Introduction
Breast carcinoma is the most prevalent female
malignance and the cause of a majority of cancer-related deaths
among women worldwide (1).
According to the status of estrogen receptor (ER), progesterone
receptor (PR) and human epidermal growth factor 2 (Her2), breast
cancer can be classified into four main distinct molecular
subtypes, including luminal, Her2 overexpression, basal type and
normal-like (2). Although
significant achievement has been made in therapeutic strategies,
the clinical outcome of breast cancer patients remains
unsatisfactory due to recurrence, metastasis or
chemotherapy-resistance (1).
Breast cancer stem cells (CSCs), which have extraordinary ability
of self-renewal, proliferation and generation of heterogenic
lineages of tumor cells, account for recurrence and metastasis of
breast cancer (3).
CD44, a multi-structural and multi-functional
transmembrane glycoprotein, participates in the regulation of many
cellular processes including cell division, adhesion and migration
through binding with its main ligand the hyaluronic acid (HA)
(4,5). It has been demonstrated that CD44
also plays essential roles in tumorigenesis (6), invasion and metastasis, as well as
therapy-response (6–8). It promotes carcinogenesis by
acceleration of proliferation or suppression of apoptosis, partly
resulting from stimulation of Ras-Raf-Mek-Erk-cyclin D1 signaling
and phosphoinositide 3-kinase (PI3K)-Akt pathway, respectively
(6). CD44, a well-known CSC
marker, is involved in the complex process of
epithelial-mesenchymal transition (EMT), which stimulates tumor
invasion and metastasis (5,9).
Epidermal growth factor receptor (EGFR), belonging
to the protein kinase superfamily, plays a critical role in cell
proliferation (10). It has been
demonstrated that EGFR is aberrantly expressed in a variety of
epithelial tumor types, indicating that it might be implicated in
the etiology and progression of these cancers, including lung
carcinoma, head and neck squamous cell carcinoma and breast cancer
(11,12). Positive staining of EGFR protein
was observed in ~45% of breast cancer (13). EGFR expression was found mainly in
basal-like carcinoma, and tended to be inversely associated with
hormone receptor (ER, PR) levels (13). Combined status of positive EGFR and
negative ER often portended a significantly worse clinical outcome
(13). In consideration of its
established role in cancer cell proliferation and survival, EGFR is
an effective target for cancer treatment and relative drugs against
EGFR have been put into clinical use. For instance, small molecule
inhibitors, such as gefitinib and erlotinib, targeted the
intracellular ATP-binding site in the tyrosine kinase domain of
EGFR (14–16). However, acquired resistance
eventually developed in nearly all patients through second mutation
of EGFR or activation of parallel signals (17). Various agents and strategies have
been developed to overcome EGFR-TKIs resistance. For example,
AST1306, an irreversible small molecular blocker of EGFR, HER2 and
HER4, achieved promising antitumor activity even in patients
previously treated with HER2 inhibitor (18).
CD44 and EGFR were both enriched in basal-like
breast cancer and exerted favorable effects on breast tumor
progression (13,19). Therefore, we hypothesized that EGFR
upregulated the expression of CD44, contributing to CSCs and
mesenchymal phenotypes. To test this hypothesis, we conducted a
combined analysis of available published microarray data and
immunohistochemistry analysis on tissue microarrays (TMA).
Furthermore, causative linkage between CD44 and EGFR in breast
cancer cell lines was also investigated.
Materials and methods
Breast cancer tissue microarray and
immunohistochemistry detection
To evaluate the association between CD44 protein
abundance and breast cancer risks, tumor grade, as well as the
status of ER, PR, Her2 and EGFR in patients with breast cancer,
commercially available tissue microarray (TMA) slides (BR1502, US
Biomax, Inc, Rockville, MD, USA) containing histologically
confirmed tissues were purchased for immunohistochemistry (IHC)
analysis. This microarray contains 150 samples including 10 normal
breast tissues and 140 breast cancer samples. Due to tissue
rejection, the actual number of samples enrolled was 125 including
5 normal tissues and 120 breast tumor samples. Among these 120
breast cancer samples, there were 16 grade I, 90 grade II and 14
grade III samples, and we classified grade I and grade II into
low-grade tumors and grade III was high-grade. A total of 4, 75, 24
and 17 cases were Tis, T1, T2, T3 and T4, respectively. For lymph
node metastasis status, there were a total of 88, 16, 11 and 5
tumors for N0, N1, N2 and N3, respectively. For ER status, the
number of ER-negative cases was 41 and that of ER-positive ones was
79. The number of ER-negative cases was 64 and PR-positive 56. The
number of HER2-negative and positive were 48 and 72, respectively.
The original status of EGFR staining was displayed as ‘−’, ‘+’,
‘++’ and ‘+++’. We classified 109 cases of EGFR stain ‘−’ and ‘+’
as low EGFR and 11 cases of ‘++’ and ‘+++’ into high EGFR
group.
Specific primary antibody against CD44 (polyclonal
rabbit antibody, 1:200; ProteinTech Group) was used for IHC with a
2-step protocol. Immunohistochemical staining was performed as
previously described (20).
Tissues in BR1502, embedded in paraffin, were baked at 60°C for 1 h
and then deparaffinised and hydrated through a series of xylenes
and alcohols. Following antigen retrieval, slides were incubated in
3% H2O2 diluted in methanol for 30 min to
block the activity of endogenous peroxidases. Non-specific binding
of antibodies was blocked with 2.5% horse normal serum (20 min).
Next, slides were incubated overnight with primary antibody at 4°C.
Hematoxylin was used for nuclear counterstaining.
Analysis and quantification of
staining
Two evaluators, blinded to the clinical data,
reviewed the immunoreactivity for CD44 protein under a light
microscope. For quantification, all stains were evaluated at ×200
magnifications and at least 3 areas for each core were counted. The
protein expression was scored independently according to the
intensity of cellular staining and the proportion of stained tumor
cells. The staining intensity was scored as 0 (no staining), 1
(weak staining, light yellow), 2 (moderate staining, yellow brown)
and 3 (strong staining, brown). The proportions of stained tumor
cells were classified as 0 (≤5% positive cells), 1 (6–25% positive
cells), 2 (26–50% positive cells) and 3 (≥51% positive cells). The
multiplication for intensity and proportion scores was utilized to
represent the level of CD44 protein abundance (21). According to the final staining
score, 1–3 was grouped to low expression, while >4 was
classified into high expression.
Cell culture and drug treatment
Breast cancer cell lines, including MDA-MB-231 and
HBL100 cells, were cultured in Dulbecco's modified Eagle's medium
(DMEM) with 10% fetal bovine serum (FBS, Life Technologies, Inc.).
These two cell lines were seeded into two 6 cm culture dishes and
grew to 70–80% confluence before incubation with erlotinib (Sigma)
at 10 μM for 24 h. The cells were harvested for the following
experiments.
Migration and invasion assay
Migration and invasion assays were performed as
previously described (22).
Generally, 2×104 MDA-MB-231 and HBL100 cells treated
with erlotinib or vehicle control were suspended in DMEM without
fetal bovine serum and seeded on an 8-μm pore size Transwell filter
insert (Corning Inc., Corning, NY, USA) coated with diluted
Matrigel (BD Biosciences, Bedford, MA, USA). Lower chamber was
filled with DMEM supplemented with 10% FBS as a chemo-attractant.
After incubation for 18 h at 37°C, invaded cells were stained with
0.5% crystal violet solution mixed with 4% para-formaldehyde and
counted by light microscopy (×200). Breast cancer cell invasion
ability with or without erlotinib treatment was quantitatively
measured by BD coated invasion system.
Western blot detection
Cells were washed with cold PBS, scraped into RIPA
buffer and centrifuged. The cell lysates were subjected to 10%
SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF)
hybridization transfer membrane. The primary antibodies used were
as follows: CD44 (ProteinTech group), EGFR (Santa Cruz
Laboratories), Snail (Cell Signaling Technology), p-EGFR (Santa
Cruz Laboratories), p-AKT (Cell Signaling Technology), p-ERK (Santa
Cruz Laboratories), KLF4 (Santa Cruz Laboratories), c-Myc (Santa
Cruz Laboratories), Slug (Cell Signaling Technology), Snail (Cell
Signaling Technology), Vinculin (Santa Cruz Laboratories).
Secondary staining and detection were carried out in accordance
with standard protocols (23).
Analysis of gene expression data
GSE42568, available through the published Gene
Expression Omnibus (GEO) databases and containing 17 normal cases
and 104 breast cancer patients, was analyzed to evaluate the
association between CD44 expression and breast tumor risks,
histological grade, EGFR, KRT5, KRT17 and FOXA1.
Additionally, the association between CD44 mRNA level and the
progression-free survival (PFS) of patients with
HER2-overexpressing and basal-like breast cancer was analyzed on
public data of KM PLOTTER.
Statistical analysis
Correlation analyses of CD44 with EGFR,
KRT5, KRT6 and KRT17 were performed using SPSS 20
statistical software (SPSS Inc., Chicago, IL, USA). The Student's
t-test was applied to evaluate the differences in groups as
appropriate and the significance level was set at p<0.05. The
association between CD44 expression and the clinicopathological
parameters was evaluated by the χ2 test. A two-tailed
p-value of <0.05 was considered statistically significant.
Analysis of prognosis was conducted with the Kaplan-Meier method
and the log-rank test.
Results
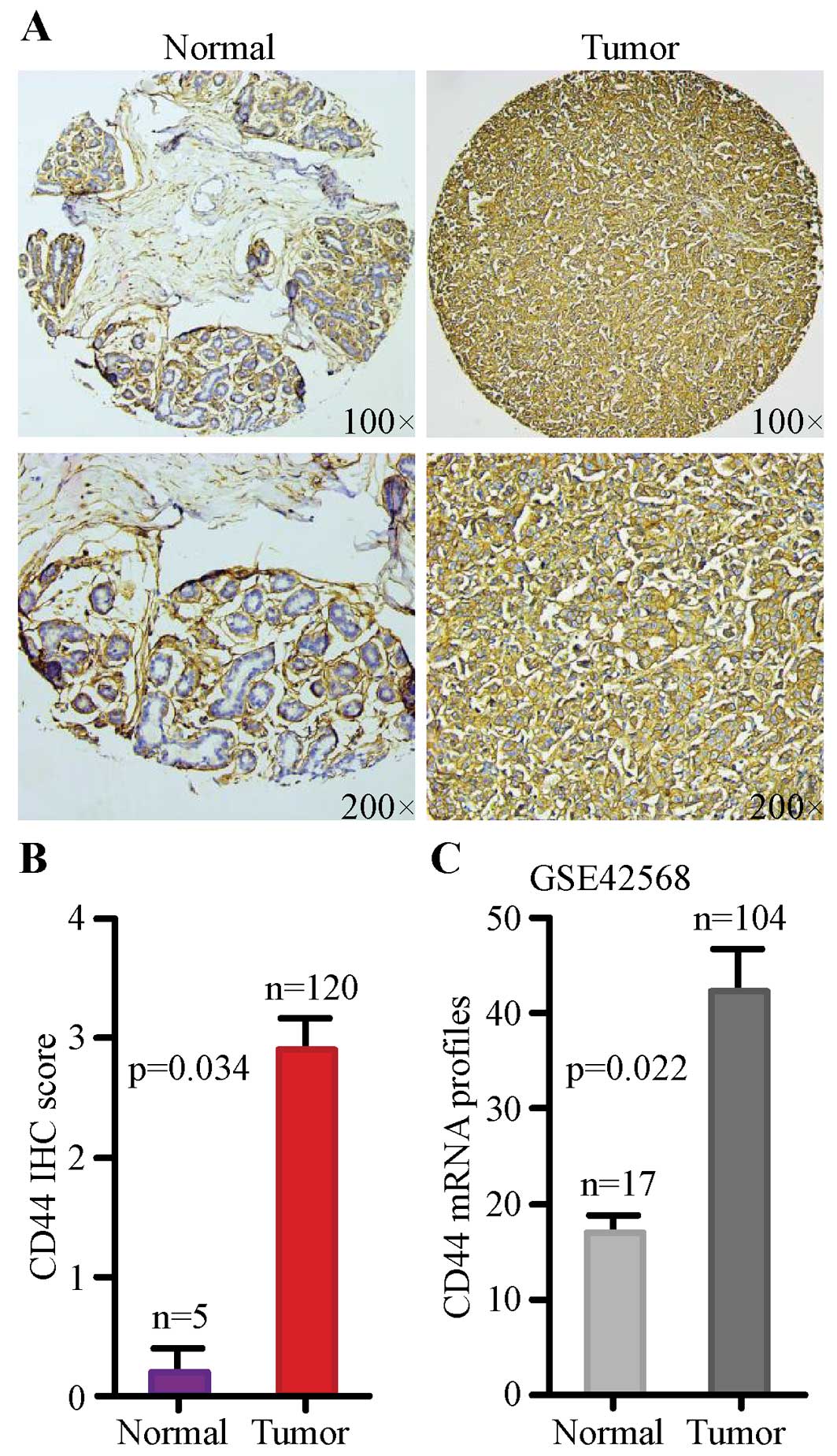
CD44 expression is increased in breast
cancer in comparison with normal breast
In order to evaluate CD44 protein level between
normal breast tissues and malignant tissues, we analyzed a TMA
containing 120 informative patients with breast cancer by IHC. CD44
was mainly detected on the membrane of breast cancer cells.
Representative images of immunohistochemical staining for
non-cancerous and cancerous tissues are shown in Fig. 1A. Next, we examined the potential
association of CD44 protein abundance with breast cancer risks by
using semi-quantitative criteria. The result indicated that protein
abundance of CD44 was significantly higher in breast cancer tissues
compared with normal tissues (Fig.
1B, p=0.034).
In order to assess whether the mRNA transcription of
CD44 is consistent with the protein expression, GSE42568 was
assessed. The mRNA level of CD44 in breast cancer was
remarkably enhanced compared with normal breast tissue (Fig. 1C, p=0.022). Together, our results
suggested that expression of CD44 was significantly
upregulated at both protein and mRNA level in breast cancer tissues
when compared with normal tissue.
High level of CD44 was associated with
histological grade of human breast cancer
To further explore the correlation between CD44
protein abundance and histological grade, representative images of
immunohistochemical staining for low-grade and high-grade cancer
tissues are shown in Fig. 2A.
Comparison of IHC scores suggested that CD44 protein abundance was
greatly elevated in high-grade breast cancer tissues (Fig. 2B, p=0.005).
In addition, we also evaluated the mRNA expression
of CD44 in both low-grade and high-grade tumors in GSE42568,
and the results showed that the mRNA expression of CD44 was
significantly enhanced in high-grade tumors in comparison with
low-grade group (Fig. 2C,
p=0.044). Our results suggested that high level of CD44
tended to be associated with high histological grade in breast
cancer.
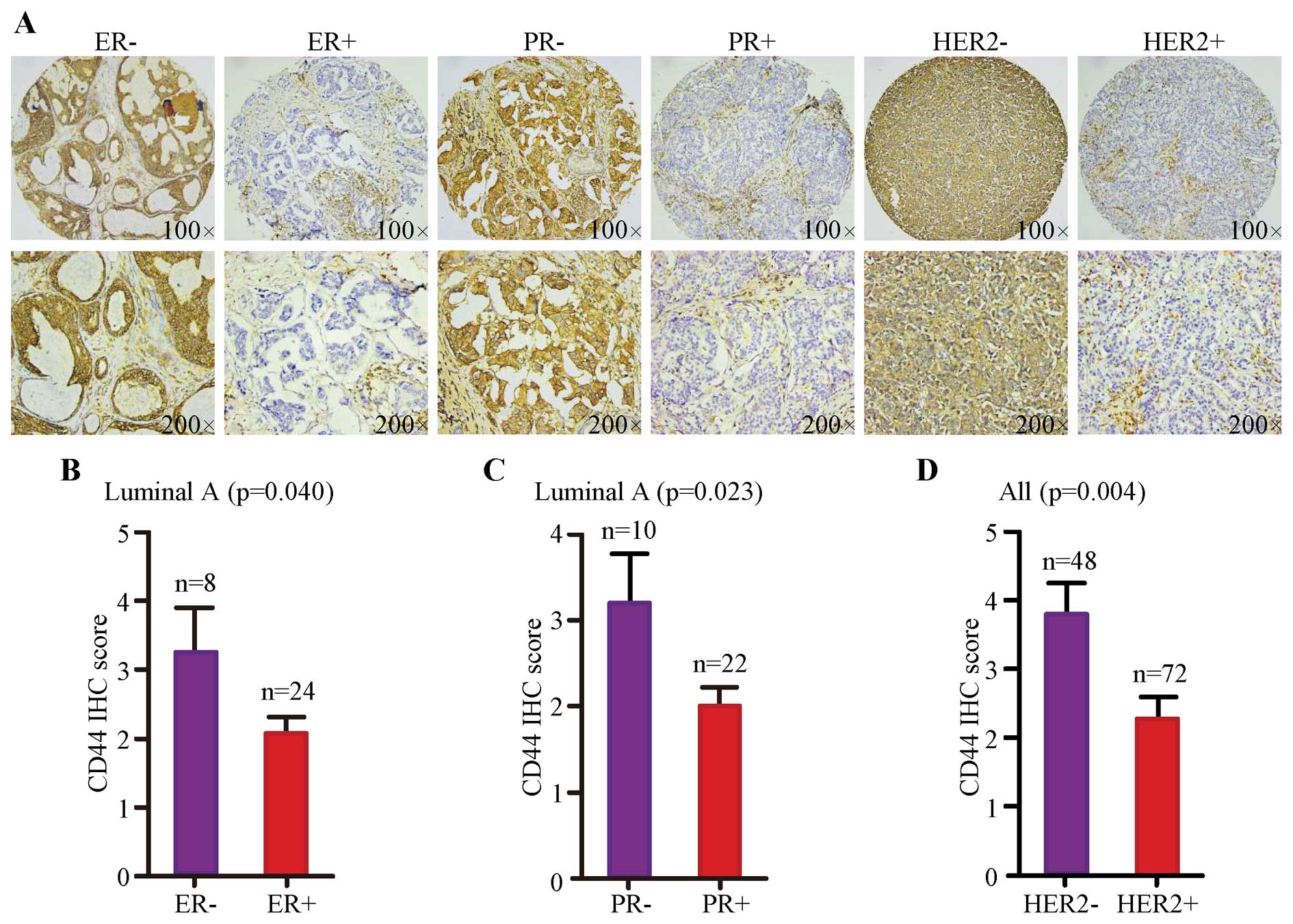
CD44 protein abundance tends to be
associated with molecular subtype of breast cancer
To assess whether there was any association between
CD44 protein abundance and the status of ER, PR and HER2, we
combined IHC results with that information provided by TMA.
Representative images of immunohistochemical staining for negative
and positive status of ER, PR and HER2 are, respectively, shown in
Fig. 3A. Statistical analysis on
IHC score revealed that higher level of CD44 was significantly
correlated with lower status of ER (Fig. 3B, p=0.040) and PR (Fig. 3C, p=0.023) in luminal A subtype as
well as lower expression of HER2 (Fig.
3D, p=0.004).
Additionally, the correlation between CD44 and EGFR
was analyzed. Representative images for CD44 staining in low EGFR
and high EGFR samples are shown in Fig. 4A. Semi-quantitative result
indicated that CD44 scores were higher in high EGFR cases (Fig. 4B, p=0.004). Besides, analysis on
public gene expression data indicated that CD44 expression
was significantly correlated with that of EGFR at mRNA level
(Fig. 4C, p<0.001). Then, the
correlation between CD44 mRNA level and basal cytokeratin
markers KRT5 and KRT17, as well as luminal marker
FOXA1 were analyzed in GSE42568. The results indicated that
CD44 mRNA expression was positively associated with the
expression of KRT5 (Fig.
4D, p=0.005) and KRT17 (Fig. 4E, p=0.006), but inversely
correlated with FOXA1 (Fig.
4F, p<0.001).
High CD44 mRNA transcription predicts
rapid progression
Public KM PLOTTER datasets were employed to evaluate
the effects of CD44 mRNA level on the PFS of patients with
HER2 overexpression and basal-like breast cancer. The results
showed that both HER2-overexpressing (Fig. 5A, p=0.007) and basal-like patients
(Fig. 5B, p=0.035) with greater
amount of CD44 mRNA tended to have shortened time to cancer
progression, indicating that CD44 exerted favorable effects
on progression of patients with these two subtypes.
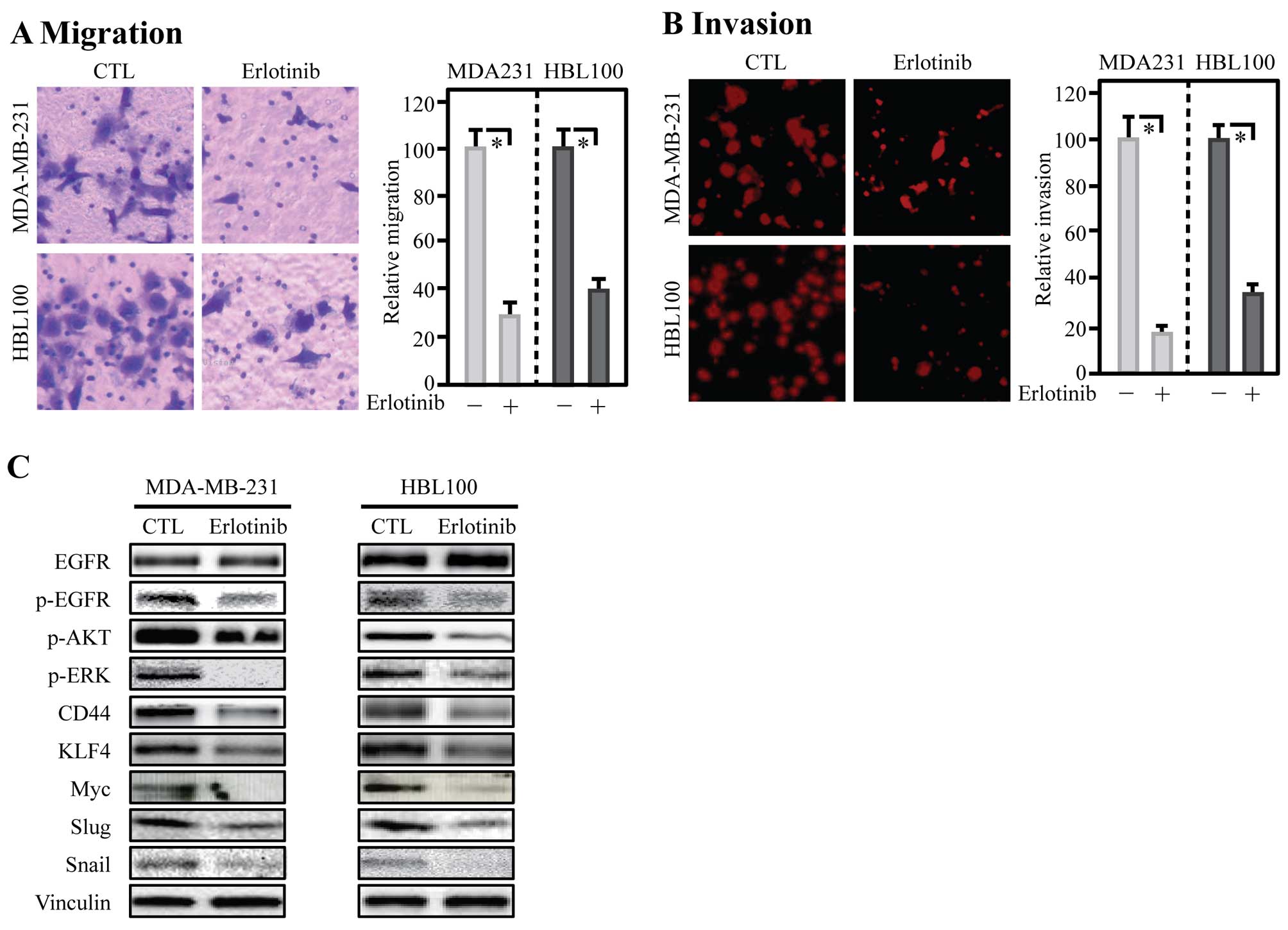
Inhibition of EGFR decreased invasion
with reduced expression of CD44, EMT and CSC-related genes
To evaluate whether EGFR regulated CD44, a small
molecular inhibitor of EGFR, erlotinib, was incubated with breast
cancer cells for 24 h, then the ability of migration and invasion
was assessed. The results of transwell assay showed that inhibition
of EGFR markedly reduced migration of MDA-MB-231 and HBL100 cells
(Fig. 6A). Similarly, invasion
capability of these two cell lines was also impaired by erlotinib
(Fig. 6B). To detect the abundance
of CD44 and related proteins after erlotinib treatment, total
protein was extracted and analyzed by western blotting. The results
showed that inhibition of EGFR with erlotinib in breast cancer
MDA-MB-231 and HBL100 cell lines significant downregulated p-AKT,
p-ERK, CD44, KLF4, Myc, Slug and Snail in these cells (Fig. 6C).
Discussion
Overwhelming number of studies have been carried out
to explore the roles of CD44 in cancer. As a well-known marker of
CSCs, CD44 promotes carcinogenesis of diverse tumor types,
including colorectal (24),
pancreatic (25) and breast cancer
(26). It has been demonstrated
that CD44 is a powerful regulator of EMT process as well as tumor
invasion, migration and therapy-resistance (5). We provided previously the fact that
CD44 was enhanced in breast cancer and closely linked to tumor
grade (27). Our study further
indicated that higher CD44 protein abundance tended to be parallel
with the status of ER and PR, while be negatively associated with
HER2 status. Finally, blockade of EGFR activity by tyrosine kinase
inhibitor (TKI) induced a reduction of CD44 and a series of markers
of CSCs and EMT. Thus, we drew a conclusion that CD44 served
downstream of EGFR to function in the progression of breast tumor.
Identification of clinically relevant genes may be explored as a
molecular biomarker to aid precision therapy (28).
Consistent with previous studies, our research also
offered evidence that CD44 was remarkably upregulated in breast
tumor tissues in comparison with normal counterparts at both
protein and mRNA levels, indicating that CD44 was involved in
tumorigenesis of breast cancer. Breast cancer cells with ectopic
expression of the cleaved intracellular domain of CD44 (CD44ICD)
displayed potent ability of tumorigenesis and showed enhanced
metastatic potential in mouse models, compared with the control
cells (29). Aberrant expression
and nuclear accumulation of CD44ICD played a pivotal role in
transcriptional activation and nuclear localization of stemness
factors such as Nanog, Sox2 and Oct4, which are the stimulating
effects of CD44 on the initiation of breast cancer (29). Our previous meta-analysis on human
breast cancer tissues demonstrated that greater mRNA amount of CD44
tended to be linked to higher histological grade (27). Consistently, our
immunohistochemistry analysis showed higher CD44 protein abundance
was also parallel with histological grade, which provided more
evidence to support that CD44 was positively associated with breast
tumor grade as well as might have unfavorable impact on breast
cancer cell differentiation. It is consistent with previous
analysis of 448 primary breast tumors by McFarlane, et al
showing that breast tumors with high CD44 protein abundance tended
to be high-grade (30).
CD44, a widely recognized CSC marker and an
important upregulator of EMT process, not only stimulated
carcinogenesis, but also promoted metastasis and therapy-resistance
(5). For instance, CD44 restrained
the combination of membrane-associated E-cadherin and β-catenin,
which promoted β-catenin to nuclear and then transcriptionally
activated genes that were involved in the invasion and migration of
colon cancer cells (31). Our
prognostic analysis showed that high expression of CD44 mRNA
had adverse impact on the PFS of patients with HER2 overexpressing
and basal-like breast cancer, indicating that CD44 was
capable of promoting breast tumor progression. CD44 enhanced
adhesion of breast cancer cells to endothelium and fibronection
through α5β1-integrin to facilitate metastasis of breast cancer
(32). Furthermore, ectopic
expression of CD44 played important role in the
doxorubicin-resistance of breast cancer patients, and anti-CD44 mAb
remarkably suppressed migration and invasion of breast cancer MCF-7
cells (33).
The status of ER, PR and HER2 were utilized to
divide breast cancer into four distinct subtypes, including
luminal, HER2-positive, basal-type and normal-like. Different
subgroups displayed diverse biological behavior and showed
different susceptibility to common therapeutics (2). Hereby, we explored whether there was
any significant association between CD44 protein abundance and the
status of ER, PR and HER2. Our results indicated that high content
of CD44 protein abundance was negatively associated with the status
of ER and PR in luminal A group and it also had an inverse linkage
with HER2 status. In support, the previous research got similar
conclusion that most of CD44-negative cases were observed in
luminal A subtype and the number of CD44-positive cases in
HER2-positive samples was 4.8 times less than that of HER2-negative
ones (19). KRT5 and
KRT17 are well-recognized factors for basal-like breast
cancer, while FOXA1 was enriched in luminal type by
activating ER mRNA expression (34) and maintaining ER sensitivity
(35). Thus, the positive
correlations between CD44 with KRT5 and KRT17
as well as inverse association with FOXA1 suggested that
CD44 was enriched in basal-like breast cancer, which was consistent
with the finding in our meta-analysis (27).
Activated EGFR stimulated signal transduction
pathways that were involved in the regulation of several cell
functions such as cell proliferation and motility (36), including the Ras-ERKs (10) and PI3K-AKT pathway (37), both of which were growth-promoting
signaling cascades. It has been well demonstrated that
dysregulation of EGFR plays key roles in the malignant
transformation and tumor development of diverse tumors such as lung
cancer (38), oral cancer
(39), hepatocarcinoma (40), and gastrointestinal (41), and breast cancer (42). By receptor and ligand
overexpression and deficiency of specific phosphatases, EGFR
promoted carcinogenesis (43).
EGFR combined with DNA-PK in the nucleus to enhance DNA-PK activity
and DNA repair (44), which
resulted in radiotherapy-resistance.
In primary head and neck squamous cell carcinoma,
the expression patterns of CD44 and EGFR were overlapping and were
significantly connected (45).
Although CD44 and EGFR have great impact on the initiation and
progression of breast cancer, whether there was any connection
between CD44 and EGFR still lacks direct evidence in breast cancer.
Our study of immunohistochemistry analysis on TMA and correlation
analysis on GSE42568 supported that the expression of CD44 was
positively correlated with EGFR. Blocking activity of EGFR with
erlotinib impaired the ability of migration and invasion of breast
cancer cells. Further analysis by western blotting displayed that
blockade of EGFR resulted in the downregulation of p-AKT, p-ERK and
CD44. Besides, CSCs proteins KLF4, Myc and mesenchymal protein Slug
and Snail were also reduced by inhibition of EGFR. These supported
that high EGFR activity in basal type might drive CSC property and
EMT phenotype. EMT process is controlled by both transcriptional
and post-transcriptional regulation (46). The detailed mechanism by which
EGFR-TKIs reverse EMT needs further classification. The EGFR-TKI
reduced migration and invasion of in vitro cultured breast
cancer cells. Ongoing study on xenograft mouse model will test
whether EGFR-TKI inhibits in vivo invasion, thereby reducing
metastasis. It has been reported that
CD44+/CD24− CSCs are resistant to
conventional chemotherapy, but sensitive to Cdk2 inhibitor
(47), suggesting that patients
with CD44 high expression may need personalized therapeutic
strategy.
In conclusion, this study demonstrated the
expression of CD44 was upregulated in breast cancer and was closely
correlated with high grade. More importantly, our research
indicated that EGFR exert effects on the initiation and progression
of breast cancer through upregulating the level of CD44 and related
markers of cancer stem cells and EMT. Identification of possible
interaction between CD44 and EGFR could provide better
understanding of the development of breast cancer as well as help
improve therapy strategy for breast cancer patients.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (NSFC) nos. 81572608 and 81172422
(K.W.), 81502209 (N.H.) and 81441087 (L.Z.).
Abbreviations:
|
CD44
|
cluster of differentiation 44
|
|
EGFR
|
epidermal growth factor receptor
|
|
TMAs
|
tissue microarrays
|
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
HER2
|
human epidermal growth factor
receptor-2
|
|
CSCs
|
cancer stem cells
|
|
HA
|
hyaluronic acid
|
|
PI3K
|
phosphoinositide 3-kinase
|
|
EMT
|
epithelial-mesenchymal transition
|
|
IHC
|
immunohistochemistry
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
PVDF
|
polyvinylidene fluoride
|
|
PFS
|
progression-free survival
|
|
TKI
|
tyrosine kinase inhibitor
|
|
CD44ICD
|
intracellular domain of CD44
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Makki J: Diversity of breast carcinoma:
Histological subtypes and clinical relevance. Clin Med Insights
Pathol. 8:23–31. 2015. View Article : Google Scholar
|
|
3
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells - perspectives on current status and future directions:
AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Götte M and Yip GW: Heparanase,
hyaluronan, and CD44 in cancers: A breast carcinoma perspective.
Cancer Res. 66:10233–10237. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu H, Tian Y, Yuan X, Wu H, Liu Q, Pestell
RG and Wu K: The role of CD44 in epithelial-mesenchymal transition
and cancer development. Onco Targets Ther. 8:3783–3792. 2015.
|
|
6
|
Zöller M: CD44: Can a cancer-initiating
cell profit from an abundantly expressed molecule? Nat Rev Cancer.
11:254–267. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei X, Xu M, Wei Y, Huang F, Zhao T, Li X,
Feng R and Ye BH: The addition of rituximab to CHOP therapy alters
the prognostic significance of CD44 expression. J Hematol Oncol.
7:342014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lv L, Liu HG, Dong SY, Yang F, Wang QX,
Guo GL, Pan YF and Zhang XH: Upregulation of CD44v6 contributes to
acquired chemoresistance via the modulation of autophagy in colon
cancer SW480 cells. Tumour Biol. Jan 9–2016.(Epub ahead of print).
View Article : Google Scholar
|
|
9
|
Erb U, Megaptche AP, Gu X, Büchler MW and
Zöller M: CD44 standard and CD44v10 isoform expression on leukemia
cells distinctly influences niche embedding of hematopoietic stem
cells. J Hematol Oncol. 7:292014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Veale D, Ashcroft T, Marsh C, Gibson GJ
and Harris AL: Epidermal growth factor receptors in non-small cell
lung cancer. Br J Cancer. 55:513–516. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Weichselbaum RR, Dunphy EJ, Beckett MA,
Tybor AG, Moran WJ, Goldman ME, Vokes EE and Panje WR: Epidermal
growth factor receptor gene amplification and expression in head
and neck cancer cell lines. Head Neck. 11:437–442. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klijn JG, Berns PM, Schmitz PI and Foekens
JA: The clinical significance of epidermal growth factor receptor
(EGF-R) in human breast cancer: A review on 5232 patients. Endocr
Rev. 13:3–17. 1992.PubMed/NCBI
|
|
14
|
Wakeling AE, Guy SP, Woodburn JR, Ashton
SE, Curry BJ, Barker AJ and Gibson KH: ZD1839 (Iressa): An orally
active inhibitor of epidermal growth factor signaling with
potential for cancer therapy. Cancer Res. 62:5749–5754.
2002.PubMed/NCBI
|
|
15
|
Hidalgo M, Siu LL, Nemunaitis J, Rizzo J,
Hammond LA, Takimoto C, Eckhardt SG, Tolcher A, Britten CD, Denis
L, et al: Phase I and pharmacologic study of OSI-774, an epidermal
growth factor receptor tyrosine kinase inhibitor, in patients with
advanced solid malignancies. J Clin Oncol. 19:3267–3279.
2001.PubMed/NCBI
|
|
16
|
Sun W, Yuan X, Tian Y, Wu H, Xu H, Hu G
and Wu K: Non-invasive approaches to monitor EGFR-TKI treatment in
non-small-cell lung cancer. J Hematol Oncol. 8:952015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Niu FY and Wu YL: Novel agents and
strategies for overcoming EGFR TKIs resistance. Exp Hematol Oncol.
3:22014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Cao J, Li J, Zhang Y, Chen Z,
Peng W, Sun S, Zhao N, Wang J, Zhong D, et al: A phase I study of
AST1306, a novel irreversible EGFR and HER2 kinase inhibitor, in
patients with advanced solid tumors. J Hematol Oncol. 7:222014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gudadze M, Kankava K, Mariamidze A,
Mosidze T and Burkadze G: Distribution of CD44/CD24 positive cells
in ductal invasive carcinoma of breast of different grade and
molecular subtype. Georgian Med News. 222:50–57. 2013.PubMed/NCBI
|
|
20
|
Liu Y, Zhou R, Yuan X, Han N, Zhou S, Xu
H, Guo M, Yu S, Zhang C, Yin T, et al: DACH1 is a novel predictive
and prognostic biomarker in hepatocellular carcinoma as a negative
regulator of Wnt/β-catenin signaling. Oncotarget. 6:8621–8634.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xie JW, Chen PC, Zheng CH, Li P, Wang JB,
Lin JX, Lu J, Chen QY, Cao LL, Lin M, et al: Evaluation of the
prognostic value and functional roles of CD44v6 in gastric cancer.
J Cancer Res Clin Oncol. 141:1809–1817. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han N, Yuan X, Wu H, Xu H, Chu Q, Guo M,
Yu S, Chen Y and Wu K: DACH1 inhibits lung adenocarcinoma invasion
and tumor growth by repressing CXCL5 signaling. Oncotarget.
6:5877–5888. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chu Q, Han N, Yuan X, Nie X, Wu H, Chen Y,
Guo M, Yu S and Wu K: DACH1 inhibits cyclin D1 expression, cellular
proliferation and tumor growth of renal cancer cells. J Hematol
Oncol. 7:732014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du L, Wang H, He L, Zhang J, Ni B, Wang X,
Jin H, Cahuzac N, Mehrpour M, Lu Y, et al: CD44 is of functional
importance for colorectal cancer stem cells. Clin Cancer Res.
14:6751–6760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang D, Zhu H, Liu Y, Liu Q, Xie X, Zhou
Y, Zhang L, Zhu Y, Zhang Z and Su Z: The low chamber pancreatic
cancer cells had stem-like characteristics in modified transwell
system: Is it a novel method to identify and enrich cancer
stem-like cells? BioMed Res Int. 2014:7603032014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xie G, Yao Q, Liu Y, Du S, Liu A, Guo Z,
Sun A, Ruan J, Chen L, Ye C, et al: IL-6-induced
epithelial-mesenchymal transition promotes the generation of breast
cancer stem-like cells analogous to mammosphere cultures. Int J
Oncol. 40:1171–1179. 2012.
|
|
27
|
Xu H, Tian Y, Yuan X, Liu Y, Wu H, Liu Q,
Wu GS and Wu K: Enrichment of CD44 in basal-type breast cancer
correlates with EMT, cancer stem cell gene profile, and prognosis.
Onco Targets Ther. 9:431–444. 2016.PubMed/NCBI
|
|
28
|
Smith AD, Roda D and Yap TA: Strategies
for modern biomarker and drug development in oncology. J Hematol
Oncol. 7:702014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cho Y, Lee HW, Kang HG, Kim HY, Kim SJ and
Chun KH: Cleaved CD44 intracellular domain supports activation of
stemness factors and promotes tumorigenesis of breast cancer.
Oncotarget. 6:8709–8721. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McFarlane S, Coulter JA, Tibbits P,
O'Grady A, McFarlane C, Montgomery N, Hill A, McCarthy HO, Young
LS, Kay EW, et al: CD44 increases the efficiency of distant
metastasis of breast cancer. Oncotarget. 6:11465–11476. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cho SH, Park YS, Kim HJ, Kim CH, Lim SW,
Huh JW, Lee JH and Kim HR: CD44 enhances the epithelial-mesenchymal
transition in association with colon cancer invasion. Int J Oncol.
41:211–218. 2012.PubMed/NCBI
|
|
32
|
McFarlane S, McFarlane C, Montgomery N,
Hill A and Waugh DJ: CD44-mediated activation of α5β1-integrin,
cortactin and paxillin signaling underpins adhesion of basal-like
breast cancer cells to endothelium and fibronectin-enriched
matrices. Oncotarget. 6:36762–36773. 2015.PubMed/NCBI
|
|
33
|
Uchino M, Kojima H, Wada K, Imada M, Onoda
F, Satofuka H, Utsugi T and Murakami Y: Nuclear beta-catenin and
CD44 upregulation characterize invasive cell populations in
non-aggressive MCF-7 breast cancer cells. BMC Cancer. 10:4142010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bernardo GM, Lozada KL, Miedler JD,
Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE,
Kaestner KH, et al: FOXA1 is an essential determinant of ERalpha
expression and mammary ductal morphogenesis. Development.
137:2045–2054. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kouros-Mehr H, Slorach EM, Sternlicht MD
and Werb Z: GATA-3 maintains the differentiation of the luminal
cell fate in the mammary gland. Cell. 127:1041–1055. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mitsudomi T and Yatabe Y: Epidermal growth
factor receptor in relation to tumor development: EGFR gene and
cancer. FEBS J. 277:301–308. 2010. View Article : Google Scholar
|
|
37
|
Okano J, Gaslightwala I, Birnbaum MJ,
Rustgi AK and Nakagawa H: Akt/protein kinase B isoforms are
differentially regulated by epidermal growth factor stimulation. J
Biol Chem. 275:30934–30942. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cattaneo F, Iaccio A, Guerra G, Montagnani
S and Ammendola R: NADPH-oxidase-dependent reactive oxygen species
mediate EGFR transactivation by FPRL1 in WKYMVm-stimulated human
lung cancer cells. Free Radic Biol Med. 51:1126–1136. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Brusevold IJ, Tveteraas IH, Aasrum M,
Ødegård J, Sandnes DL and Christoffersen T: Role of LPAR3, PKC and
EGFR in LPA-induced cell migration in oral squamous carcinoma
cells. BMC Cancer. 14:4322014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tveteraas IH, Müller KM, Aasrum M, Ødegård
J, Dajani O, Guren T, Sandnes D and Christoffersen T: Mechanisms
involved in PGE2-induced transactivation of the epidermal growth
factor receptor in MH1C1 hepatocarcinoma cells. J Exp Clin Cancer
Res. 31:722012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yoshida K, Fujino H, Otake S, Seira N,
Regan JW and Murayama T: Induction of cyclooxygenase-2 expression
by prostaglandin E2 stimulation of the prostanoid EP4 receptor via
coupling to Gαi and transactivation of the epidermal growth factor
receptor in HCA-7 human colon cancer cells. Eur J Pharmacol.
718:408–417. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zajac M, Law J, Cvetkovic DD, Pampillo M,
McColl L, Pape C, Di Guglielmo GM, Postovit LM, Babwah AV and
Bhattacharya M: GPR54 (KISS1R) transactivates EGFR to promote
breast cancer cell invasiveness. PLoS One. 6:e215992011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kondapaka SB, Fridman R and Reddy KB:
Epidermal growth factor and amphiregulin up-regulate matrix
metalloproteinase-9 (MMP-9) in human breast cancer cells. Int J
Cancer. 70:722–726. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dittmann K, Mayer C, Kehlbach R and
Rodemann HP: Radiation-induced caveolin-1 associated EGFR
internalization is linked with nuclear EGFR transport and
activation of DNA-PK. Mol Cancer. 7:692008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Leinung M, Ernst B, Döring C, Wagenblast
J, Tahtali A, Diensthuber M, Stöver T and Geissler C: Expression of
ALDH1A1 and CD44 in primary head and neck squamous cell carcinoma
and their value for carcinogenesis, tumor progression and cancer
stem cell identification. Oncol Lett. 10:2289–2294. 2015.PubMed/NCBI
|
|
46
|
Guo F, Parker Kerrigan BC, Yang D, Hu L,
Shmulevich I, Sood AK, Xue F and Zhang W: Post-transcriptional
regulatory network of epithelial-to-mesenchymal and
mesenchymal-to-epithelial transitions. J Hematol Oncol. 7:192014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Opyrchal M, Salisbury JL, Iankov I, Goetz
MP, McCubrey J, Gambino MW, Malatino L, Puccia G, Ingle JN, Galanis
E, et al: Inhibition of Cdk2 kinase activity selectively targets
the CD44+/CD24−/low stem-like subpopulation
and restores chemosensitivity of SUM149PT triple-negative breast
cancer cells. Int J Oncol. 45:1193–1199. 2014.PubMed/NCBI
|