 13:37–50. 2013.
View Article : Google Scholar
|
|
26
|
Bedford MT and Clarke SG: Protein arginine
methylation in mammals: Who, what, and why. Mol Cell. 33:1–13.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao WW, Xiao RQ, Peng BL, Xu HT, Shen HF,
Huang MF, Shi TT, Yi J, Zhang WJ, Wu XN, et al: Arginine
methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene
activation. Proc Natl Acad Sci USA. 112:E3327–E3336. 2015.
View Article : Google Scholar
|
|
28
|
Kao J, Salari K, Bocanegra M, Choi YL,
Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar
AF, et al: Molecular profiling of breast cancer cell lines defines
relevant tumor models and provides a resource for cancer gene
discovery. PLoS One. 4:e61462009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin WW and Karin M: A cytokine-mediated
link between innate immunity, inflammation, and cancer. J Clin
Invest. 117:1175–1183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shirakawa T, Kako K, Shimada T, Nagashima
Y, Nakamura A, Ishida J and Fukamizu A: Production of free
methylarginines via the proteasome and autophagy pathways in
cultured cells. Mol Med Rep. 4:615–620. 2011.PubMed/NCBI
|
|
31
|
Dou Z, Xu C, Donahue G, Shimi T, Pan JA,
Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al: Autophagy
mediates degradation of nuclear lamina. Nature. 527:105–109. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Klionsky DJ, Abdalla FC, Abeliovich H,
Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M,
Agostinis P, Aguirre-Ghiso JA, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy. Autophagy.
8:445–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun L, Liu N, Liu S, Xia W, Liu M, Li L
and Gao J: Beclin-1-independent autophagy mediates programmed
cancer cell death through interplays with endoplasmic reticulum
and/or mitochondria in colbat chloride-induced hypoxia. Am J Cancer
Res. 5:2626–2642. 2015.PubMed/NCBI
|
|
34
|
Huang R, Xu Y, Wan W, Shou X, Qian J, You
Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, et al:
Deacetylation of nuclear LC3 drives autophagy initiation under
starvation. Mol Cell. 57:456–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Blommaart EF, Krause U, Schellens JP,
Vreeling-Sindelarova H and Meijer AJ: The phosphatidylinositol
3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in
isolated rat hepatocytes. Eur J Biochem. 243:240–246. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang B, Chen Y, Zhao Y and Bruick RK:
JMJD6 is a histone arginine demethylase. Science. 318:444–447.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang H, Huang ZQ, Xia L, Feng Q,
Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst
P, et al: Methylation of histone H4 at arginine 3 facilitating
transcriptional activation by nuclear hormone receptor. Science.
293:853–857. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Buchert M, Burns CJ and Ernst M: Targeting
JAK kinase in solid tumors: Emerging opportunities and challenges.
Oncogene. May 18–2015.(Epub ahead of print). View Article : Google Scholar : 2015. PubMed/NCBI
|
|
39
|
Bowman T, Garcia R, Turkson J and Jove R:
STATs in oncogenesis. Oncogene. 19:2474–2488. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hirahara K, Onodera A, Villarino AV,
Bonelli M, Sciumè G, Laurence A, Sun HW, Brooks SR, Vahedi G, Shih
HY, et al: Asymmetric action of STAT transcription factors drives
transcriptional outputs and cytokine specificity. Immunity.
42:877–889. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mowen KA, Tang J, Zhu W, Schurter BT,
Shuai K, Herschman HR and David M: Arginine methylation of STAT1
modulates IFNalpha/beta-induced transcription. Cell. 104:731–741.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhu W, Mustelin T and David M: Arginine
methylation of STAT1 regulates its dephosphorylation by T cell
protein tyrosine phosphatase. J Biol Chem. 277:35787–35790. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Melen K, Kinnunen L and Julkunen I:
Arginine/lysine-rich structural element is involved in
interferon-induced nuclear import of STATs. J Biol Chem.
276:16447–16455. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sugawara K, Suzuki NN, Fujioka Y,
Mizushima N, Ohsumi Y and Inagaki F: The crystal structure of
microtubule-associated protein light chain 3, a mammalian homologue
of Saccharomyces cerevisiae Atg8. Genes Cells. 9:611–618. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pei B, Zhao M, Miller BC, Véla JL,
Bruinsma MW, Virgin HW and Kronenberg M: Invariant NKT cells
require autophagy to coordinate proliferation and survival signals
during differentiation. J Immunol. 194:5872–5884. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kanayama M, He YW and Shinohara ML: The
lung is protected from spontaneous inflammation by autophagy in
myeloid cells. J Immunol. 194:5465–5471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schlie K, Westerback A, DeVorkin L,
Hughson LR, Brandon JM, MacPherson S, Gadawski I, Townsend KN, Poon
VI, Elrick MA, et al: Survival of effector CD8+ T cells
during influenza infection is dependent on autophagy. J Immunol.
194:4277–4286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Michaud M, Martins I, Sukkurwala AQ,
Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot
G, et al: Autophagy-dependent anticancer immune responses induced
by chemotherapeutic agents in mice. Science. 334:1573–1577. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Konno H, Konno K and Barber GN: Cyclic
dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to
prevent sustained innate immune signaling. Cell. 155:688–698. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guo ML, Liao K, Periyasamy P, Yang L, Cai
Y, Callen SE and Buch S: Cocaine mediated microglial activation
involves the ER stress-autophagy axis. Autophagy. 11:995–1009.
2015. View Article : Google Scholar
|
|
51
|
Deretic V: Autophagy as an innate immunity
paradigm: Expanding the scope and repertoire of pattern recognition
receptors. Curr Opin Immunol. 24:21–31. 2012. View Article : Google Scholar :
|
|
52
|
Chang KH, Sengupta A, Nayak RC, Duran A,
Lee SJ, Pratt RG, Wellendorf AM, Hill SE, Watkins M, Gonzalez-Nieto
D, et al: p62 is required for stem cell/progenitor retention
through inhibition of IKK/NF-κB/Ccl4 signaling at the bone marrow
macrophage-osteoblast niche. Cell Rep. 9:2084–2097. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS,
Kwon HJ, Kim HM, Kim DH and Yoon SY: Autophagy in microglia
degrades extracellular β-amyloid fibrils and regulates the NLRP3
inflammasome. Autophagy. 10:1761–1775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu L, Yang M, Kang R, Dai Y, Yu Y, Gao F,
Wang H, Sun X, Li X, Li J, et al: HMGB1-DNA complex-induced
autophagy limits AIM2 inflammasome activation through RAGE. Biochem
Biophys Res Commun. 450:851–856. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Meunier E, Dick MS, Dreier RF, Schürmann
N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki
N, Takeda K, et al: Caspase-11 activation requires lysis of
pathogen-containing vacuoles by IFN-induced GTPases. Nature.
509:366–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wildenberg ME, Vos AC, Wolfkamp SC,
Duijvestein M, Verhaar AP, Te Velde AA, van den Brink GR and Hommes
DW: Autophagy attenuates the adaptive immune response by
destabilizing the immunologic synapse. Gastroenterology.
142:1493–1503 e1496. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lévy J, Cacheux W, Bara MA, L'Hermitte A,
Lepage P, Fraudeau M, Trentesaux C, Lemarchand J, Durand A, Crain
AM, et al: Intestinal inhibition of Atg7 prevents tumour initiation
through a microbiome-influenced immune response and suppresses
tumour growth. Nat Cell Biol. 17:1062–1073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Colotta F, Allavena P, Sica A, Garlanda C
and Mantovani A: Cancer-related inflammation, the seventh hallmark
of cancer: Links to genetic instability. Carcinogenesis.
30:1073–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|