Introduction
The major causes of treatment failure for patients
with leukemia are drug resistance and metastasis and traditional
therapies cannot eradicate all aggressive leukemia cells. New
chemotherapy drugs and hematopoietic stem cell transplantation
technology have improved the remission and disease-free survival
rate of leukemia patients, but still fail to prevent drug
resistance and metastasis (1). One
generally accepted theory is that leukemia is maintained by
leukemia stem cells (LSCs) (2),
which are quiescent and do not respond well to cell cycle-specific
cytotoxic agents used to treat leukemia. It is therefore impossible
to eradicate leukemia unless LSCs were eliminated (3,4).
Cancer stem cells (CSCs) are tumor-initiating and
tumor-propagating cells with enhanced resistance to
chemotherapeutic drugs. Vice versa, tumor cells resistant to
chemotherapeutic drugs have been shown to display certain
phenotypes and characteristics of CSCs (5). For example, colon cancer cells
treated with chemotherapy drugs exhibit CSC phenotypes (6). It is therefore speculated that
drug-tolerant or drug-surviving tumor cells may be enriched in
CSCs. In recent years, some CSC properties such as multidrug
resistance (7,8) and high migration ability have been
employed to isolate CSCs (9).
Theoretically, such strategies for investigating LSCs may well be
worth the efforts since drug resistance and metastasis are very
common and represent the worst clinical outcome in leukemia
patients.
Some studies suggest that the phenotype of LSCs is
CD34+CD38− (10,11).
Sox2, Oct4, Nanog, C-myc and Klf4 play essential roles in stem cell
maintenance (12–14). Research from the Tang laboratory
(14,15) has demonstrated that Nanog may also
play an important role in the self-renewal of CSCs. Lessard et
al (16) have reported that
Bmi-1 is a key factor in maintaining the self-renewal of LSCs
although it is not required for the initial leukemia
development.
Both CXC chemokine receptor 4 (CXCR4) and cell
adhesion ligand receptor CD44 have been implicated in leukemia
recurrence and metastasis. CXCR4 regulates tumor cell homing and
migration (17) and has been
implicated in maintaining prostate cancer stem-like cells and in
the drug resistance of breast cancer stem-like cells to tamoxifen
(18,19). Moreover, CXCR4 has become a new
target for the treatment of LSCs (20). On the other hand, the role of CD44
in LSCs seems to be controversial. Some studies suggest that CD44
contributes to the drug-resistance of LSCs to chemotherapeu-tics
and promotes leukemia development (21), consistent with the reported
involvement of CD44 in maintaining the stemness characteristics and
tumor metastasis of prostate CSCs (22). On the other hand, there are also
studies suggesting that CD44 (23)
may also inhibit growth of tumor cells. Therefore, it will be
worthwhile to explore the functions of CXCR4 and CD44 in the
invasion and metastasis of leukemic cells or LSCs.
The aim of this study is to investigate the
biological properties of chemotherapy-resistant leukemic cells with
an ultimate goal of developing novel therapeutic strategies against
the relapse of leukemia. Through our experiments we demonstrated
that hMDSCs-MOLT4 cells possess many CSC-like phenotypes and
properties.
Materials and methods
Cell lines and cell cultures
Human leukemia cell lines K562 (Chronic Myelogenous
Leukemia cell line), HL60 (Acute Promyelocytic Leukemia cell line),
MOLT4 (T-cell acute lymphocytic leukemia cell line, T-ALL) were
obtained from ATCC and cultured in RPMI-1640 medium (Hyclone,
Logan, UT, USA) supplemented with 10% FBS (Life Technologies, Grand
Island, NY, USA), penicillin (100 U/ml) and streptomycin (100
μg/ml). All cells were cultured in a humidified incubator at 37ºC
with 5% CO2. Only cells in the logarithmic phase of
growth were used for experiments.
MTT assays
Cells re-suspended in RPMI-1640 supplemented with
10% FBS were added into 96-well plates at 5000 cells/well in 100 μl
medium. Doxorubicin (DOX) was then added to cells at final
concentrations of 0.05, 0.1, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4,
1.6, and 1.8 μg/ml. Each drug concentration was set up in
triplicate wells with no-drug wells as controls. After incubation
at 37ºC with 5% CO2 for 24 h, 10 μl MTT solutions were
added to each well for additional 4 h. Cells in the plates were
centrifuged at 1000 rpm for 20 min and then 100 μl DMSO was added
to each well to dissolve any precipitate. Finally, cells were
placed on a shaker at low speed for 20 min to fully dissolve the
crystals and absorbance values were obtained by reading at
OD570 on a micro-plate reader.
Establishment of drug-surviving cells
(DSCs)
HL60, K562, or MOLT4 cells were treated with DOX at
the concentrations of 1.05 μg/ml, 1.35 μg/ml, or 0.6 μg/ml,
respectively, for 48 h, according to the IC50
concentrations of DOX on these cells. The surviving cells were
termed DSCs and used for subsequent experiments.
Establishment of high migration
drug-surviving (short-term) MOLT4 cells (hMDSCs-MOLT4)
Briefly, 500 μl culture medium with 20% FBS was
added in 24-well plate lower chamber, and 100 μl MOLT4 cell
suspension (5×105/ml) added in the Transwell chambers.
The pore size of Transwell is 0.8 μm. Cells were incubated at 37ºC
with 5% CO2 for 24 h. Then the chemotherapeutic drug DOX
at 0.6 μg/ml was added to the lower chambers for 72 h. The
surviving cells were termed hMDSCs-MOLT4 and used for subsequent
experiments. The percentage of imputs is ~18.3%.
Immunofluorescence
Cells were plated and cultured on sterilized
polylysine-coated coverslips for 15 min followed by washing in PBS.
To label the CXCR4 protein, cells were first fixed in 4%
paraformaldehyde in PBS for 10 min, permeabilized in 0.1% Triton
X-100 for 3 min at room temperature, followed by incubation with an
anti-CXCR4 antibody at 4ºC overnight. After washing in PBS, samples
were incubated with an FITC-linked secondary antibody in the dark
at room temperature for 30 min and then glass slides mounted in 90%
glycerol. Images were visualized and acquired with an
epifluorescence microscope.
Flow cytometry
Cells were harvested through centrifugation and
washed once with cold PBS. Then the cells were incubated with
FITC-anti-CD34, FITC-anti-CD38, PE-anti-CD44, PE-anti-CXCR4
(R&D Systems; Minneapolis, MN, USA), or PE-anti-P-gp
(eBioscience, San Diego, CA, USA) antibodies in the dark at 4ºC for
30 min, followed by washing in ice-cold PBS and finally
re-suspended in 500 μl PBS for flow cytometric analysis (Beckman,
Miami, FL, USA). As control, cells were stained with the matching
isotype control Abs.
RNA preparation, reverse
transcriptase-polymerase chain reaction (RT-PCR) and real-time
quantitative PCR
Total RNA was isolated from MOLT4 cells and
hMDSCs-MOLT4 using TRIzol reagent (Invitrogen, Merelbeke, Belgium)
according to the manufacturer's instructions. The concentration and
purity of total RNA were determined using spectrophotometry. RT was
carried out with 500 ng of total RNA from each sample using the RNA
PCR kit (Promega, Madison, WI, USA). Target mRNAs analysed included
those coding for P-gp and GAPDH. PCR products (10 μl) were
separated on 3% agarose gels, and the gel imaging system (Vilber
Lourma, Marne-la-Vallée, France) was used to scan the gel and
quantify the levels of expression. Amplification of GAPDH was used
as the control. As to the real-time quantitative PCR, All-in-One™
qPCR mix (GeneCopoeia, Rockville, MD, USA) and the primers for
Sox2, Oct4, c-Myc, Klf4, Nanog, and Bmi-1 were used to assess their
relative mRNA quantities. The Roter-Gene software was employed to
derive amplification and melting curves, and to calculate the
ratios of expression of target mRNAs in hMDSCs compared with MOLT-4
cells using the 2−ΔΔCT method.
Western blotanalysis
MOLT4 cells and hMDSCs-MOLT4 were harvested and
washed with PBS once and lysed in RIPA buffer. Protein content was
then determined by the BCA assay. The extracted proteins were
separated in a 12% SDS-polyacrylamide gel electrophoresis and
transferred to PVDF membranes. The membranes were first blocked
with 5% (w/v) nonfat dry milk (NFDM) in TBST and then probed with
the indicated primary antibodies with gentle shaking at 4ºC
overnight. After washing three times (5 min per time), the
membranes were incubated with the HRP-conjugated secondary
antibodies for 2 h. The signals were detected using an enhanced
chemiluminescence detection kit (Thermo Scientific, Rockford, IL,
USA). Western blot analysis was performed using the following
antibodies: anti-Sox2 (R&D Systems), anti-Oct4 (R&D
Systems), anti-c-Myc (R&D Systems), anti-Bmi-1 (R&D
Systems), anti-Klf4 (Abcam) and anti-Nanog (Abcam) antibodies,
anti-GAPDH (Protech) and secondary antibodies were obtained from
Guge Biology Company.
Measurement of cell proliferation by Cell
Counting Kit-8 (CCK-8)
Cells at 1.5×104/ml were added to 96-well
plates in 100 μl/well. Each condition was run in 6 repeats and the
plates were cultured in regular CO2 incubator. At 24, 48
and 72 h, 10 μl of CCK-8 solution was added to each well for an
additional 3 h of culture before termination. Cell numbers were
deduced by reading absorbance at OD450 on a microplate
reader.
Tumor formation ability of MOLT-4 and
hMDSCs-MOLT4 in vivo
SCID (severe combined immunodeficiency) mice were
obtained from Huafukang Institute (Beijing, China) and raised in
Individual Ventilated Cages in Wuhan University Center for Animal
Experiment/Animal Biosafty Level III laboratory. Experiments using
animal were approved by the Medical Ethics Committee of Wuhan
University School of Medicine. Under sterile conditions, MOLT-4
cells and hMDSCs-MOLT4 cells were injected subcutaneously into the
SCID mice (1.5×106 per mouse). Animals were observed and
weighed, and tumors measured and weighed every other day. Tumor
volume was estimated using the formula V=1/2 (length ×
width2) × Mice were sacrificed after the tumors grew for
7 days, and tumor tissue sections were analyzed by hematoxylin and
eosin (H&E) and immunohistochemistry staining.
H&E and immunohistochemical
staining
Tumor tissues from all xenografts were processed
following the routine procedure after 24 h fixation. Tumor tissue
sections were stained with H&E. For immunohistochemistry, all
samples were incubated at room temperature and washed with PBS.
Tumor tissue sections of 4 μm on glass slides were deparaffinized
and rehydrated. Antigen retrieval was performed in a pressure
cooker at 110ºC for 5 min in retrieval buffer. Endogenous
peroxidase activity was blocked with 3% hydrogen peroxide. Tumor
tissue sections were then incubated with anti-CXCR4 or CD44
antibody (Abcam, USA) for 30 min. Immunocomplexes were detected
after incubation with HRP (horseradish peroxidase) polymer for 30
min followed by incubation with DAB plus substrate for 10 min.
Normal IgG from the same species of the primary antibody and
diluted to match the concentration of the primary antibody was used
as the negative control. Images were analyzed using Image-Pro Plus
6.0 software by obtaining positive integrated optical density (IOD.
The average IOD value of all photos in each group was used to
represent the IOD of the group and expressed as the mean ± SD.
Statistical analysis
All experiments were repeated at least three times.
Experimental data were expressed as mean ± SD, using SPSS17.0
statistical software for data analysis of variance (ANOVA) and
Student's t-test. P<0.05 was considered statistically
significant.
Results
Response of three leukemia cell lines to
doxorubicin (DOX)
We first used MTT assays to evaluate the effects of
DOX on three leukemia cell lines, i.e., HL60, K562, and MOLT4. We
determined the IC50 of HL60, K562, and MOLT4 cells to
DOX treatment to be 1.05, 1.35 and 0.6 μg/ml, respectively. In all
subsequent experiments, we used DOX at the above IC50
concentrations to treat cells over 48 h followed by collecting
drug-surviving cells.
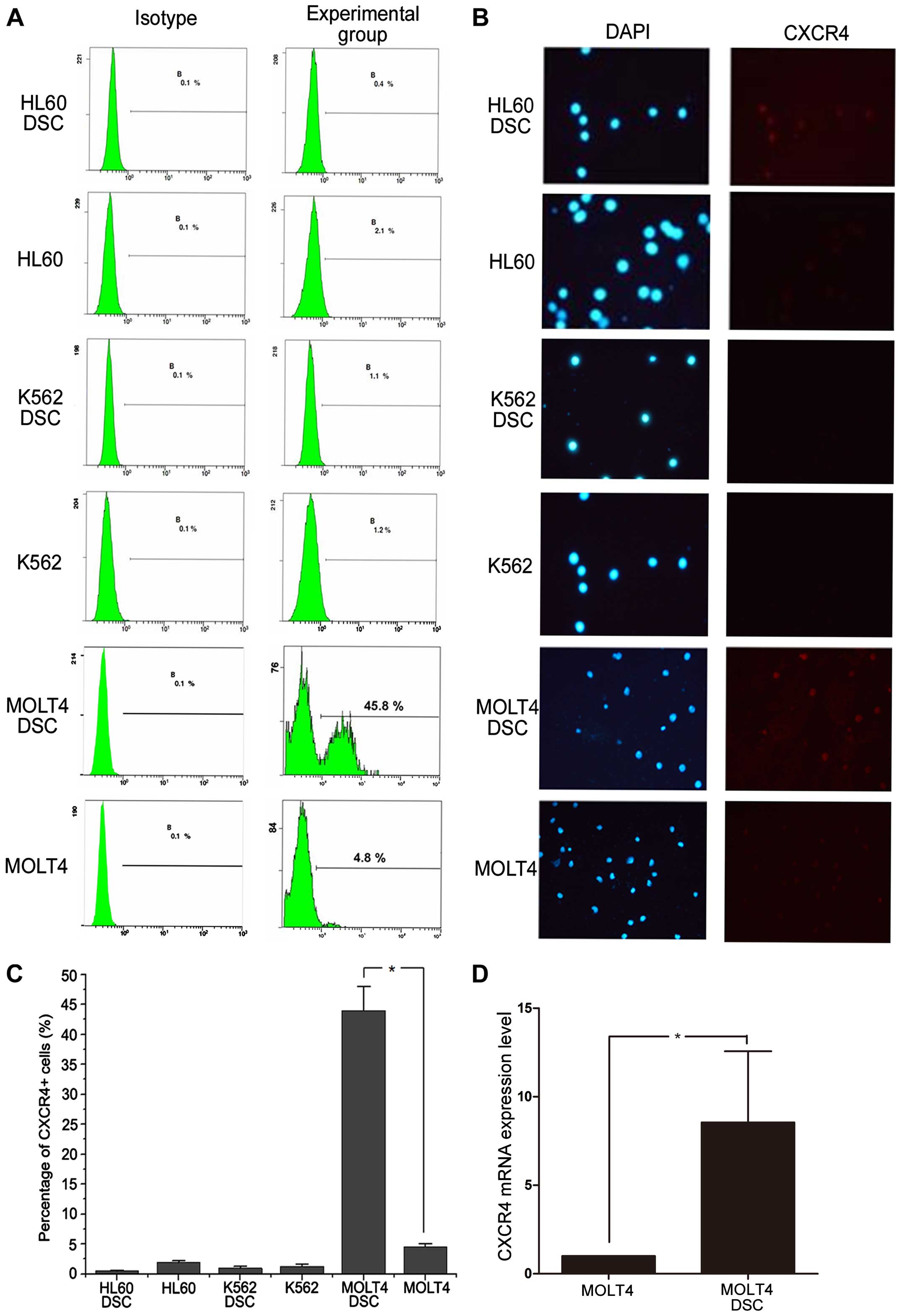
Analysis of CXCR4 expression in
drug-surviving cells
CXCR4 and its ligand CXCL12 are highly expressed on
acute promyelocytic leukemia stem cells and involved in regulating
the migration of LSCs. We used flow cytometry (FCM) and
immunofluorescence microscopy to assess the CXCR4 expression levels
in HL60, K562, and MOLT4 cells and their drug-surviving cells
(DSCs). There was no significant difference in CXCR4 expression
between drug-surviving HL60 and K562 cells and their corresponding
parental cells (Fig. 1A–C).
However, compared with the parental cells, drug-surviving MOLT4
cells expressed much higher levels of the stem cell surface marker
CXCR4 (45.8% vs. 4.8%; P<0.05; Fig.
1A and C). In addition, we detected CXCR4 mRNA expression in
MOLT4 cells and MOLT4 DSCs. The result showed increased CXCR4 mRNA
expression in hMDSCs-MOLT4 vs. MOLT4 cells (n=5, P<0.05)
(Fig. 1D).
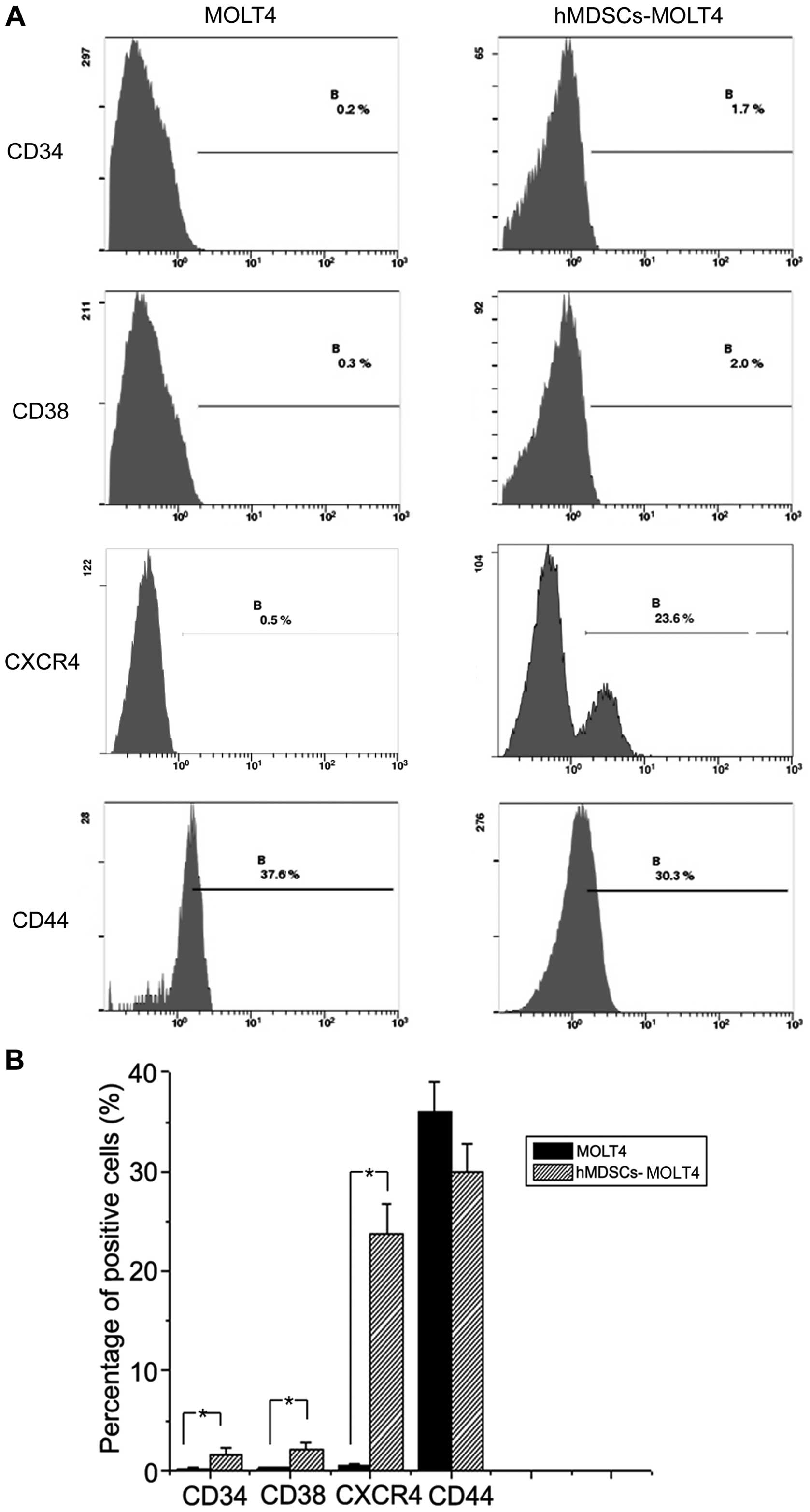
Stem cell-like properties of hMDSCs-MOLT4
cells
Next, we employed flow cytometry to examine the
expression levels of several stem cell related markers including
CD34, CD38, CXCR4, and CD44 in hMDSC-MOLT4 cells. The results
showed that the expression levels of CD34 and CD38 on MOLT4 and
hMDSC-MOLT4 cells were 0.2% vs. 1.7%, and 0.3% vs. 2%, respectively
(Fig. 2), suggesting upregulation
of both molecules in hMDSC-MOLT4 cells. CXCR4 expression went up
significantly, from 0.5% in MOLT4 cells to 23.3% in hMDSC-MOLT4
cells (n=5, P<0.05) (Fig.
2).
In contrast to the upregulation of the above three
molecules, CD44, one of the cell adhesion molecule (CAM) family
members associated with proliferation of some tumor cells
(including multiple myeloma and AMl cells) and with drug
resistance, did not show significant alterations. In fact, its
expression showed a slight decrease, from 37.5% in MOLT4 cells to
30.3% in hMDSC-MOLT4 cells (Fig.
2).
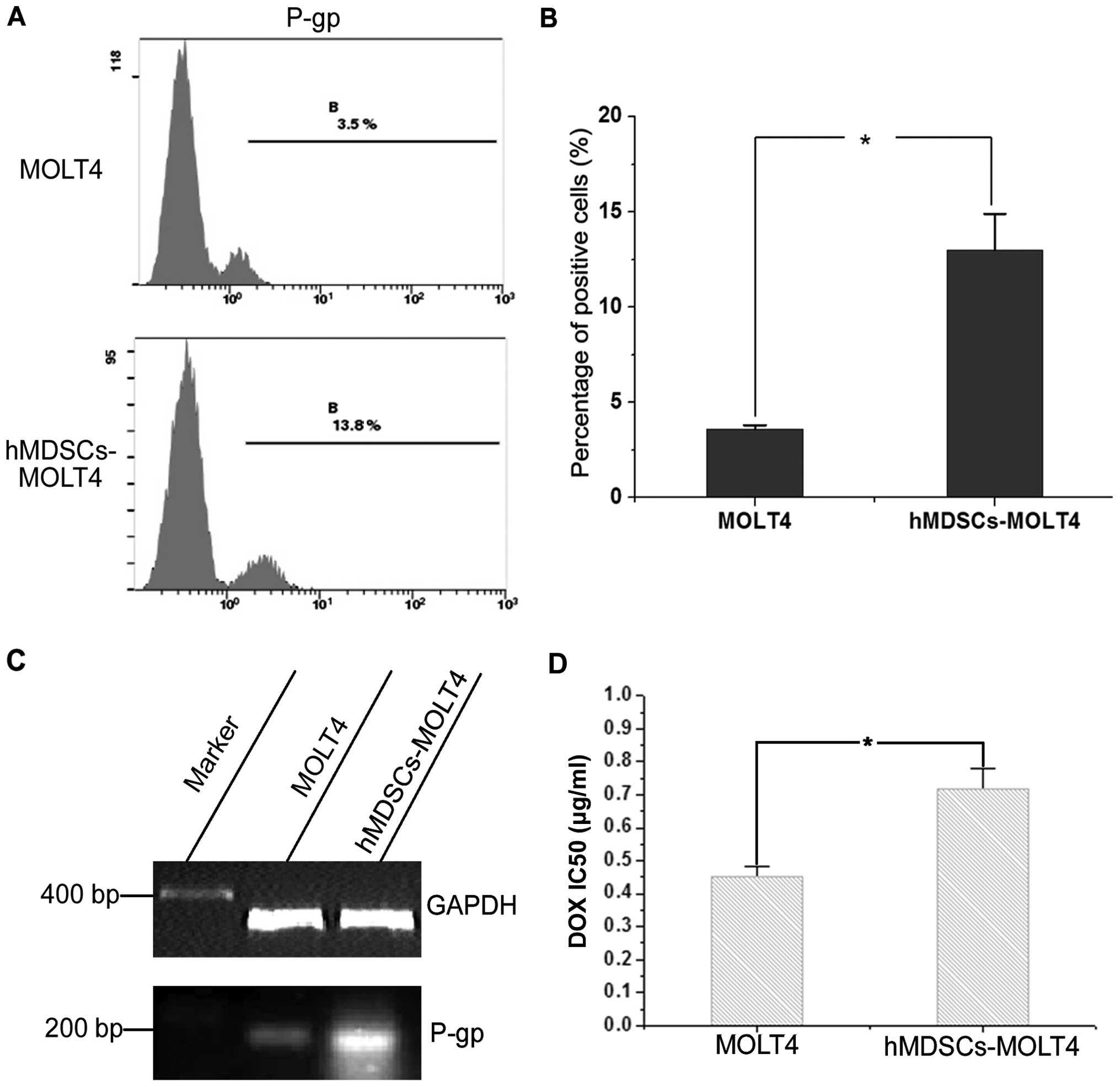
Increased drug resistance in hMDSCs-MOLT4
cells
P-glycoprotein (P-gp) is one of the cell surface
pump proteins that mediate drug resistance (24). We first used FCM to assess P-gp
protein expression in MOLT4 and hMDSCs-MOLT4 cells. The results
revealed increased P-gp in MOLT4 (3.5%) vs. hMDSCs-MOLT4 (13.8%)
cells (n=6, P<0.05) (Fig. 3A and
B). Consistent with the FCM data, RT-PCR analysis also
demonstrated increased P-gp mRNA expression in hMDSCs-MOLT4 cells
in comparison to MOLT4 cells (Fig.
3C). In addition, MTT assays showed that the IC50
values of parental MOLT4 cells and hMDSCs-MOLT4 cells were 0.451
and 0.718, respectively (n=6, P<0.05) (Fig. 3D), indicating enhanced drug
resistance of hMDSCs-MOLT4 cells.
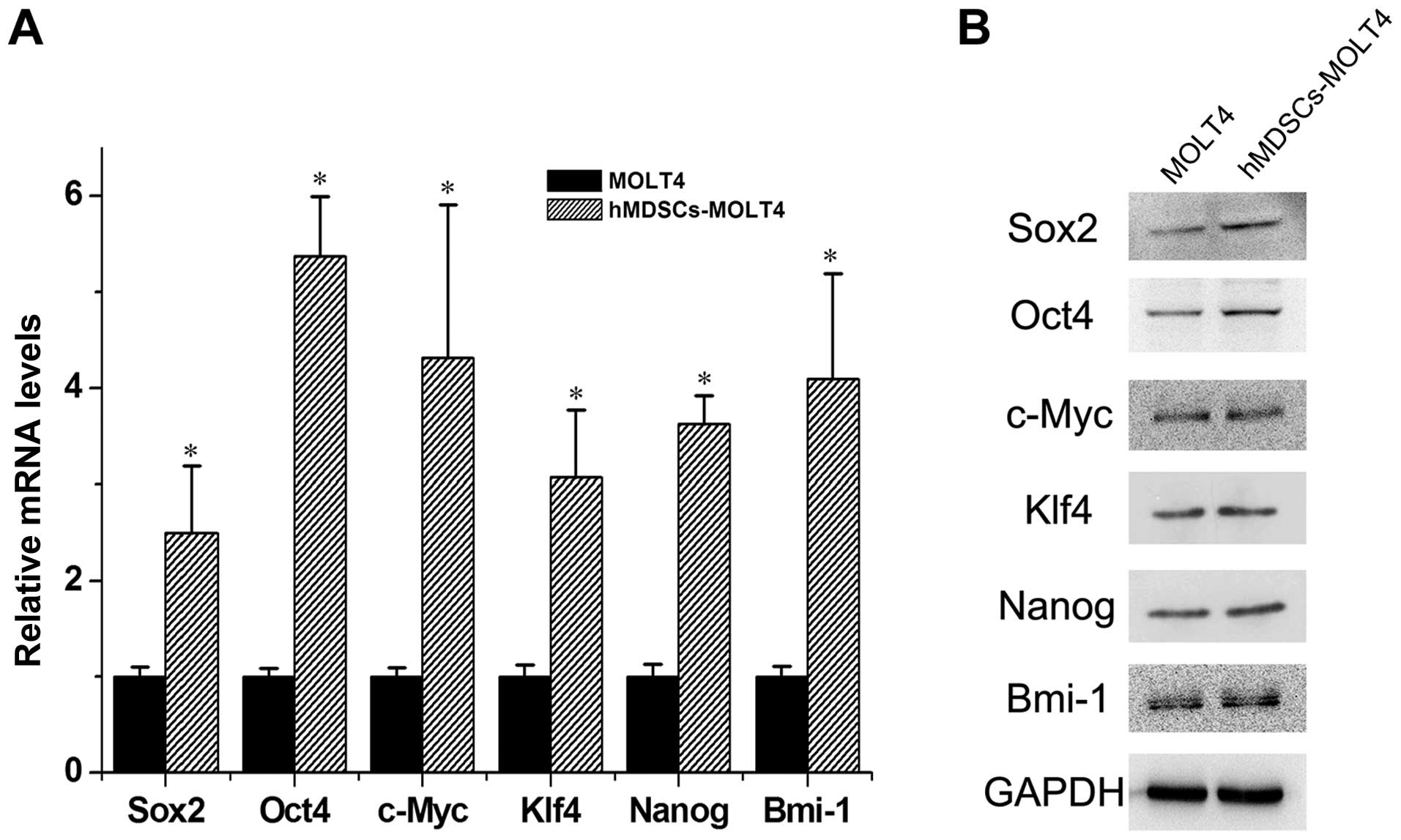
Increased expression of stem cell-related
molecules in hMDSCs-MOLT4 cells
Subsequently, we carried out real-time quantitative
RT-PCR and western blot analyses to assess the expression levels of
Sox2, Oct4, c-Myc, Klf4, Nanog, and Bmi-1 in MOLT4 vs. hMDSCs-MOLT4
cells. The results revealed ~2.5-, 5.4-, 4.3-, 3.1-, 3.6- and
4.1-fold higher mRNAs of Sox2, Oct4, c-Myc, Klf4, Nanog, and Bmi-1
in hMDSCs-MOLT4 cells relative to those in MOLT4 cells (Fig. 4A). The protein level of Sox2, Oct4,
Klf4 and Nanog were also upregulated in hMDSCs-MOLT4 cells
(Fig. 4B).
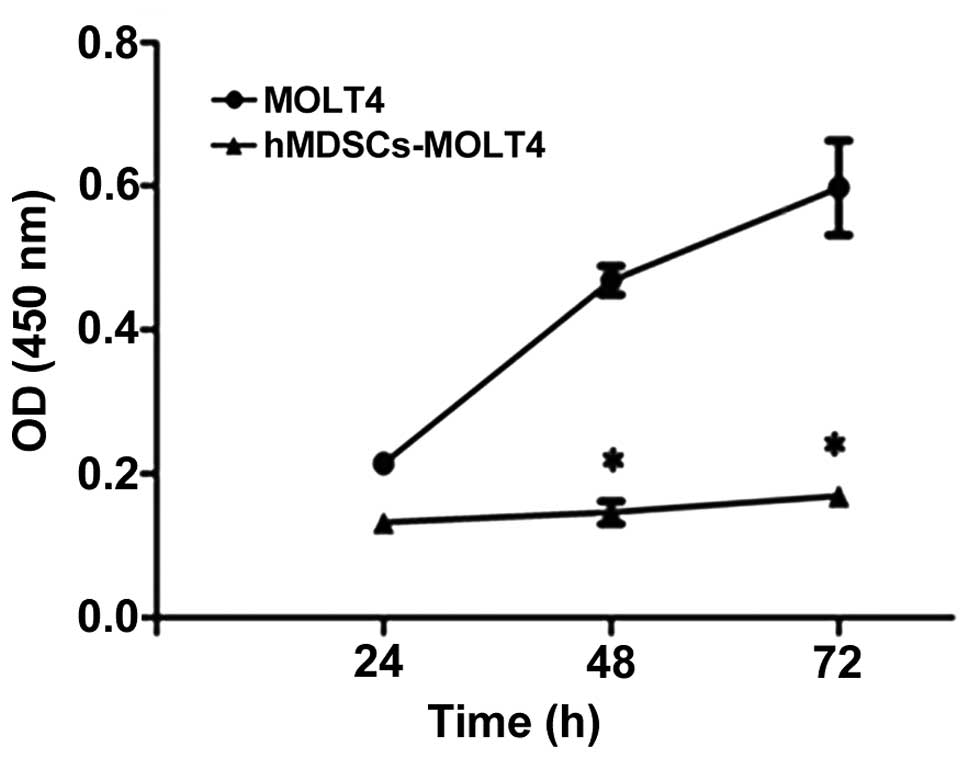
Decreased proliferation of hMDSCs-MOLT4
cells
We use the CCK8 assay to determine the relative cell
proliferation. In this assay, CCK-8 is reduced to formazan by some
intracellular dehydrogenase enzymes released by the mitochondria in
viable tumor cells. The 450-nm absorbance is positively associated
with the cell number. Our results revealed decreased proliferation
in hMDSCs-MOLT4 cells in comparison to the parental cells (Fig. 5).
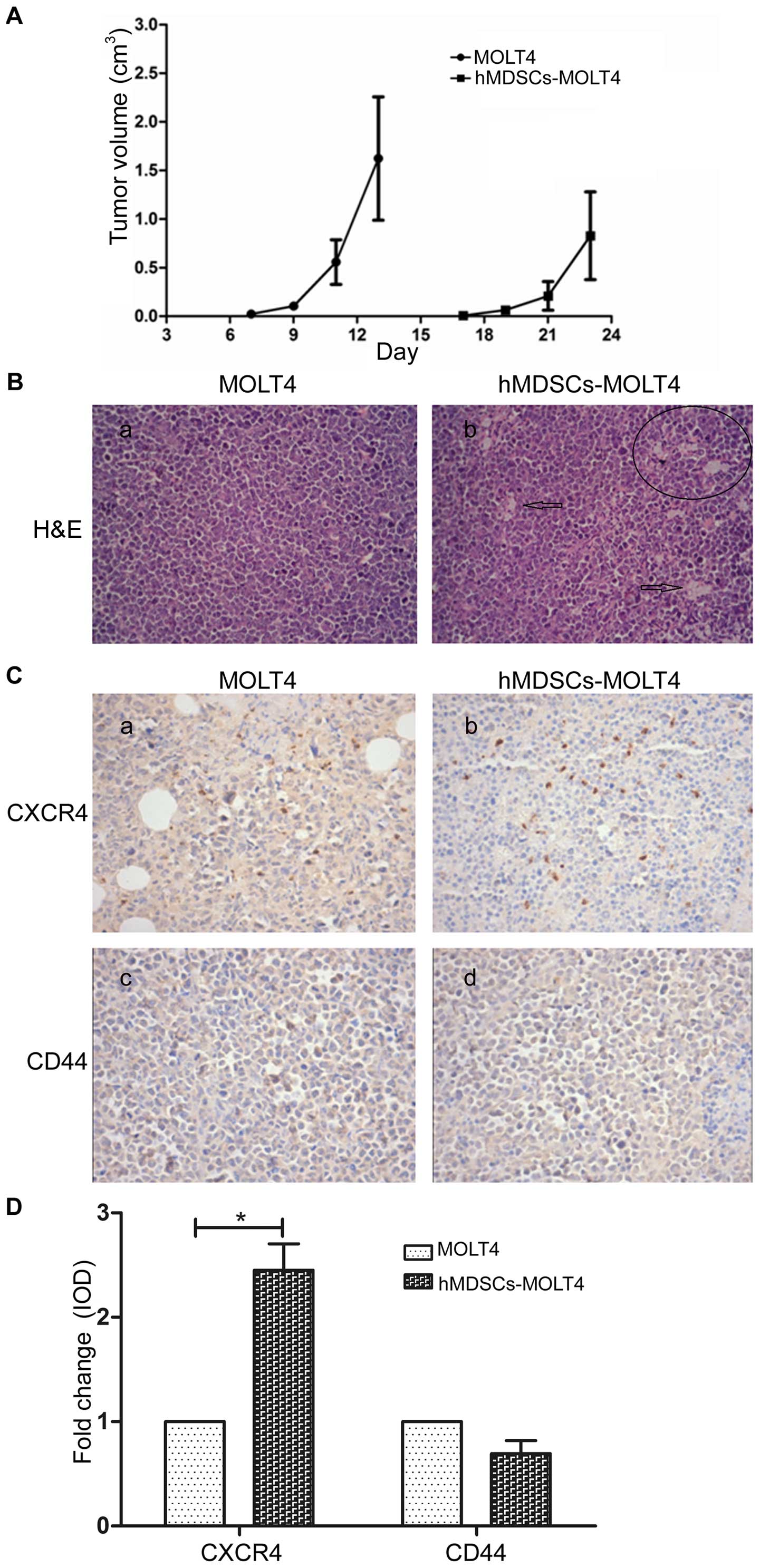
Tumorigenicity of MOLT4 and hMDSCs-MOLT4
cells
We then determined the tumorigenic potential of
MOLT4 and hMDSCs-MOLT4 cells by injecting equal numbers of the two
cell types subcutaneously in SCID mice. We did not observe any
differences in the groups of mice with respect to hair coat, body
weights and overall animal well being. However, the injected
hMDSCs-MOLT4 cells developed much smaller and slower-growing tumors
than MOLT4 cells (Fig. 6A). The
group injected with MOLT4 cells developed tumors as early as 7 days
post-injection whereas the group injected with hMDSCs-MOLT4 cells
did not develop tumors until 17 days after tumor cell injections.
Histological evaluations by H&E staining of tumor tissues
indicated that the hMDSCs-MOLT4 tumors appeared to have more
abundant connective tissues and blood vessels (i.e., tumor stroma)
(Fig. 6B).
Finally, we analyzed, by IHC, the expression of
CXCR4 and CD44 in tumor tissue sections. The results demonstrated
that tumors derived from hMDSCs-MOLT4 cells showed significantly
higher levels of CXCR4 (n=9, P<0.01; Fig. 6C and D) but slight lower levels of
CD44 compared to tumors derived from MOLT4 cells (Fig. 6C and D).
Discussion
Some evidence has demonstrated that LSCs contribute
to treatment failure. However, few studies have focused on the
relationship of drug surviving leukemia cells and LSCs, not to
mention the high migration drug surviving cells. Herein, we found
that, compared with the parental cells, drug-surviving (short-term)
MOLT4 cells expressed much higher levels of the stem cell surface
marker CXCR4 which also related to the tumor metastasis. Then we
have paid close attention to these hMDSCs-MOLT4 cells. For the
first time, we explore stemness, drug resistance and metastasis in
hMDSCs-MOLT4 cells.
It has been reported that CD34, CD44, Sox2, Oct4,
Nanog are the markers of cancer stem/progenitor cells and play an
important role in maintaining their ‘stemness’ (25). C-Myc and KLF4 can maintain the
stemness of stem cells and promote tumor formation (26). Bmi-1 is related to the self-renewal
of leukemia stem cells (16).
Another study showed that overexpression of C-Myc, Bmi-1, Oct4,
Nanog in precancerous and cancerous cells may initiate oncogenic
epithelial-mesenchymal transition and tumorigenesis, which plays
important roles in the genesis of cancer stem cells (CSCs),
malignant tumor initiation and progression, cancer metastasis, and
drug resistance (27). In this
study, hMDSC-MOLT4 cells have some characteristics of leukemia
stem-like cells with high expression of CD34, CXCR4, SOX2, OCT4,
C-Myc, KLF4, Nanog and BMI-1.
The emergence of resistance to chemotherapy by tumor
cells, when combined with metastasis, is the primary driver of
mortality in cancer patients (28). The hMDSCs-MOLT4 cells, possessing
some stemness of LSCs, highly expressed P-gp and demonstrated
enhanced drug resistance of these cells. As is well known,
metastasis is one of the primary biological characteritics of
malignant tumors and the most important factor for the prognosis.
We found that the expression of CXCR4, which is related to tumor
cell homing and migration, was significantly higher in hMDSCs-MOLT4
cells than in MOLT4 cells. We also confirmed the high expression
CXCR4 in tumor sections in vivo.
In addition, hMDSCs-MOLT4 cells seem to have a
strong invasive potential in vivo, evidenced by strong
interstitial and vascular tissues in tumor tissue sections. At the
same time, the results showed that hMDSCs-MOLT4 cells exhibit
decreased proliferation ability in vitro and in vivo.
In line with our results, Stiehl et al reported that
leukemia stem cells have a lower proliferative activity than other
mitotic cell types and their replication is the rate-limiting
process during expansion of leukemic cells. Moreover, slower
cycling LSCs are the potential risk leading to relapse (29). Therefore, we speculate that
hMDSCs-MOLT4 cells show similar characteristics with LSCs.
In this study, we also paid attention to another
molecule, i.e., CD44. Researchers have reported that its high
expression level is often related to proliferation and drug
resistance in some tumor cells whereas some other studies presented
opposite results (22,23). We observed a slight decrease in
CD44 expression in hMDSCs-MOLT4 cells in vitro and in
vivo. In addition, we also observed that slightly decreased
expression of CD44 was correlated with decreased proliferation of
hMDSCs-MOLT4 cells in vivo and in vitro. Herein, we
did not find the closed relationship between CD44 molecules and
migration and drug resistance.
Prud'homme (30)
suggests that CXCR4 and its ligand CXCL12 are metastasis-related
key target molecules in the therapy of CSCs. Carter et al
propose that imatinib combined with other drugs that target
self-renewal and induce apoptosis could enhance the effect of
imatinib on CMl cells (31). Our
studies on hMDSCs-MOLT4 cells suggest that targeting stemness
factors such as Oct4, Nanog, Sox2, Klf4 and CXCR4 may represent
plausible options for eliminating T-ALL stem-like cells.
Furthermore, an appropriate combination of drugs targeting the
above mentioned stem cell factors and conventional chemotherapeutic
agents may be a promising new treatment strategy. Finally,
hMDSCs-MOLT4 cells may be used as a good research tool for finding
novel drugs that could target cancer stem-like cells and prove to
be effective in managing the relapse and metastasis of T-ALL.
In conclusion, the present study demonstrated that
hMDSCs-MOLT4 cells exhibited strong drug resistance and certain
cancer stem cell-like characteristics. For the first time, we
proposed that stemness factors such as Sox2, Oct4, Klf4, Nanog and
CXCR4 may be used as targets for eliminating T-ALL stem-like cells.
It indicated that high migration drug surviving leukemia cells in
T-ALL patients were likely to play a similar role to that of LSCs,
and were probably important in the relapse of T-ALL. These findings
provide new possibilities in understanding the relationship between
high migration drug surviving leukemia cells and LSCs, and may
present a new research direction for T-ALL relapse.
Acknowledgements
This work was supported by National Natural Science
Foundation of China (no. 81400121, 81270607, 81541027) and Ph.D.
Independent Research Program of Wuhan University (no. 410500106).
We thank Ms. Weihuang Liu for FCM analysis.
Abbreviations:
|
CSCs
|
cancer stem cells
|
|
CXCR4
|
CXC chemokine receptor 4
|
|
DOX
|
doxorubicin
|
|
DSCs
|
drug-surviving cells
|
|
hMDSCs-MOLT4
|
high migration drug-surviving
(short-term) MOLT4 cells
|
|
IOD
|
integrated optical density
|
|
IVC
|
individual ventilated cages
|
|
LSCs
|
leukemia stem cells
|
|
P-gp
|
P-glycoprotein
|
References
|
1
|
Vyas P, Appelbaum FR and Craddock C:
Allogeneic hematopoietic cell transplantation for acute myeloid
leukemia. Biol Blood Marrow Transplant. 21:8–15. 2015. View Article : Google Scholar
|
|
2
|
Lang F, Wojcik B and Rieger MA: Stem cell
hierarchy and clonal evolution in acute lymphoblastic leukemia.
Stem Cells Int. 2015:1371642015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krause DS and Van Etten RA: Right on
target: Eradicating leukemic stem cells. Trends Mol Med.
13:470–481. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lane SW, Scadden DT and Gilliland DG: The
leukemic stem cell niche: Current concepts and therapeutic
opportunities. Blood. 114:1150–1157. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schulenburg A, Blatt K, Cerny-Reiterer S,
Sadovnik I, Herrmann H, Marian B, Grunt TW, Zielinski CC and Valent
P: Cancer stem cells in basic science and in translational
oncology: Can we translate into clinical application? J Hematol
Oncol. 8:162015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dallas NA, Xia L, Fan F, Gray MJ, Gaur P,
van Buren G II, Samuel S, Kim MP, Lim SJ and Ellis LM:
Chemoresistant colorectal cancer cells, the cancer stem cell
phenotype, and increased sensitivity to insulin-like growth
factor-I receptor inhibition. Cancer Res. 69:1951–1957. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khan IN, Al-Karim S, Bora RS, Chaudhary AG
and Saini KS: Cancer stem cells: A challenging paradigm for
designing targeted drug therapies. Drug Discov Today. 20:1205–1216.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li HZ, Yi TB and Wu ZY: Suspension culture
combined with chemotherapeutic agents for sorting of breast cancer
stem cells. BMC Cancer. 8:1352008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu YM, Li XF, Liu H and Wu XL:
Ultrasound-targeted micro-bubble destruction-mediated
downregulation of CD133 inhibits epithelial-mesenchymal transition,
stemness and migratory ability of liver cancer stem cells. Oncol
Rep. 34:2977–2986. 2015.PubMed/NCBI
|
|
10
|
Becker MW and Jordan CT: Leukemia stem
cells in 2010: Current understanding and future directions. Blood
Rev. 25:75–81. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kornblau SM, Qutub A, Yao H, York H, Qiu
YH, Graber D, Ravandi F, Cortes J, Andreeff M, Zhang N, et al:
Proteomic profiling identifies distinct protein patterns in acute
myelogenous leukemia CD34+CD38− stem-like
cells. PLoS One. 8:e784532013. View Article : Google Scholar
|
|
12
|
Amaya CN and Bryan BA: Enrichment of the
embryonic stem cell reprogramming factors Oct4, Nanog, Myc, and
Sox2 in benign and malignant vascular tumors. BMC Clin Pathol.
15:182015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin X, Zhang BH, Zheng SS, Gao DM, Qiu SJ,
Wu WZ and Ren ZG: Coexpression of gene Oct4 and Nanog initiates
stem cell characteristics in hepatocellular carcinoma and promotes
epithelial-mesenchymal transition through activation of Stat3/Snail
signaling. J Hematol Oncol. 8:232015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeter CR, Liu B, Liu X, Chen X, Liu C,
Calhoun-Davis T, Repass J, Zaehres H, Shen JJ and Tang DG: NANOG
promotes cancer stem cell characteristics and prostate cancer
resistance to androgen deprivation. Oncogene. 30:3833–3845. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeter CR, Badeaux M, Choy G, Chandra D,
Patrawala L, Liu C, Calhoun-Davis T, Zaehres H, Daley GQ and Tang
DG: Functional evidence that the self-renewal gene NANOG regulates
human tumor development. Stem Cells. 27:993–1005. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lessard J and Sauvageau G: Bmi-1
determines the proliferative capacity of normal and leukaemic stem
cells. Nature. 423:255–260. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han AR, Lee JY, Kim HJ, min WS, Park G and
Kim SH: A CXCR4 antagonist leads to tumor suppression by activation
of immune cells in a leukemia-induced microenvironment. Oncol Rep.
34:2880–2888. 2015.PubMed/NCBI
|
|
18
|
Dubrovska A, Elliott J, Salamone RJ,
Telegeev GD, Stakhovsky AE, Schepotin IB, Yan F, Wang Y, Bouchez
LC, Kularatne SA, et al: CXCR4 expression in prostate cancer
progenitor cells. PLoS One. 7:e312262012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dubrovska A, Hartung A, Bouchez LC, Walker
JR, Reddy VA, Cho CY and Schultz PG: CXCR4 activation maintains a
stem cell population in tamoxifen-resistant breast cancer cells
through AhR signalling. Br J Cancer. 107:43–52. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tavernier E, Aanei C, Solly F,
Flandrin-Gresta P, Campos L and Guyotat D: CXCR4: A new therapeutic
target of the leukaemic cell? Role of the SDF-1/CXCR4 axis in acute
myeloid leukaemia. Bull Cancer. 101:593–604. 2014.(In French).
PubMed/NCBI
|
|
21
|
Williams DA and Cancelas JA: Leukaemia:
Niche retreats for stem cells. Nature. 444:827–828. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Erb U, Megaptche AP, Gu X, Büchler MW and
Zöller M: CD44 standard and CD44v10 isoform expression on leukemia
cells distinctly influences niche embedding of hematopoietic stem
cells. J Hematol Oncol. 7:292014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Assanhou AG, Li W, Zhang L, Xue L, Kong L,
Sun H, Mo R and Zhang C: Reversal of multidrug resistance by
co-delivery of paclitaxel and lonidamine using a TPGS and
hyaluronic acid dual-functionalized liposome for cancer treatment.
Biomaterials. 73:284–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hämmerle B, Yañez Y, Palanca S, Cañete A,
Burks DJ, Castel V and Font de Mora J: Targeting neuroblastoma stem
cells with retinoic acid and proteasome inhibitor. PLoS One.
8:e767612013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Liu Q, Hou B, Zhang W, Yan M, Jia
H, Li H, Yan D, Zheng F, Ding W, et al: Concomitant targeting of
multiple key transcription factors effectively disrupts cancer stem
cells enriched in side population of human pancreatic cancer cells.
PLoS One. 8:e739422013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo P, Gao A, Zhang G, Han H and Zhou Q:
Decoding the knots of initiation of oncogenic
epithelial-mesenchymal transition in tumor progression. Curr Cancer
Drug Targets. 13:996–1011. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu A, Loutherback K, Lambert G,
Estévez-Salmerón L, Tlsty TD, Austin RH and Sturm JC: Cell motility
and drug gradients in the emergence of resistance to chemotherapy.
Proc Natl Acad Sci USA. 110:16103–16108. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Stiehl T, Baran N, Ho AD and
Marciniak-Czochra A: Cell division patterns in acute myeloid
leukemia stem-like cells determine clinical course: A model to
predict patient survival. Cancer Res. 75:940–949. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Prud'homme GJ: Cancer stem cells and novel
targets for antitumor strategies. Curr Pharm Des. 18:2838–2849.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Carter BZ, Mak DH, Cortes J and Andreeff
M: The elusive chronic myeloid leukemia stem cell: Does it matter
and how do we eliminate it? Semin Hematol. 47:362–370. 2010.
View Article : Google Scholar : PubMed/NCBI
|