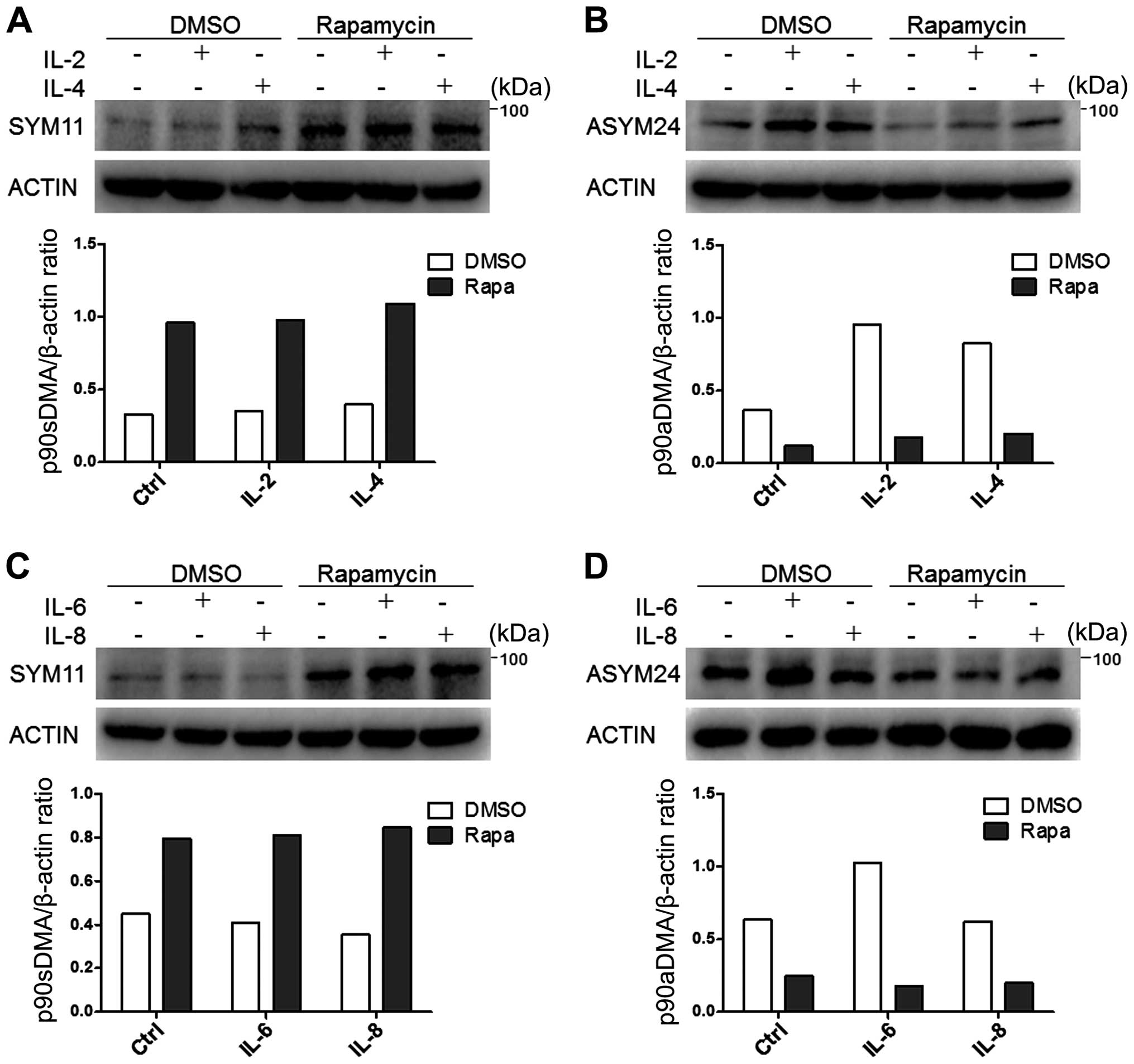
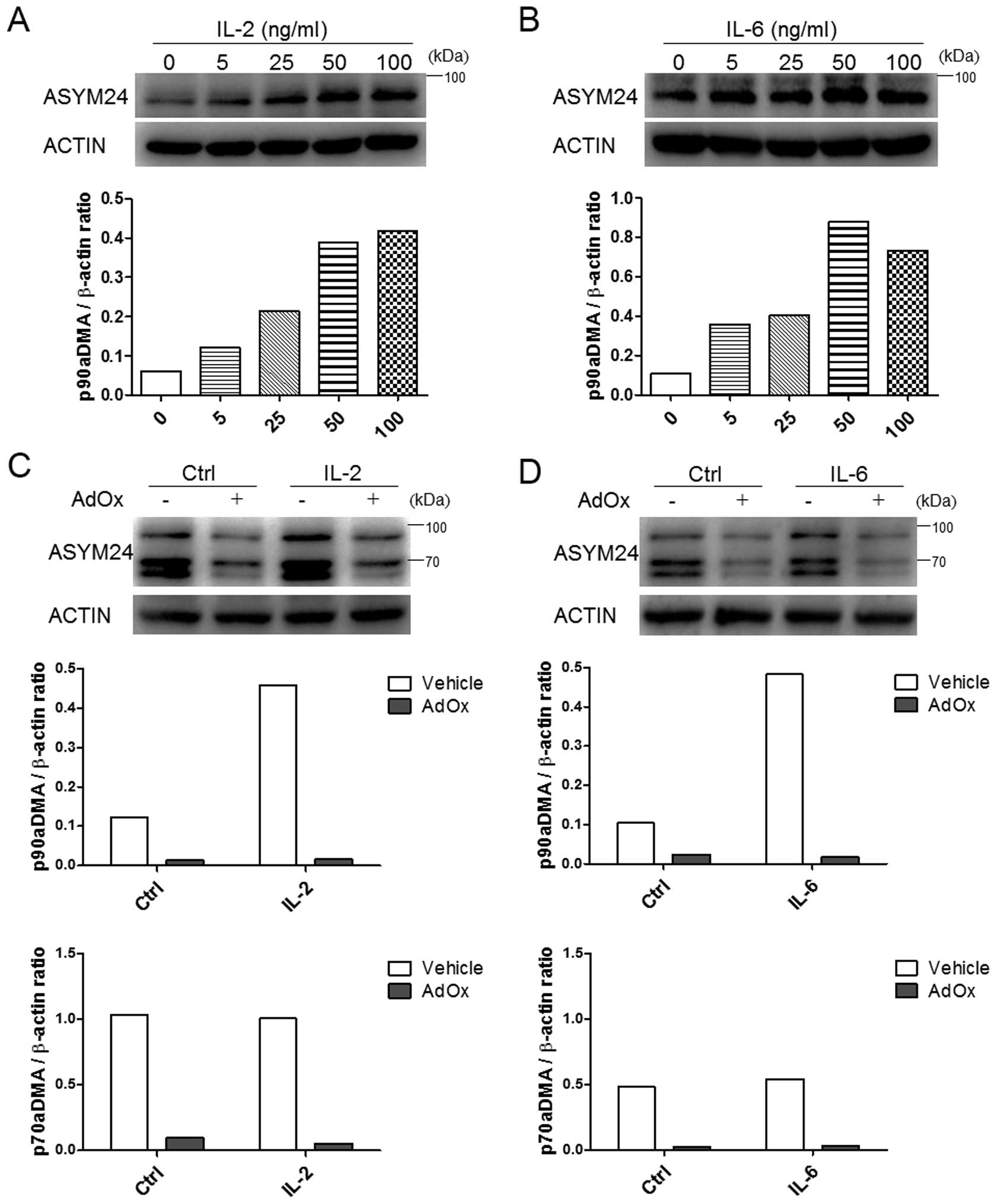
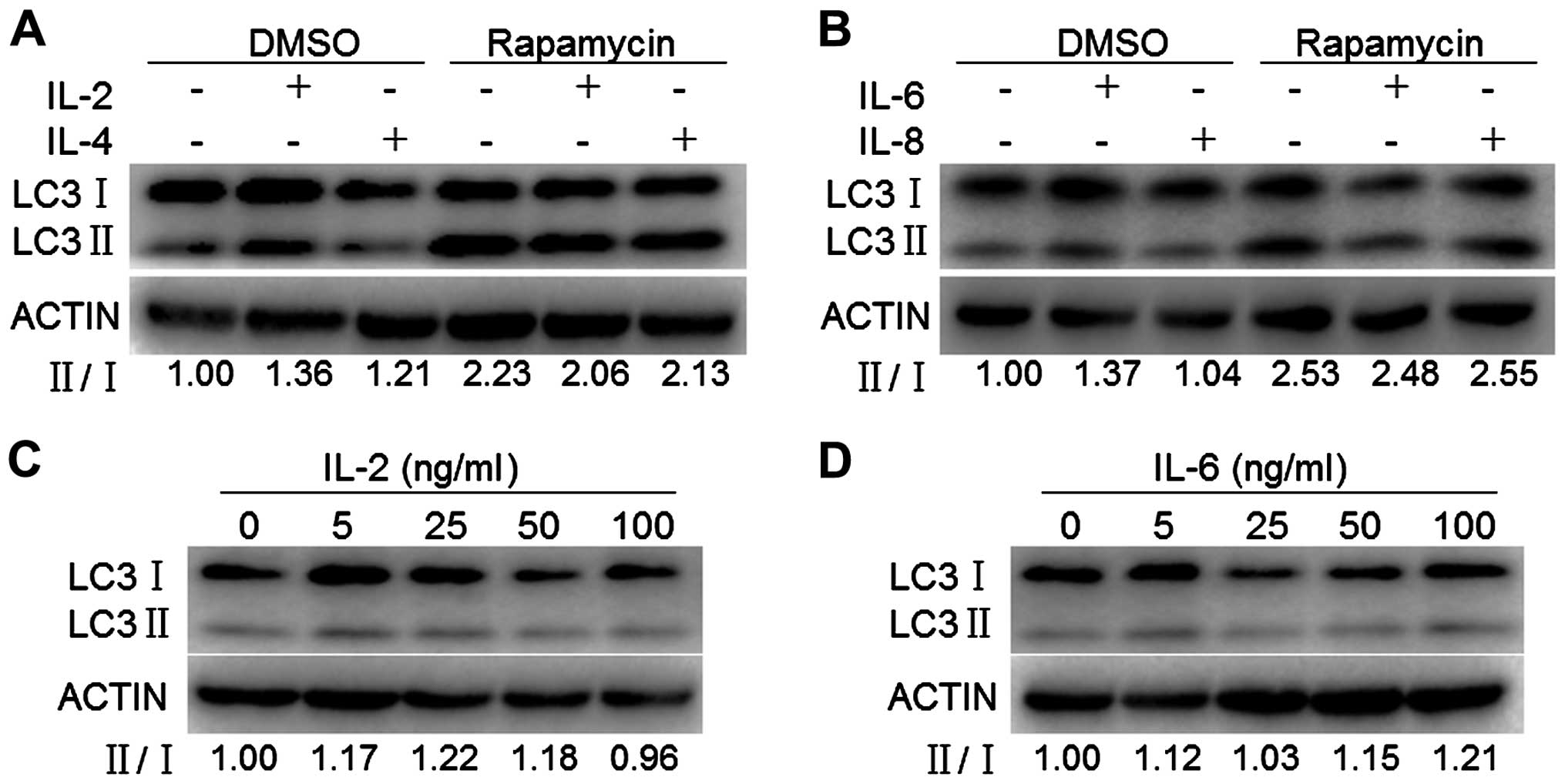
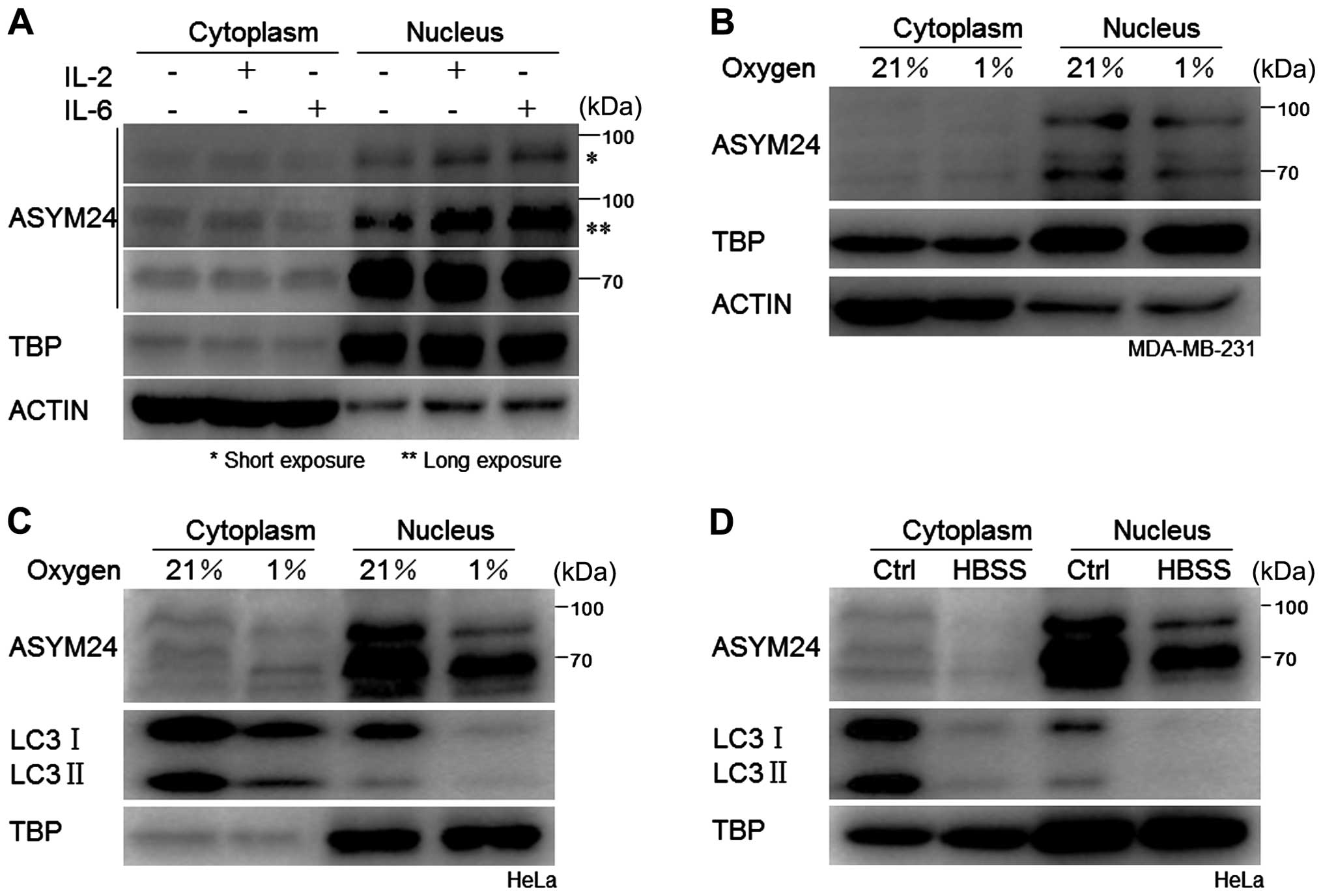
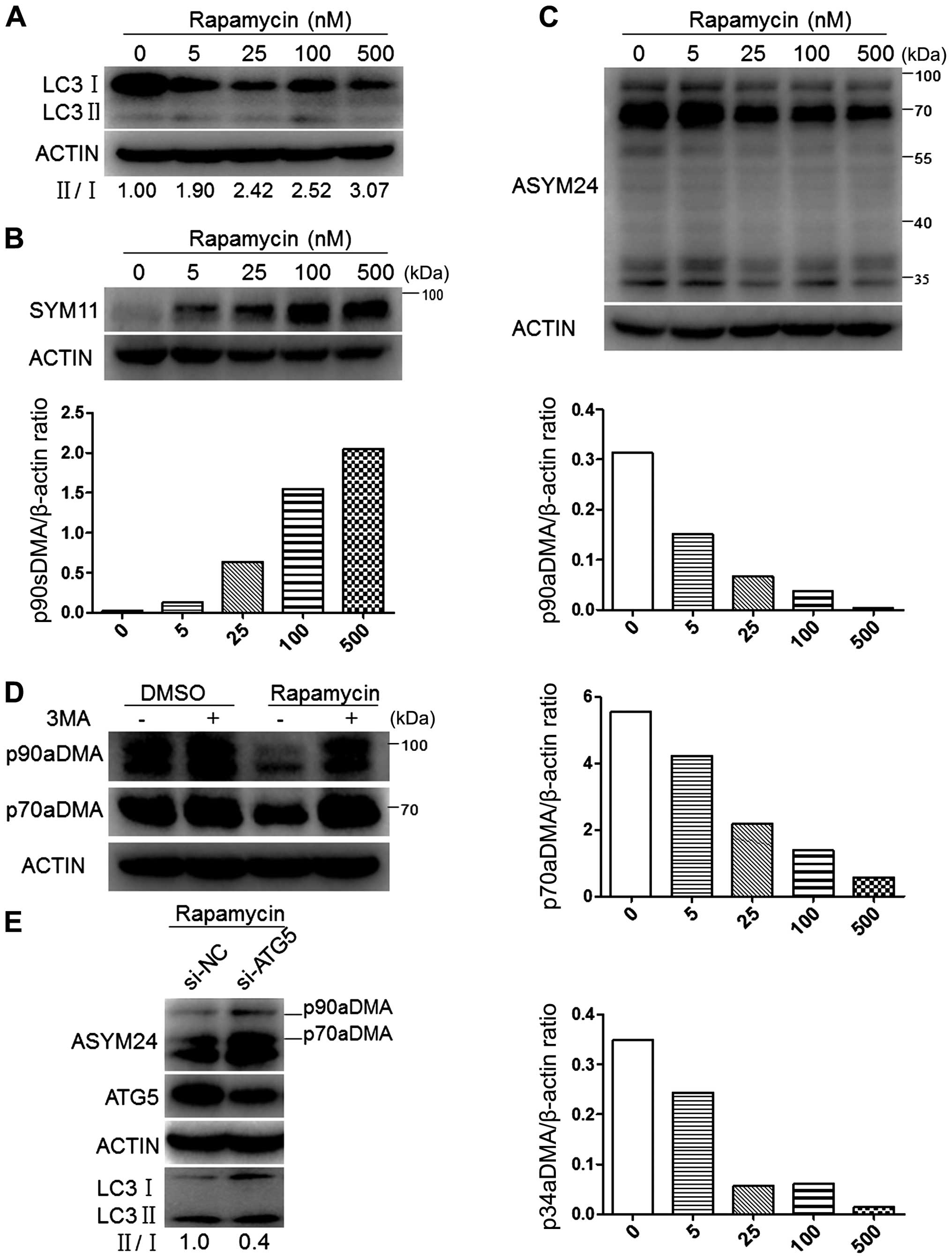
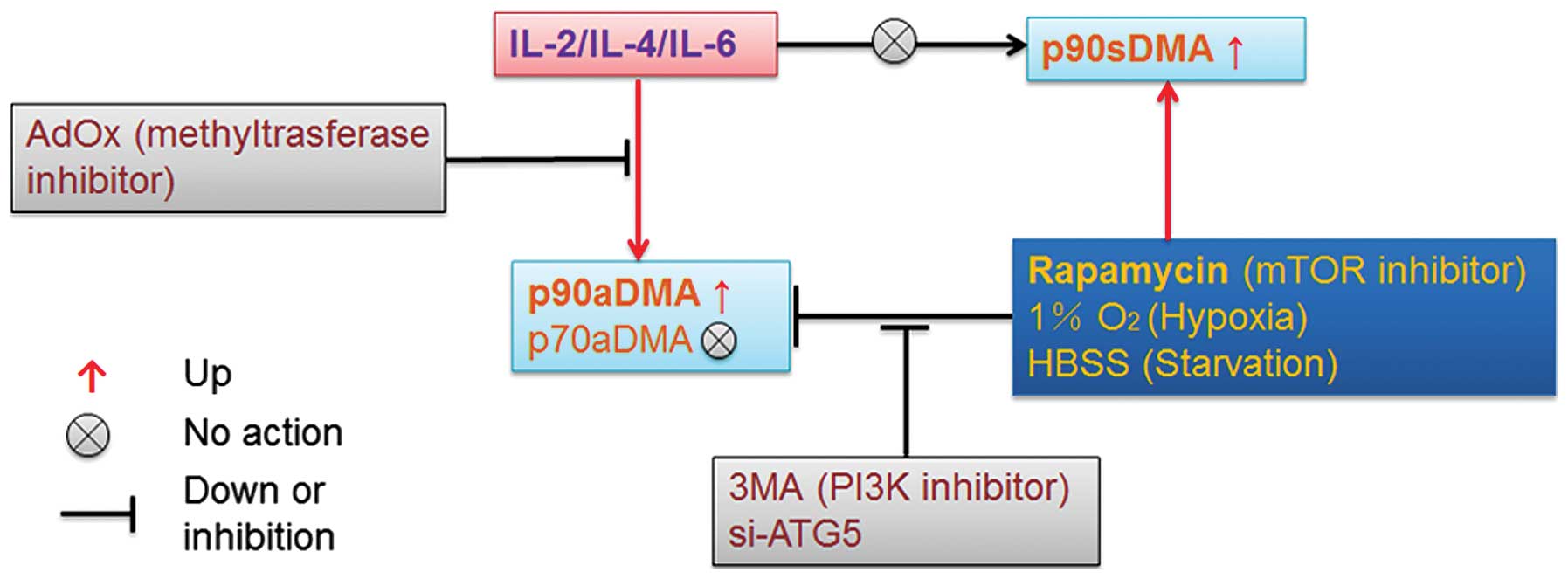
|
1
|
Semenza GL: The hypoxic tumor
microenvironment: A driving force for breast cancer progression.
Biochim Biophys Acta. S0167-4889(15)00192-5. 2015.PubMed/NCBI
|
|
2
|
Johansson A, Hamzah J and Ganss R: More
than a scaffold: Stromal modulation of tumor immunity. Biochim
Biophys Acta. S0304-419X(15)00044-X. 2015.PubMed/NCBI
|
|
3
|
Pitt LA, Tikhonova AN, Hu H, Trimarchi T,
King B, Gong Y, Sanchez-Martin M, Tsirigos A, Littman DR, Ferrando
AA, et al: CXCL12-producing vascular endothelial niches control
acute T cell leukemia maintenance. Cancer Cell. 27:755–768. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dranoff G: Cytokines in cancer
pathogenesis and cancer therapy. Nat Rev Cancer. 4:11–22. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van der Jeught K, Bialkowski L,
Daszkiewicz L, Broos K, Goyvaerts C, Renmans D, Van Lint S, Heirman
C, Thielemans K and Breckpot K: Targeting the tumor
microenvironment to enhance antitumor immune responses. Oncotarget.
6:1359–1381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosenberg SA: IL-2: The first effective
immunotherapy for human cancer. J Immunol. 192:5451–5458. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Blagosklonny MV: Hypoxia, MTOR and
autophagy: Converging on senescence or quiescence. Autophagy.
9:260–262. 2013. View Article : Google Scholar :
|
|
8
|
Kim YC and Guan K-L: mTOR: A pharmacologic
target for autophagy regulation. J Clin Invest. 125:25–32. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun L, Li T, Wei Q, Zhang Y, Jia X, Wan Z
and Han L: Upregulation of BNIP3 mediated by ERK/HIF-1α pathway
induces autophagy and contributes to anoikis resistance of
hepatocellular carcinoma cells. Future Oncol. 10:1387–1398. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mochida K, Oikawa Y, Kimura Y, Kirisako H,
Hirano H, Ohsumi Y and Nakatogawa H: Receptor-mediated selective
autophagy degrades the endoplasmic reticulum and the nucleus.
Nature. 522:359–362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stolz A, Ernst A and Dikic I: Cargo
recognition and trafficking in selective autophagy. Nat Cell Biol.
16:495–501. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wild P, McEwan DG and Dikic I: The LC3
interactome at a glance. J Cell Sci. 127:3–9. 2014. View Article : Google Scholar
|
|
13
|
Wilkinson DS, Jariwala JS, Anderson E,
Mitra K, Meisenhelder J, Chang JT, Ideker T, Hunter T, Nizet V,
Dillin A, et al: Phosphorylation of LC3 by the Hippo kinases
STK3/STK4 is essential for autophagy. Mol Cell. 57:55–68. 2015.
View Article : Google Scholar :
|
|
14
|
Xie Y, Kang R, Sun X, Zhong M, Huang J,
Klionsky DJ and Tang D: Posttranslational modification of
autophagy-related proteins in macroautophagy. Autophagy. 11:28–45.
2015. View Article : Google Scholar :
|
|
15
|
Hunter T: The age of crosstalk:
Phosphorylation, ubiquitination, and beyond. Mol Cell. 28:730–738.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lazarou M, Sliter DA, Kane LA, Sarraf SA,
Wang C, Burman JL, Sideris DP, Fogel AI and Youle RJ: The ubiquitin
kinase PINK1 recruits autophagy receptors to induce mitophagy.
Nature. 524:309–314. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Birgisdottir AB, Lamark T and Johansen T:
The LIR motif - crucial for selective autophagy. J Cell Sci.
126:3237–3247. 2013.PubMed/NCBI
|
|
18
|
Wild P, Farhan H, McEwan DG, Wagner S,
Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et
al: Phosphorylation of the autophagy receptor optineurin restricts
Salmonella growth. Science. 333:228–233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jo C, Gundemir S, Pritchard S, Jin YN,
Rahman I and Johnson GV: Nrf2 reduces levels of phosphorylated tau
protein by inducing autophagy adaptor protein NDP52. Nat Commun.
5:34962014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu J, Blenis J and Yuan J: Activation of
PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by
phosphorylating and promoting the degradation of Mad1. Proc Natl
Acad Sci USA. 105:6584–6589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li S, Yang P, Tian E and Zhang H: Arginine
methylation modulates autophagic degradation of PGL granules in C.
elegans. Mol Cell. 52:421–433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pahlich S, Zakaryan RP and Gehring H:
Protein arginine methylation: Cellular functions and methods of
analysis. Biochim Biophys Acta. 1764:1890–1903. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yagoub D, Hart-Smith G, Moecking J, Erce
MA and Wilkins MR: Yeast proteins Gar1p, Nop1p, Npl3p, Nsr1p, and
Rps2p are natively methylated and are substrates of the arginine
methyltransferase Hmt1p. Proteomics. 15:3209–3218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McBride AE and Silver PA: State of the
arg: Protein methylation at arginine comes of age. Cell. 106:5–8.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Y and Bedford MT: Protein arginine
methyltransferases and cancer. Nat Rev Cancer. 13:37–50. 2013.
View Article : Google Scholar
|
|
26
|
Bedford MT and Clarke SG: Protein arginine
methylation in mammals: Who, what, and why. Mol Cell. 33:1–13.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao WW, Xiao RQ, Peng BL, Xu HT, Shen HF,
Huang MF, Shi TT, Yi J, Zhang WJ, Wu XN, et al: Arginine
methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene
activation. Proc Natl Acad Sci USA. 112:E3327–E3336. 2015.
View Article : Google Scholar
|
|
28
|
Kao J, Salari K, Bocanegra M, Choi YL,
Girard L, Gandhi J, Kwei KA, Hernandez-Boussard T, Wang P, Gazdar
AF, et al: Molecular profiling of breast cancer cell lines defines
relevant tumor models and provides a resource for cancer gene
discovery. PLoS One. 4:e61462009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin WW and Karin M: A cytokine-mediated
link between innate immunity, inflammation, and cancer. J Clin
Invest. 117:1175–1183. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shirakawa T, Kako K, Shimada T, Nagashima
Y, Nakamura A, Ishida J and Fukamizu A: Production of free
methylarginines via the proteasome and autophagy pathways in
cultured cells. Mol Med Rep. 4:615–620. 2011.PubMed/NCBI
|
|
31
|
Dou Z, Xu C, Donahue G, Shimi T, Pan JA,
Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, et al: Autophagy
mediates degradation of nuclear lamina. Nature. 527:105–109. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Klionsky DJ, Abdalla FC, Abeliovich H,
Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M,
Agostinis P, Aguirre-Ghiso JA, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy. Autophagy.
8:445–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun L, Liu N, Liu S, Xia W, Liu M, Li L
and Gao J: Beclin-1-independent autophagy mediates programmed
cancer cell death through interplays with endoplasmic reticulum
and/or mitochondria in colbat chloride-induced hypoxia. Am J Cancer
Res. 5:2626–2642. 2015.PubMed/NCBI
|
|
34
|
Huang R, Xu Y, Wan W, Shou X, Qian J, You
Z, Liu B, Chang C, Zhou T, Lippincott-Schwartz J, et al:
Deacetylation of nuclear LC3 drives autophagy initiation under
starvation. Mol Cell. 57:456–466. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Blommaart EF, Krause U, Schellens JP,
Vreeling-Sindelarova H and Meijer AJ: The phosphatidylinositol
3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in
isolated rat hepatocytes. Eur J Biochem. 243:240–246. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang B, Chen Y, Zhao Y and Bruick RK:
JMJD6 is a histone arginine demethylase. Science. 318:444–447.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang H, Huang ZQ, Xia L, Feng Q,
Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst
P, et al: Methylation of histone H4 at arginine 3 facilitating
transcriptional activation by nuclear hormone receptor. Science.
293:853–857. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Buchert M, Burns CJ and Ernst M: Targeting
JAK kinase in solid tumors: Emerging opportunities and challenges.
Oncogene. May 18–2015.(Epub ahead of print). View Article : Google Scholar : 2015. PubMed/NCBI
|
|
39
|
Bowman T, Garcia R, Turkson J and Jove R:
STATs in oncogenesis. Oncogene. 19:2474–2488. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hirahara K, Onodera A, Villarino AV,
Bonelli M, Sciumè G, Laurence A, Sun HW, Brooks SR, Vahedi G, Shih
HY, et al: Asymmetric action of STAT transcription factors drives
transcriptional outputs and cytokine specificity. Immunity.
42:877–889. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mowen KA, Tang J, Zhu W, Schurter BT,
Shuai K, Herschman HR and David M: Arginine methylation of STAT1
modulates IFNalpha/beta-induced transcription. Cell. 104:731–741.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhu W, Mustelin T and David M: Arginine
methylation of STAT1 regulates its dephosphorylation by T cell
protein tyrosine phosphatase. J Biol Chem. 277:35787–35790. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Melen K, Kinnunen L and Julkunen I:
Arginine/lysine-rich structural element is involved in
interferon-induced nuclear import of STATs. J Biol Chem.
276:16447–16455. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sugawara K, Suzuki NN, Fujioka Y,
Mizushima N, Ohsumi Y and Inagaki F: The crystal structure of
microtubule-associated protein light chain 3, a mammalian homologue
of Saccharomyces cerevisiae Atg8. Genes Cells. 9:611–618. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pei B, Zhao M, Miller BC, Véla JL,
Bruinsma MW, Virgin HW and Kronenberg M: Invariant NKT cells
require autophagy to coordinate proliferation and survival signals
during differentiation. J Immunol. 194:5872–5884. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kanayama M, He YW and Shinohara ML: The
lung is protected from spontaneous inflammation by autophagy in
myeloid cells. J Immunol. 194:5465–5471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schlie K, Westerback A, DeVorkin L,
Hughson LR, Brandon JM, MacPherson S, Gadawski I, Townsend KN, Poon
VI, Elrick MA, et al: Survival of effector CD8+ T cells
during influenza infection is dependent on autophagy. J Immunol.
194:4277–4286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Michaud M, Martins I, Sukkurwala AQ,
Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot
G, et al: Autophagy-dependent anticancer immune responses induced
by chemotherapeutic agents in mice. Science. 334:1573–1577. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Konno H, Konno K and Barber GN: Cyclic
dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to
prevent sustained innate immune signaling. Cell. 155:688–698. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guo ML, Liao K, Periyasamy P, Yang L, Cai
Y, Callen SE and Buch S: Cocaine mediated microglial activation
involves the ER stress-autophagy axis. Autophagy. 11:995–1009.
2015. View Article : Google Scholar
|
|
51
|
Deretic V: Autophagy as an innate immunity
paradigm: Expanding the scope and repertoire of pattern recognition
receptors. Curr Opin Immunol. 24:21–31. 2012. View Article : Google Scholar :
|
|
52
|
Chang KH, Sengupta A, Nayak RC, Duran A,
Lee SJ, Pratt RG, Wellendorf AM, Hill SE, Watkins M, Gonzalez-Nieto
D, et al: p62 is required for stem cell/progenitor retention
through inhibition of IKK/NF-κB/Ccl4 signaling at the bone marrow
macrophage-osteoblast niche. Cell Rep. 9:2084–2097. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS,
Kwon HJ, Kim HM, Kim DH and Yoon SY: Autophagy in microglia
degrades extracellular β-amyloid fibrils and regulates the NLRP3
inflammasome. Autophagy. 10:1761–1775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu L, Yang M, Kang R, Dai Y, Yu Y, Gao F,
Wang H, Sun X, Li X, Li J, et al: HMGB1-DNA complex-induced
autophagy limits AIM2 inflammasome activation through RAGE. Biochem
Biophys Res Commun. 450:851–856. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Meunier E, Dick MS, Dreier RF, Schürmann
N, Kenzelmann Broz D, Warming S, Roose-Girma M, Bumann D, Kayagaki
N, Takeda K, et al: Caspase-11 activation requires lysis of
pathogen-containing vacuoles by IFN-induced GTPases. Nature.
509:366–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wildenberg ME, Vos AC, Wolfkamp SC,
Duijvestein M, Verhaar AP, Te Velde AA, van den Brink GR and Hommes
DW: Autophagy attenuates the adaptive immune response by
destabilizing the immunologic synapse. Gastroenterology.
142:1493–1503 e1496. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lévy J, Cacheux W, Bara MA, L'Hermitte A,
Lepage P, Fraudeau M, Trentesaux C, Lemarchand J, Durand A, Crain
AM, et al: Intestinal inhibition of Atg7 prevents tumour initiation
through a microbiome-influenced immune response and suppresses
tumour growth. Nat Cell Biol. 17:1062–1073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Colotta F, Allavena P, Sica A, Garlanda C
and Mantovani A: Cancer-related inflammation, the seventh hallmark
of cancer: Links to genetic instability. Carcinogenesis.
30:1073–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|