|
1
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allison DC, Carney SC, Ahlmann ER,
Hendifar A, Chawla S, Fedenko A, Angeles C and Menendez LR: A
meta-analysis of osteosarcoma outcomes in the modern medical era.
Sarcoma. 2012:7048722012. View Article : Google Scholar : PubMed/NCBI
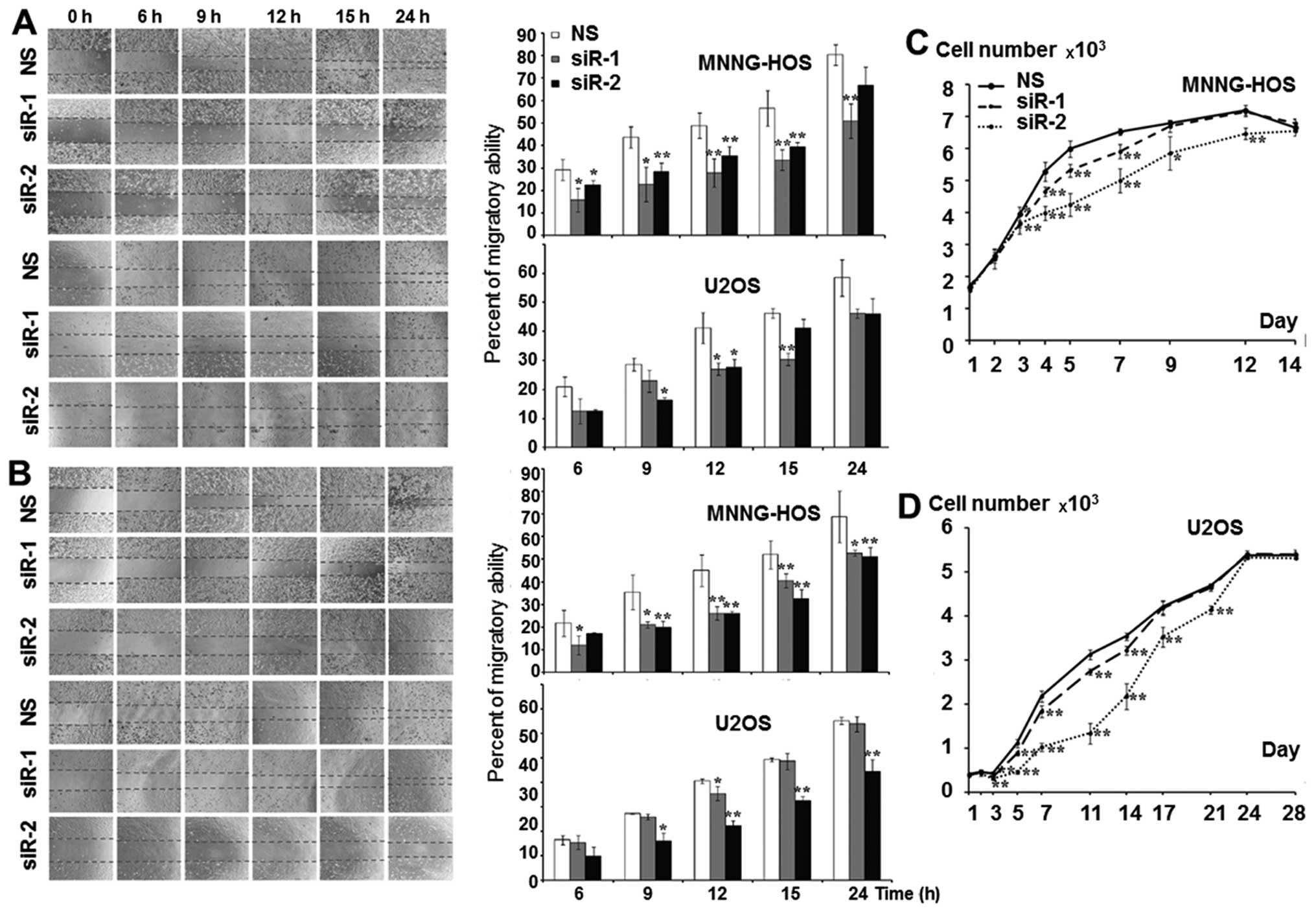
|
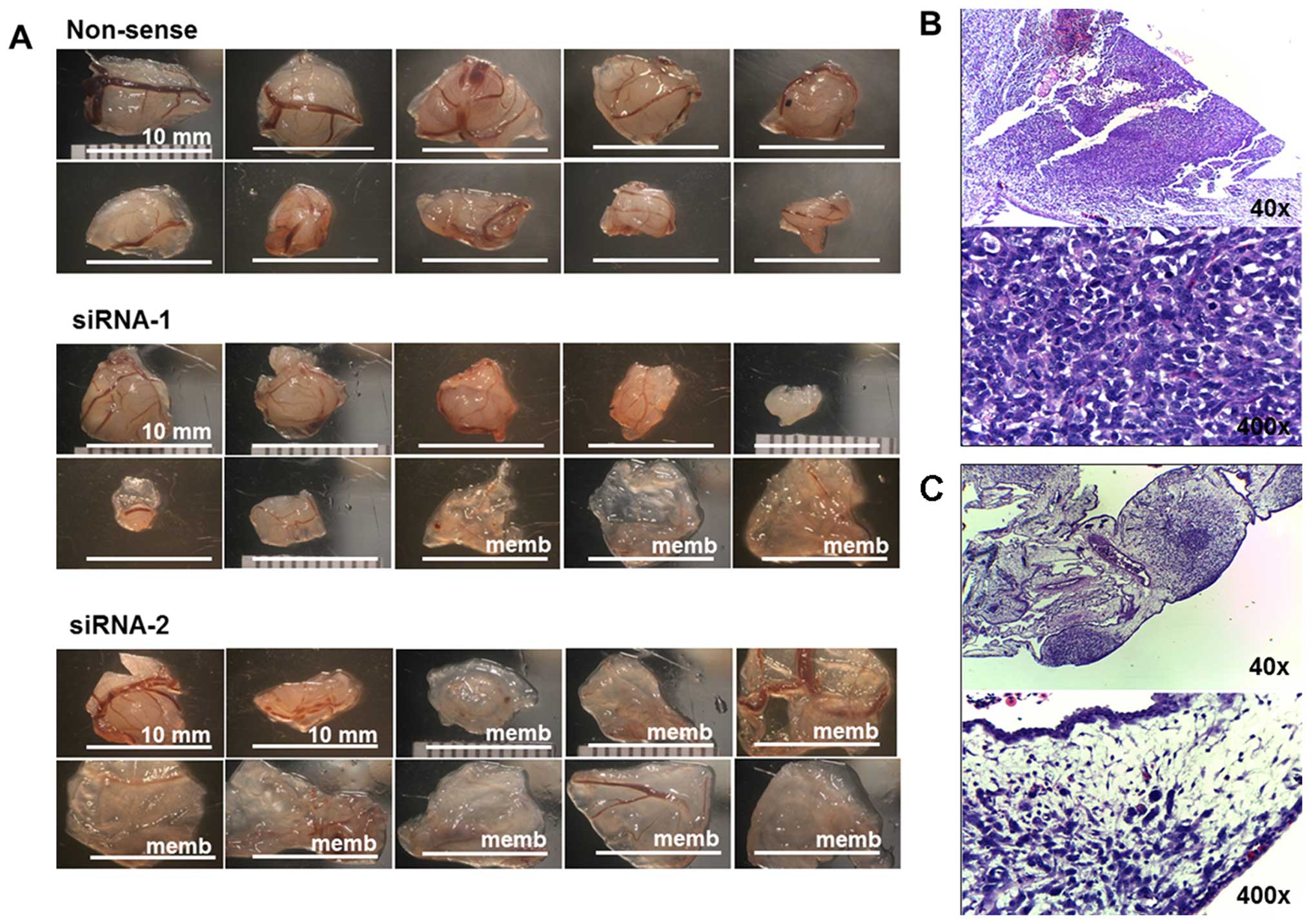
|
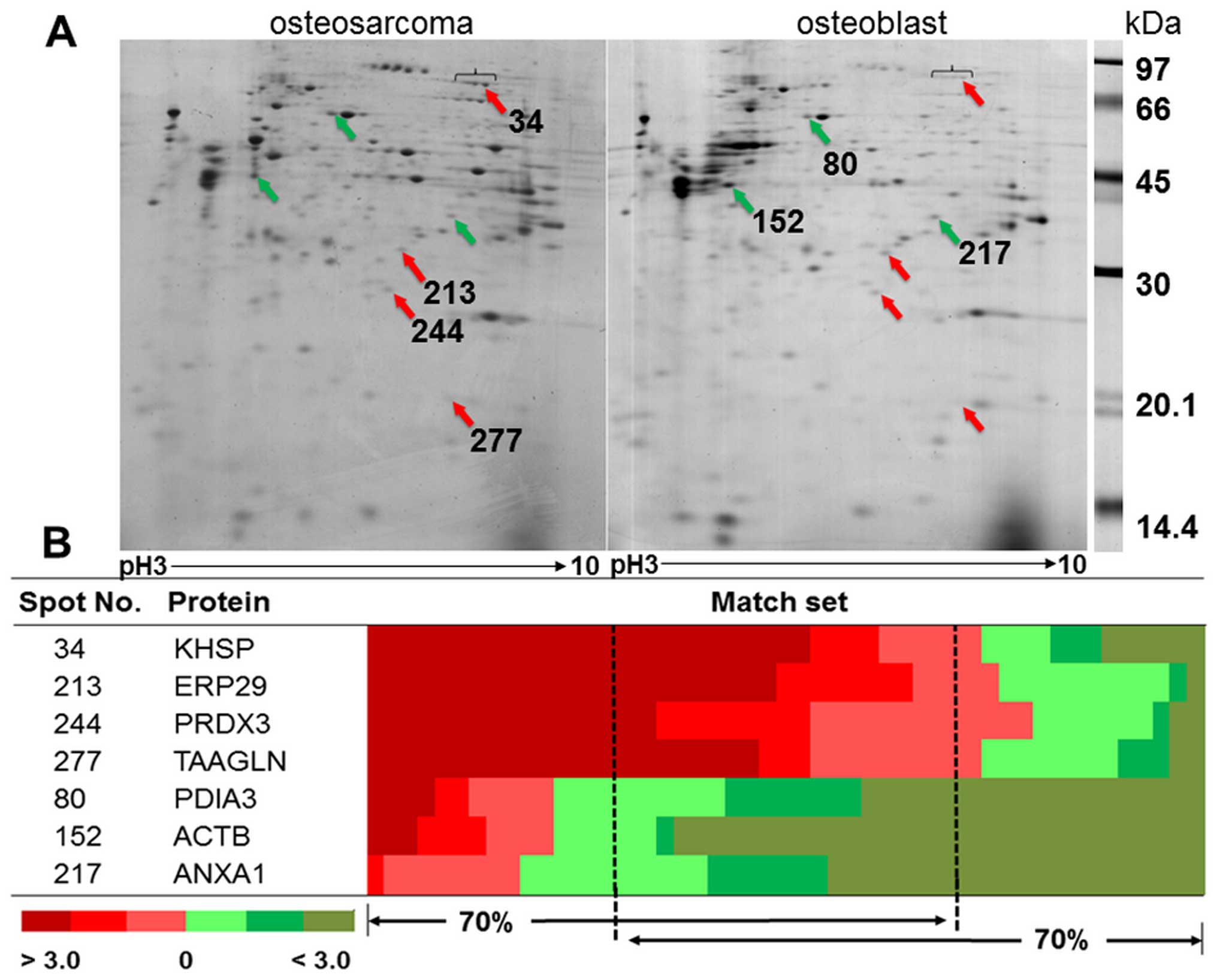
3
|
Gelberg KH, Fitzgerald EF, Hwang S and
Dubrow R: Growth and development and other risk factors for
osteosarcoma in children and young adults. Int J Epidemiol.
26:272–278. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ottaviani G and Jaffe N: The etiology of
osteosarcoma. Cancer Treat Res. 152:15–32. 2009. View Article : Google Scholar
|
|
5
|
Zhou W, Hao M, Du X, Chen K, Wang G and
Yang J: Advances in targeted therapy for osteosarcoma. Discov Med.
17:301–307. 2014.PubMed/NCBI
|
|
6
|
Jonsson KB, Frost A, Nilsson O, Ljunghall
S and Ljunggren O: Three isolation techniques for primary culture
of human osteoblast-like cells: A comparison. Acta Orthop Scand.
70:365–373. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pautke C, Schieker M, Tischer T, Kolk A,
Neth P, Mutschler W and Milz S: Characterization of osteosarcoma
cell lines MG-63, Saos-2 and U-2 OS in comparison to human
osteoblasts. Anticancer Res. 24:3743–3748. 2004.
|
|
8
|
Alameh M, Jean M, Dejesus D, Buschmann MD
and Merzouki A: Chitosanase-based method for RNA isolation from
cells transfected with chitosan/siRNA nanocomplexes for real-time
RT-PCR in gene silencing. Int J Nanomed. 5:473–481. 2010.
|
|
9
|
Martins AM, Pham QP, Malafaya PB, Sousa
RA, Gomes ME, Raphael RM, Kasper FK, Reis RL and Mikos AG: The role
of lipase and alpha-amylase in the degradation of
starch/poly(epsilon-caprolactone) fiber meshes and the osteogenic
differentiation of cultured marrow stromal cells. Tissue Eng Part
A. 15:295–305. 2009. View Article : Google Scholar
|
|
10
|
Gregory CA, Gunn WG, Peister A and Prockop
DJ: An Alizarin red-based assay of mineralization by adherent cells
in culture: Comparison with cetylpyridinium chloride extraction.
Anal Biochem. 329:77–84. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kleiner DE and Stetler-Stevenson WG:
Quantitative zymography: Detection of picogram quantities of
gelatinases. Anal Biochem. 218:325–329. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pruksakorn D, Lirdprapamongkol K,
Chokchaichamnankit D, Subhasitanont P, Chiablaem K, Svasti J and
Srisomsap C: Metabolic alteration of HepG2 in scaffold-based 3-D
culture: Proteomic approach. Proteomics. 10:3896–3904. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolt MW and Mahoney PA: High-efficiency
blotting of proteins of diverse sizes following sodium dodecyl
sulfate-polyacrylamide gel electrophoresis. Anal Biochem.
247:185–192. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weil D, Garçon L, Harper M, Duménil D,
Dautry F and Kress M: Targeting the kinesin Eg5 to monitor siRNA
transfection in mammalian cells. Biotechniques. 33:1244–1248.
2002.PubMed/NCBI
|
|
15
|
Ridley AJ, Schwartz MA, Burridge K, Firtel
RA, Ginsberg MH, Borisy G, Parsons JT and Horwitz AR: Cell
migration: Integrating signals from front to back. Science.
302:1704–1709. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Balke M, Neumann A, Kersting C,
Agelopoulos K, Gebert C, Gosheger G, Buerger H and Hagedorn M:
Morphologic characterization of osteosarcoma growth on the chick
chorioallantoic membrane. BMC Res Notes. 3:582010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rao UN, Hood BL, Jones-Laughner JM, Sun M
and Conrads TP: Distinct profiles of oxidative stress-related and
matrix proteins in adult bone and soft tissue osteosarcoma and
desmoid tumors: A proteomics study. Hum Pathol. 44:725–733. 2013.
View Article : Google Scholar
|
|
18
|
Li Y, Liang Q, Wen YQ, Chen LL, Wang LT,
Liu YL, Luo CQ, Liang HZ, Li MT and Li Z: Comparative proteomics
analysis of human osteosarcomas and benign tumor of bone. Cancer
Genet Cytogenet. 198:97–106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kawai A, Kondo T, Suehara Y, Kikuta K and
Hirohashi S: Global protein-expression analysis of bone and soft
tissue sarcomas. Clin Orthop Relat Res. 466:2099–2106. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kikuta K, Tochigi N, Saito S, Shimoda T,
Morioka H, Toyama Y, Hosono A, Suehara Y, Beppu Y, Kawai A, et al:
Peroxiredoxin 2 as a chemotherapy responsiveness biomarker
candidate in osteosarcoma revealed by proteomics. Proteomics Clin
Appl. 4:560–567. 2010.PubMed/NCBI
|
|
21
|
Kubota D, Mukaihara K, Yoshida A, Tsuda H,
Kawai A and Kondo T: Proteomics study of open biopsy samples
identifies peroxiredoxin 2 as a predictive biomarker of response to
induction chemotherapy in osteosarcoma. J Proteomics. 91:393–404.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Folio C, Mora MI, Zalacain M, Corrales FJ,
Segura V, Sierrasesúmaga L, Toledo G, San-Julián M and
Patiño-García A: Proteomic analysis of chemonaive pediatric
osteosarcomas and corresponding normal bone reveals multiple
altered molecular targets. J Proteome Res. 8:3882–3888. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ren L and Khanna C: Role of ezrin in
osteosarcoma metastasis. Adv Exp Med Biol. 804:181–201. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zubaidah RM, Tan GS, Tan SB, Lim SG, Lin Q
and Chung MC: 2-D DIGE profiling of hepatocellular carcinoma
tissues identified isoforms of far upstream binding protein (FUBP)
as novel candidates in liver carcinogenesis. Proteomics.
8:5086–5096. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malz M, Weber A, Singer S, Riehmer V,
Bissinger M, Riener MO, Longerich T, Soll C, Vogel A, Angel P, et
al: Overexpression of far upstream element binding proteins: A
mechanism regulating proliferation and migration in liver cancer
cells. Hepatology. 50:1130–1139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sinner D, Kordich JJ, Spence JR, Opoka R,
Rankin S, Lin SC, Jonatan D, Zorn AM and Wells JM: Sox17 and Sox4
differentially regulate beta-catenin/T-cell factor activity and
proliferation of colon carcinoma cells. Mol Cell Biol.
27:7802–7815. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Baou M, Norton JD and Murphy JJ: AU-rich
RNA binding proteins in hematopoiesis and leukemogenesis. Blood.
118:5732–5740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Davis-Smyth T, Duncan RC, Zheng T,
Michelotti G and Levens D: The far upstream element-binding
proteins comprise an ancient family of single-strand DNA-binding
transactivators. J Biol Chem. 271:31679–31687. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu J, Kouzine F, Nie Z, Chung HJ,
Elisha-Feil Z, Weber A, Zhao K and Levens D: The FUSE/FBP/FIR/TFIIH
system is a molecular machine programming a pulse of c-myc
expression. EMBO J. 25:2119–2130. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Squire JA, Pei J, Marrano P, Beheshti B,
Bayani J, Lim G, Moldovan L and Zielenska M: High-resolution
mapping of amplifications and deletions in pediatric osteosarcoma
by use of CGH analysis of cDNA microarrays. Genes Chromosomes
Cancer. 38:215–225. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu X, Cai ZD, Lou LM and Zhu YB:
Expressions of p53, c-MYC, BCL-2 and apoptotic index in human
osteosarcoma and their correlations with prognosis of patients.
Cancer Epidemiol. 36:212–216. 2012. View Article : Google Scholar
|
|
32
|
Shimizu T, Ishikawa T, Sugihara E,
Kuninaka S, Miyamoto T, Mabuchi Y, Matsuzaki Y, Tsunoda T, Miya F,
Morioka H, et al: c-MYC overexpression with loss of Ink4a/Arf
transforms bone marrow stromal cells into osteosarcoma accompanied
by loss of adipogenesis. Oncogene. 29:5687–5699. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gherzi R, Trabucchi M, Ponassi M, Ruggiero
T, Corte G, Moroni C, Chen CY, Khabar KS, Andersen JS and Briata P:
The RNA-binding protein KSRP promotes decay of beta-catenin mRNA
and is inactivated by PI3K-AKT signaling. PLoS Biol. 5:e52006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bikkavilli RK and Malbon CC:
Dishevelled-KSRP complex regulates Wnt signaling through
post-transcriptional stabilization of beta-catenin mRNA. J Cell
Sci. 123:1352–1362. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cai Y, Mohseny AB, Karperien M, Hogendoorn
PC, Zhou G and Cleton-Jansen AM: Inactive Wnt/beta-catenin pathway
in conventional high-grade osteosarcoma. J Pathol. 220:24–33. 2010.
View Article : Google Scholar
|
|
36
|
Trabucchi M, Briata P, Garcia-Mayoral M,
Haase AD, Filipowicz W, Ramos A, Gherzi R and Rosenfeld MG: The
RNA-binding protein KSRP promotes the biogenesis of a subset of
microRNAs. Nature. 459:1010–1014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pasero M, Giovarelli M, Bucci G, Gherzi R
and Briata P: Bone morphogenetic protein/SMAD signaling orients
cell fate decision by impairing KSRP-dependent microRNA maturation.
Cell Rep. 2:1159–1168. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nicastro G, García-Mayoral MF,
Hollingworth D, Kelly G, Martin SR, Briata P, Gherzi R and Ramos A:
Noncanonical G recognition mediates KSRP regulation of let-7
biogenesis. Nat Struct Mol Biol. 19:1282–1286. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ: MicroRNA
let-7a downregulates MYC and reverts MYC-induced growth in Burkitt
lymphoma cells. Cancer Res. 67:9762–9770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Viswanathan SR, Powers JT, Einhorn W,
Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA,
Lockhart VL, et al: Lin28 promotes transformation and is associated
with advanced human malignancies. Nat Genet. 41:843–848. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yamakoshi H, Kanoh N, Kudo C, Sato A, Ueda
K, Muroi M, Kon S, Satake M, Ohori H, Ishioka C, et al: KSRP/FUBP2
is a binding protein of GO-Y086, a cytotoxic curcumin analogue. ACS
Med Chem Lett. 1:273–276. 2010. View Article : Google Scholar : PubMed/NCBI
|