Introduction
Oral cavity carcinoma (OCC) is the most common
malignant cancer worldwide, with 28,030 new cases and 5,850 deaths
per year in the United States (1).
OCC prevalence is increasing for many reasons, including excessive
alcohol consumption, excessive smoking, human papilloma virus
infection and lack of hygiene (2).
Smoking and alcohol consumption are the main contributors to the
rising prevalence of OCC (3–6). The
5-year survival rate of OCC is 75–90% for stage I and 10–22% for
stage IV (7,8). Surgery followed by radiotherapy is
the primary treatment for early and localized advanced OCC.
However, local and regional recurrences are reported in 90% of
cases after surgery and radiotherapy treatment (9). Adjuvant chemotherapy after surgery
improved survival of OCC patients by 16% (10). Despite recent developments in
treatment, the overall survival of OCC has improved by just 5% in
the last 20 years (6,11). Therefore, treatments for OCC
clearly need to be improved; more personalized therapies and novel
molecular therapies with fewer adverse events may represent more
effective approaches (12).
To isolate potential molecular targets for the
diagnosis and/or treatment of OCC, we performed genome-wide gene
expression analyses and tissue microarray analyses of various solid
tumor tissues. We identified several oncoantigens involved in the
development and/or progression of cancer (13–36).
Cell division cycle associated 1 (CDCA1) mRNA was
overexpressed in cancer tissues, including OCC, colorectal, lung
cancer, cholangiocellular carcinoma and urological cancer (33–36).
In addition, HLA-A0201-restricted epitope peptides from the CDCA1
protein induced peptide-specific cytotoxic T lymphocytes,
implicating CDCA1 as a likely target for molecular targeted therapy
and/or immunotherapy (36). CDCA1
is a component of the Ndc80 complex, which contains two
subcomplexes: (KNTC2)-NUF2 (CDCA1) and SPC24–SPC25. The NCD80
complex is involved in chromosome segregation and the spindle
checkpoint (37). The attachment
of CDCA1 to the kinetochore outer plate generates
microtubule-dependent forces for chromosomal movement and protein
assembly at the kinetochore during the spindle checkpoint.
CDCA1 is expressed in human cancers, but its specific role
in OCC growth/survival or the clinical significance of CDCA1
protein as a tissue biomarker for OCC has not been determined
(38,39).
In the present study, we report that CDCA1 plays an
essential role in the malignant potential and survival of OCC, and
represents a promising diagnostic and prognostic tissue biomarker.
In addition, CDCA1 is a potential therapeutic target for new
molecular targeted therapies for OCC.
Materials and methods
Cell lines and clinical samples
The following cell lines were used in this study:
seven human OCC cell lines (FaDu, CAL-27, HSC2, HSC3, HSC4, Ca9-22
and SCC-9 cells); dysplasia of human oral keratinocyte (DOK) cells;
and human oral mucosa keratinocytes (HOMK). The histology and
suppliers of all cancer cells are summarized in Table I. All cells (except HOMK cells)
were grown in culture medium supplemented with 10% fetal bovine
serum (FBS) and antibiotics and were maintained at 37°C in an
atmosphere of humidified air. HOMK cells were grown in medium
supplemented with epilife defined growth supplement. Eight OCC
tissue samples purchased from Origene (Rockville, MD, USA) were
used for real-time PCR experiments.
 | Table IThe human OCC cell lines and oral
mucosa keratinocyte. |
Table I
The human OCC cell lines and oral
mucosa keratinocyte.
| Cell line | Histology | Resource
distributor |
|---|
| FaDu | Squamous cell
carcinoma of pharynx | ATCCa |
| SCC9 | Squamous cell
carcinoma of tongue | ATCCa |
| CAL-27 | Squamous cell
carcinoma of tongue | ATCCa |
| Ca9-22 | Gingival squamous
cell carcinoma | RIKEN BRCb |
| HSC2 | Squamous cell
carcinoma of mouth | RIKEN BRCb |
| HSC3 | Squamous cell
carcinoma of tongue | RIKEN BRCb |
| HSC4 | Squamous cell
carcinoma of tongue | RIKEN BRCb |
| DOK | Dysplastic oral
keratinocyte | European Collection
of Cell Cultures |
| HOMK | Human oral mucosa
keratinocyte | Cell Research
Corporation Pte Ltd |
Ninety-nine formalin-fixed primary OCC tissues (42
female, 57 male patients; median age, 66 years; age range, 28–92
years) and adjacent healthy tissues were obtained from patients
undergoing surgery at the Department of Oral and Maxillofacial
Surgery, Kumamoto University School of Medicine. The clinical stage
of these tumor samples was judged according to the Union for
International Cancer Control TNM classification. This study and the
use of all clinical materials were approved by individual
institutional ethics committees.
Quantitative real-time PCR
Total RNA was isolated from cultured cells and OCC
tissues using Maxwell 16 LEV simplyRNA tissue kit (Promega Corp.,
Madison, WI, USA). Complementary DNA was synthesized using ReverTra
Ace qPCR RT kit (Toyobo, Co., Ltd., Osaka, Japan). mRNAs were
quantified by real-time PCR using TaqMan Universal Master Mix II
and TaqMan assays on a StepOnePlus thermocycler (Applied
Biosystems) according to the manufacturer’s instructions. Each
experiment was done in triplicate. ACTB (Hs01060665_g1) was
used as an internal control and CDCA1 (Hs00230097_m1)
primers were used (Applied Biosystems, Foster City, CA, USA).
Western blotting
Cells were lysed in RIPA lysis buffer with protease
inhibitors (Thermo Fisher Scientific, Waltham, MA, USA). Proteins
separated by SDS-PAGE were transferred onto PVDF membranes
(Trans-Blot Turbo Transfer Pack; Bio-Rad Laboratories, Hercules,
CA, USA). After blocking with Block Ace solution (DS Pharma
Biomedical, Co., Ltd., Osaka, Japan), membranes were incubated with
anti-CDCA1 primary antibody (Abcam) for 1 h. Immunoreactive
proteins were incubated with horseradish peroxidase
(HRP)-conjugated secondary antibodies (GE Healthcare Life Sciences,
Chalfont, UK) for 1 h at room temperature. Protein bands were
visualized by enhanced chemiluminescence using an ImageQuant LAS
4000 mini system (GE Healthcare Biosciences).
Immunocytochemical analysis
Cultured cells were washed twice with PBS(−), fixed
in 4% paraformaldehyde solution for 10 min at 37°C, and
permeabilized in PBS(−) containing 0.1% Triton X-100 for 3 min.
Non-specific binding sites were blocked with blocking solution (CAS
Block; Invitrogen, Carlsbad, CA, USA) for 10 min at room
temperature before incubating with human CDCA1 antibody solution
for 1 h at room temperature. Alexa Fluor 488-conjugated goat
anti-rabbit secondary antibody (Molecular Probes, Eugene, OR, USA)
was added to detect endogenous CDCA1. Nuclei were stained with
4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories,
Burlingame, CA, USA). CDCA1 antibody staining was visualized with a
fluorescence microscope (BZ-X710; Keyence, Osaka, Japan).
Immunohistochemistry and tissue
microarray analysis
Tumor tissue microarrays were constructed using 99
formalin-fixed primary OCC tissues, which were removed during
curative surgery without neoadjuvant chemotherapy. Tumor tissue
microarrays were constructed according to the previously published
procedures (13). To measure CDCA1
protein expression in paraffin-embedded OCC samples, we performed
immunohistochemistry using a rabbit anti-CDCA1 antibody and the
EnVision HRP kit (DakoCytomation A/S, Glostrup, Denmark). For
antigen retrieval, slides were boiled in retrieval solution
(DakoCytomation). A rabbit polyclonal anti-CDCA1 antibody was added
to each slide after blocking endogenous peroxidase activity and
unspecific binding sites. After primary antibody incubation,
sections were incubated with an HRP-labeled anti-rabbit IgG
secondary antibody (DakoCytomation). Substrate-chromogen was added
to visualize labeled proteins and the specimens were counterstained
with hematoxylin. The intensity of staining within each tumor
tissue core was mostly homogeneous, therefore, the intensity of
CDCA1 staining was semi-quantitatively evaluated by two independent
investigators without prior knowledge of the clinicopathological
data. Expression was recorded as absent, weak or strong.
Statistical analysis
We used contingency tables to correlate
clinicopathological variables, (e.g., gender, age, region and
pathological TNM stage), with CDCA1 protein expression levels
determined by tissue microarray analysis. Survival curves were
generated on the basis of the date of surgery or diagnosis to the
time of death, or to the last follow-up observation. Kaplan-Meier
curves were calculated for each relevant variable and for CDCA1
expression in oral tumors. Differences in survival times among
patient subgroups were analyzed using the log-rank test. Univariate
and multivariate analyses were performed with the Cox proportional
hazard regression model to determine associations between
clinicopathological variables and cancer-related mortality.
RNA interference assay
We transfected siRNAs (Sigma-Aldrich, St. Louis, MO,
USA) into HSC4 and Ca9-22 cells, using Lipofectamine 2000 reagent
(Invitrogen). The target sequences of the synthetic
oligonucleotides for RNA interference were as follows: control 1
(luciferase: LUC) 5′-CGUACG CGGAAUACUUCGATT-3′; control 2 (EGFP)
5′-GAAGCAG CACGACUUCUUCTT-3′; siRNA-CDCA1-#1, 5′-AAGAUG
CUGCUGAAAGGGAGATT-3′; siRNA-CDCA1-#2, 5′-GAA GUCAUGUUCCACAUUTT-3′.
Five days after transfection, cell proliferation was evaluated by
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
(Dojindo Molecular Technologies) and colony formation assays.
Flow cytometry
The cell cycle of OCC cells was analyzed using a
Cycletest Plus DNA reagent kit (BD Biosciences, San Jose, CA, USA).
Ca9-22 and HSC4 cells were transfected with si-CDCA1 or si-LUC.
Seventy-two hours after siRNA transfection, 5×105
cells/ml were collected for measuring DNA ploidy. Cell cycle
analysis was performed within 3 h using a BD FACSVerse flow
cytometer. The DNA content of cells selected from at least 20,000
ungated cells was measured.
Live cell imaging
HSC4 cells were seeded into 35-mm glass dishes in
RPMI containing 10% FBS. To investigate apoptosis 48 h after
transfecting CDCA1 siRNA or control siRNA into HSC4 cells, we
performed time-lapse imaging to detect the apoptotic cells using
EVOS FL Auto cell imaging system (Life Technologies, Carlsbad, CA,
USA). We captured images every 20 min for up to 96 h.
Results
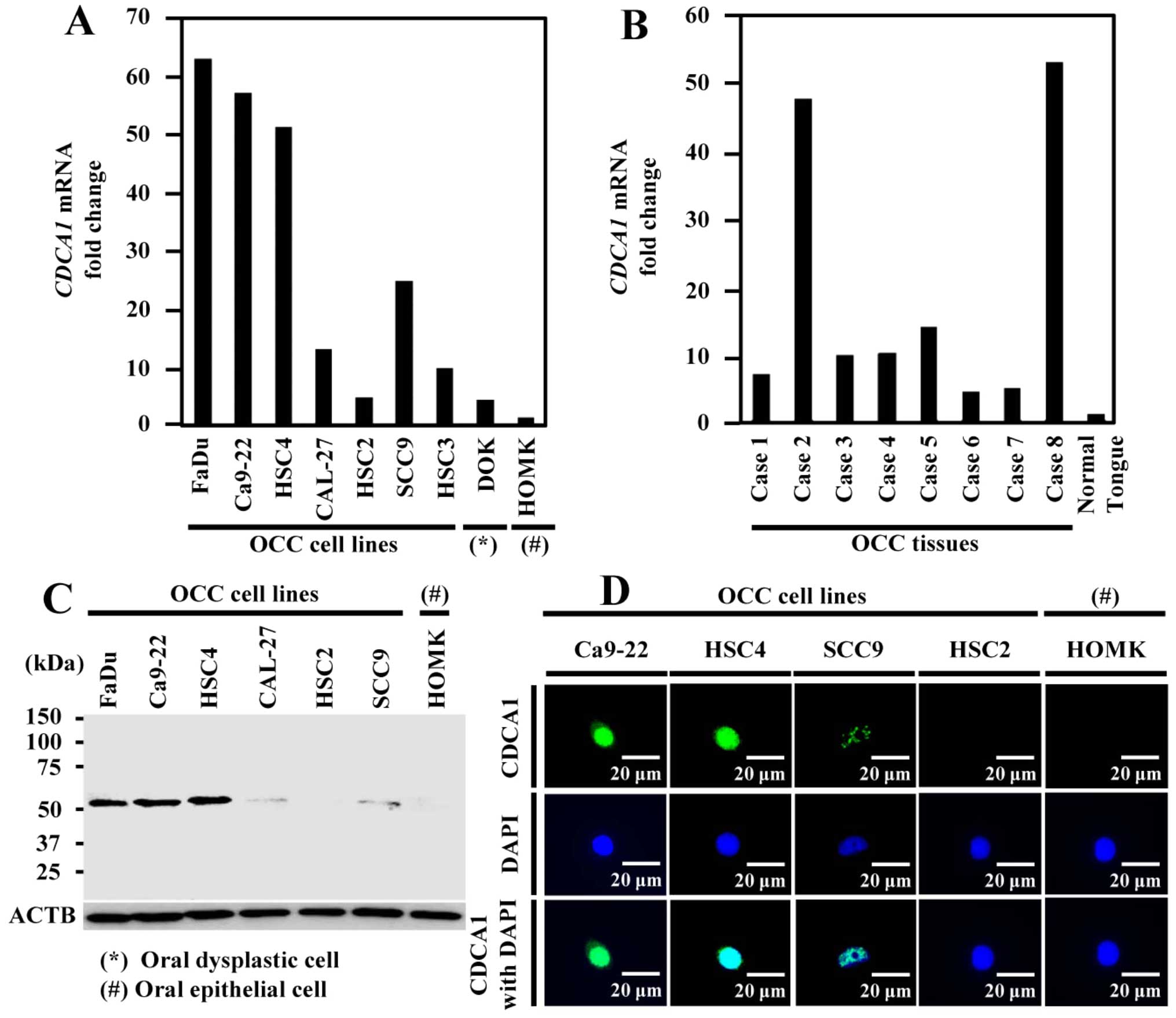
Expression of CDCA1 in OCC cells and
tissues. CDCA1
mRNA was upregulated in six of the seven OCC cell
lines, but expression was hardly detectable in healthy oral
epithelial cells by real-time PCR (Fig. 1A). We confirmed increased
CDCA1 expression in all eight OCC tissues by real-time PCR
and very low expression in healthy tongue tissue (Fig. 1B). In addition, CDCA1 protein was
strongly expressed in five out of six OCC cell lines using western
blotting (Fig. 1C). Furthermore,
immunocytochemical staining showed that CDCA1 protein was mainly
located in nucleus of Ca9-22, HSC4 and SCC9 cells, but no
expression was detected in HSC2 and human oral mucosa keratinocytes
(HOMK) cells (Fig. 1D).
Association of CDCA1 expression with OCC
prognosis
Immunohistochemical analysis using tissue
microarrays of 99 OCC samples demonstrated that CDCA1 staining was
mainly in the cell nucleus and cytoplasm of cancer cells. Strong
CDCA1 staining was observed in 39 OCC tissues (39.4%), weak
staining in 28 tissues (28.3%) and absent staining in 32 cases
(32.3%). In contrast, the adjacent healthy tongue epithelial
tissues were not stained (Fig.
2A). Next, we assessed the association of CDCA1 protein
expression with the clinical parameters. Age factor (higher in ≥65
years; P=0.0063 by Fisher’s exact test), pT factor (higher in
T3–T4; P=0.0112 by Fisher’s exact test), and pN factor (higher in
N1 and N2; P=0.0282 by Fisher’s exact test) were significantly
related to strong CDCA1 expression (Table II). Furthermore, strong CDCA1
expression was also significantly correlated with shorter survival
compared with weak/absent CDCA1 expression (P=0.0244 by log-rank
test; Fig. 2B). We also performed
univariate analysis to investigate the correlation between patient
prognosis and other factors, including age (<65 vs. ≥65 years),
gender (female vs. male), region (tongue vs. other), pT
classification (T1+T2 vs. T3+T4), pN classification (N0 vs. N1+N2)
and CDCA1 expression status (strong vs. weak/absent). Among these
parameters, strong CDCA1 expression (P=0.0310), advanced pT stage
(P=0.0080) and advanced pN stage (P=0.0474) were significantly
associated with poor prognosis (Table III).
 | Table IIAssociation of CDCA1 protein
expression in OCC tissues with the patient characteristics
(n=99). |
Table II
Association of CDCA1 protein
expression in OCC tissues with the patient characteristics
(n=99).
| Parameters | Total (99) | CDCA1 protein | P-value strong
positive vs. weak/absent |
|---|
|
|---|
| Strong expression
(39) | Weak expression
(28) | Absent expression
(32) |
|---|
| Gender |
| Male | 57 | 22 | 18 | 17 | >0.9999 |
| Female | 42 | 17 | 10 | 15 | |
| Age (years) |
| <65 | 40 | 11 | 13 | 16 | 0.0063a |
| ≥65 | 59 | 28 | 15 | 16 | |
| Region |
| Tongue | 50 | 18 | 21 | 11 | 0.5406 |
| Othersb | 49 | 21 | 7 | 21 | |
| pT factor |
| T1+T2 | 79 | 26 | 27 | 26 | 0.0112a |
| T3+T4 | 20 | 13 | 1 | 6 | |
| pN factor |
| N0 | 82 | 28 | 25 | 29 | 0.0282a |
| N1+N2 | 17 | 11 | 3 | 3 | |
 | Table IIICox’s proportional hazards model
analysis of prognostic factors in patients with OCC. |
Table III
Cox’s proportional hazards model
analysis of prognostic factors in patients with OCC.
| Variables | Hazards ratio | 95% CI |
Unfavorable/favorable | P-value |
|---|
| Univariate
analysis |
| CDCA1
expression | 2.939 | 1.103–7.830 | Strong vs.
weak/absent | 0.0310b |
| Age (years) | 2.812 | 0.914–8.648 | ≥65/<65 | 0.0713 |
| Gender | 1.667 | 0.643–4.324 | Male/female | 0.2935 |
| Region | 1.607 | 0.61–4.235 |
Tongue/othersa | 0.3372 |
| T-factor | 3.718 | 1.408–9.821 | T3+T4/T1+T2 | 0.0080b |
| N-factor | 2.902 | 1.012–8.321 | N1+N2/N0 | 0.0474b |
| Multivariate
analysis |
| CDCA1
expression | 2.388 | 0.840–6.790 | Strong vs.
weak/absent | 0.1025 |
| T-factor | 2.862 | 0.910–9.001 | T3+T4/T1+T2 | 0.072 |
| N-factor | 1.283 | 0.359–4.583 | N1+N2/N0 | 0.7013 |
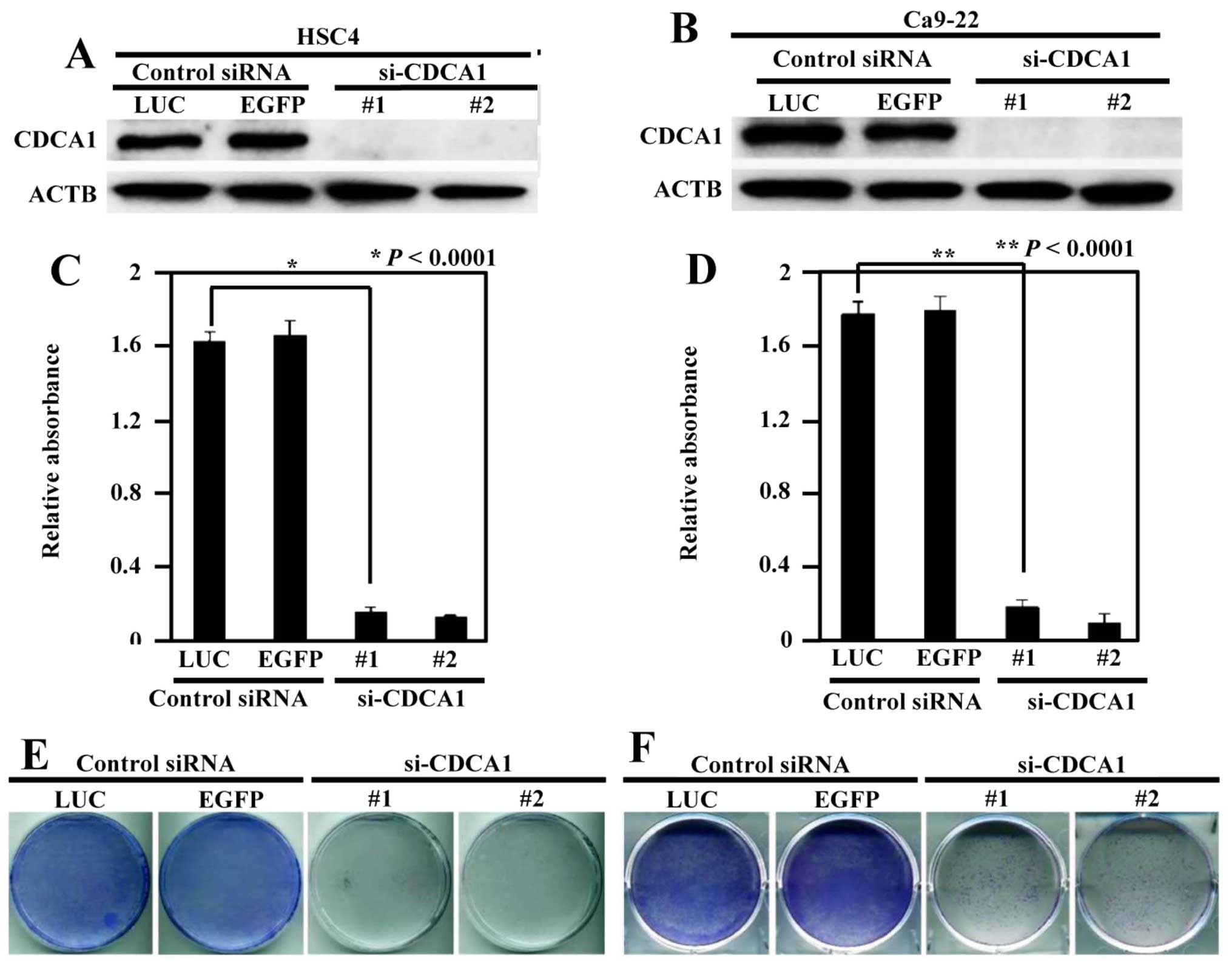
Inhibition of the growth of OCC cells by
knockdown of CDCA1 expression
To assess whether upregulation of CDCA1 played a
significant role in the growth of OCC cells, we transfected CDCA1
siRNAs (si-CDCA1-#1 and si-CDCA1-#2) and control siRNAs (si-LUC and
si-EGFP) into HSC4 and Ca9-22 cells. CDCA1 protein expression was
reduced by siRNA-mediated knockdown (Fig. 3A and B). Suppression of CDCA1
protein expression significantly inhibited cell viability in HSC4
and Ca9-22 cells compared with controls (P<0.0001 in both cell
types) (Fig. 3C and D). In
addition, colony formation assays demonstrated that reduced CDCA1
expression decreased the growth of HSC4 and Ca9-22 cells compared
with controls (Fig. 3E and F).
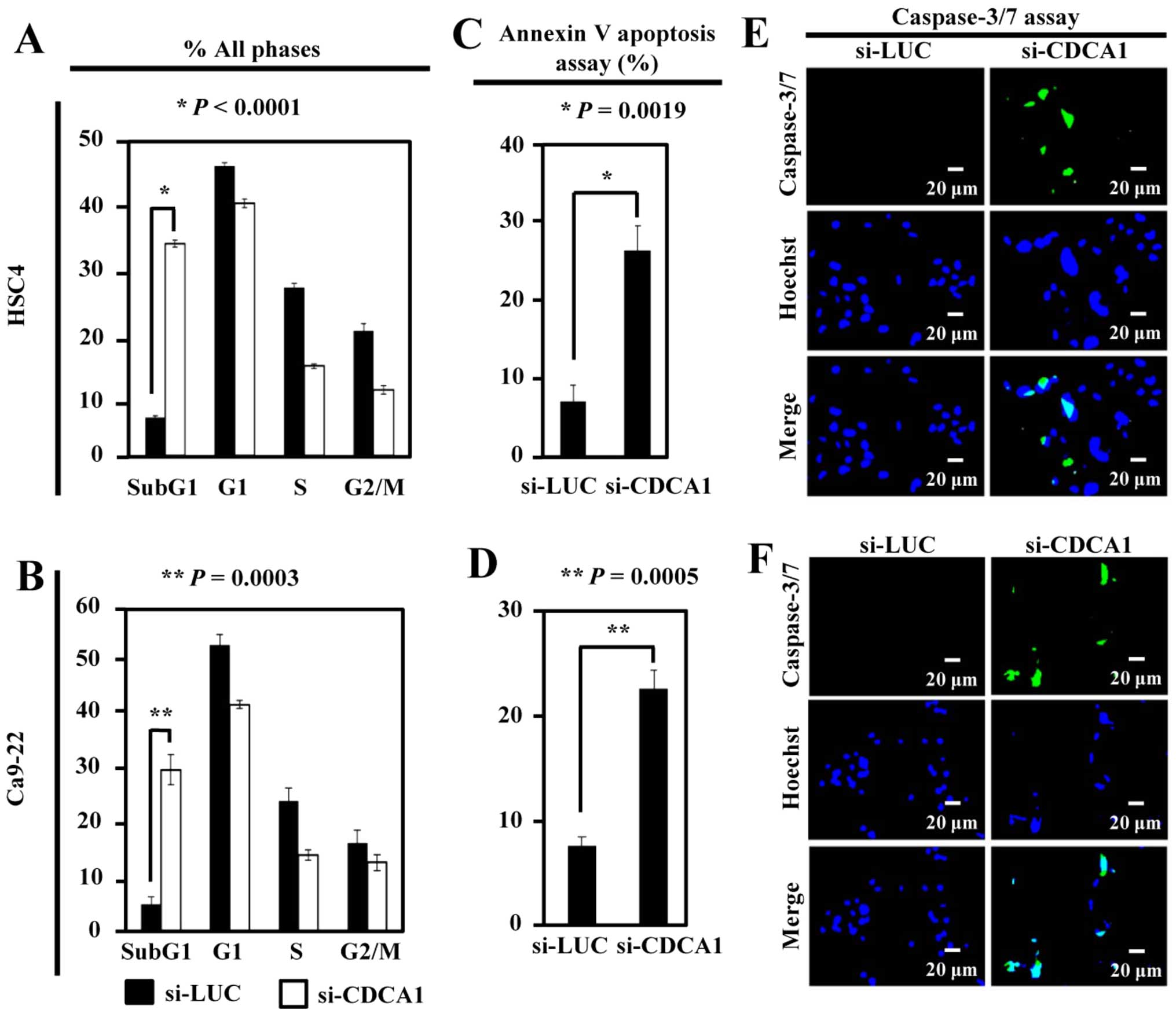
To analyze the effects of CDCA1 on the cell cycle,
flow cytometric analysis of HSC4 and Ca9-22 cells was performed 72
h after siRNA transfection. The proportion of cells in the sub-G1
phase was significantly higher in si-CDCA1-transfected HSC4 and
Ca9-22 cells than in controls (P<0.0001 and P=0.0003,
respectively) (Fig. 4A and B). The
number of apoptotic HSC4 and Ca9-22 cells was significantly higher
after siRNA-mediated CDCA1 knockdown than in control cells
(P=0.0019 and P=0.0005, respectively) (Fig. 4C and D). In addition, the
caspase-3/-7 green assay demonstrated increased activation of
caspase-3/-7 in OCC cells transfected with si-CDCA1 compared with
control siRNAs (Fig. 4E and
F).
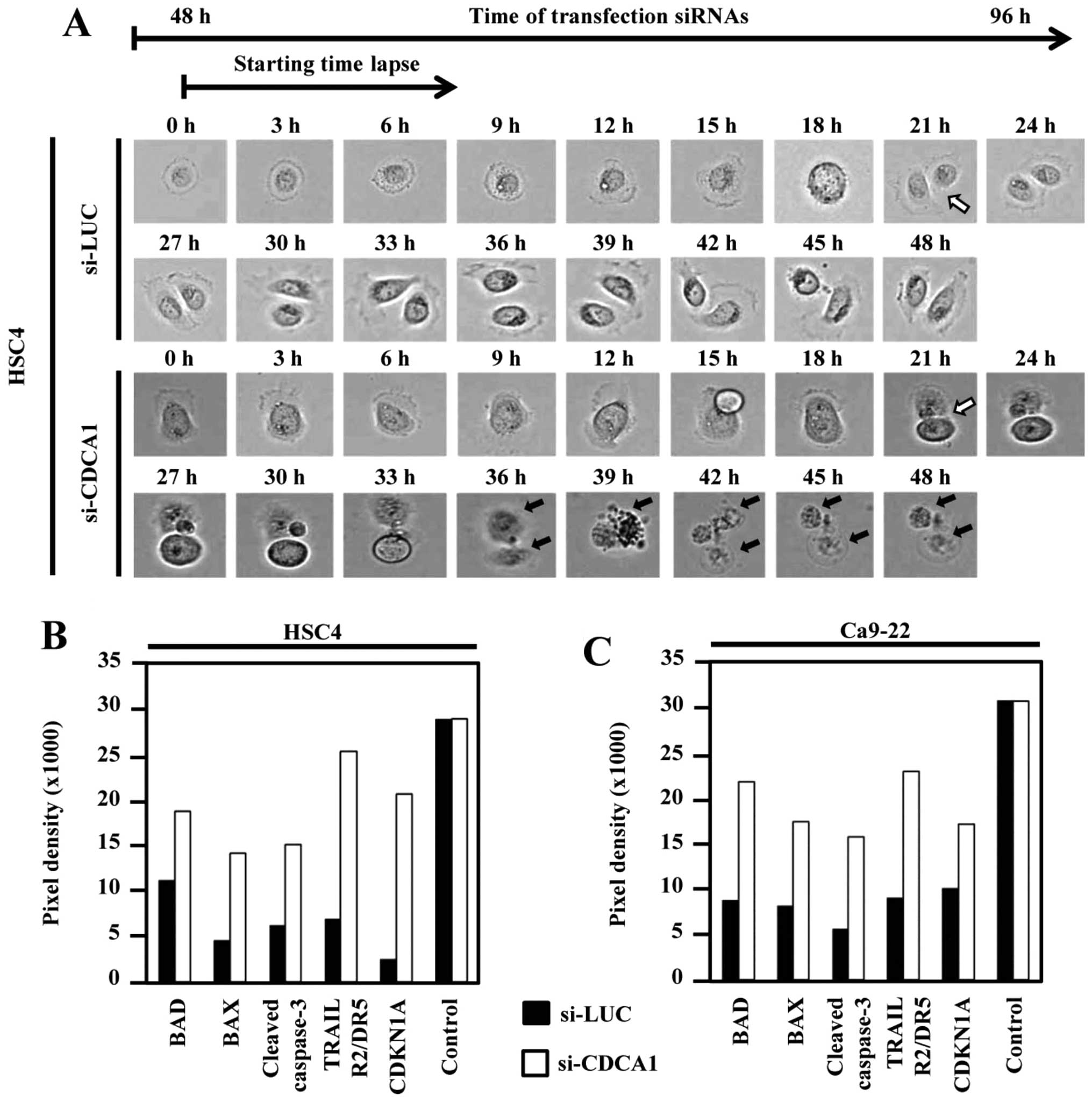
To further clarify the effect of CDCA1 knockdown on
cellular morphology and cell cycle, we performed live cell imaging
of HSC4 cells transfected with CDCA1 siRNA using time-lapse
microscopy. Dynamic HSC4 cell growth and survival were observed up
to 96 h after siRNA transfection. Mitotic cells were observed 69 h
after control-siRNA (si-LUC) transfection in HSC4 cells (white
arrow). Daughter cells transfected with si-CDCA1 died (black arrow)
after cell division, whereas cells transfected with control siRNAs
survived (Fig. 5A). In addition,
apoptosis-related proteins, including Bad, Bax, cleaved caspase-3,
TRAIL R2/DR5 and cyclin-dependent kinase inhibitor 1A (CDKN1A) were
enhanced in cells transfected with si-CDCA1 compared with control
siRNAs (Fig. 5B and C).
Discussion
Molecular targeted therapies for OCC are currently
being developed. They may provide better efficacy with fewer
side-effects by specifically targeting cancer-related mechanisms.
Molecular targeted drugs such as cetuximab (Erbitux) have now been
approved by the Food and Drug Administration as an initial
treatment of late-stage OCC in combination with chemotherapy.
Current anticancer agents can prolong the overall survival of OCC
patients, but they have limited efficacy and induce drug
resistance. Therefore, new therapeutic target drugs for OCC are
essential for improving the clinical outcome for patients.
In the present study, CDCA1 was overexpressed in OCC
tissues and CDCA1 overexpression was significantly associated with
poor prognosis. Furthermore, suppressing CDCA1 reduced OCC cell
growth and promoted apoptosis. cBioportal for Cancer Genomics
database (http://www.cbioportal.org/) revealed
that CDCA1 upregulation correlated significantly with poorer
prognosis for head and neck cancer, thus, independently supported a
prognostic potential of CDCA1 as a biomarker for OCC. To examine
mechanism of CDCA1 activation in OCC, we collected comparative
genome hybridization and genome sequencing data for CDCA1
(http://cancer.sanger.ac.uk/cosmic/gene/). Missense
mutations in CDCA1 were detected in 0.78% of OCC tissues
(2/255 cases), but no CDCA1 gene amplification or
translocation was reported. Therefore, CDCA1 overexpression could
be involved in an important epigenetic mechanism contributing to
OCC.
CDCA1 is a highly conserved component of a nuclear
division cycle complex and is categorized as an oncoantigen. We
showed that suppression of CDCA1 significantly inhibited OCC cell
growth and induced apoptosis in OCC cells, but the specific
mechanisms remain unclear. According to our findings and previous
studies, possible mechanisms exist for CDCA1-mediated regulation of
OCC growth and survival: i) stable spindle microtubule-kinetochore
attachment in mitotic prophase of highly proliferative cancer
cells; ii) regulation of apoptosis pathways. CDCA1 protein belongs
to the NDC80 complex, which contains CDCA1, HEC1, SPC24 and SPC25.
CDCA1-HEC1 heterodimers interact with the plus ends of spindle
microtubules and SPC24–SPC25 heterodimers anchor the complex into
the kinetochore (40). CDCA1 plays
a pivotal role in stable spindle microtubule-kinetochore attachment
and stable microtubule localization of centromere-associated
protein E (CENP-E) (41).
Decreased CDCA1 expression inhibited kinetochore attachment to
spindle microtubules, resulting in aberrant chromosome segregation,
prolonged mitotic blockade and cell death (42–45).
In the early phase of mitosis, after kinetochore attachment to
spindle microtubules, this complex is stable and has a half-life of
several minutes, which is essential for the next phase of mitosis.
However, the complex is unstable and has a half-life of
approximately 10 sec when binding of the kinetochore to
microtubules is impaired. This leads to abnormal cell division,
resulting in dysfunctional daughter cells that eventually die. In
agreement with previous findings, CDCA1 knockdown in OCC
cells resulted in impaired growth and apoptosis in cells.
Downregulation of CDCA1 increased the expression of
apoptosis-related proteins in OCC cells, including Bad, Bax,
cleaved caspase-3 and TRAIL R2/DR5. The data suggested the
dysregulation of appropriate cell division and subsequent induction
of apoptosis of OCC cells through various mechanisms such as
mitochondrial-dependent and caspase-dependent pathways.
In summary, CDCA1 is likely to play a significant
role in OCC carcinogenesis. Importantly, CDCA1 is associated with
growth and survival of OCC cells. Since CDCA1 was not expressed in
healthy tissues, apart from the testis (33), it could be a highly specific cancer
biomarker. In addition, targeting CDCA1 will lead to the
development of new type of therapeutic agents for OCC, such as
immunotherapies as well as molecular targeted drugs (46).
Acknowledgements
The present study was supported in part by a
Grant-in-Aid for Scientific Research (B) and Grant-in-Aid for
Scientific Research on Innovative Areas from The Japan Society for
the Promotion of Science (JSPS KAKENHI grant no. JP: 15H04761 and
16H06277). Y.D. is a member of the Shiga Cancer Treatment Project
supported by Shiga Prefecture (Japan).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen PC, Kuo C, Pan CC and Chou MY: Risk
of oral cancer associated with human papillomavirus infection,
betel quid chewing, and cigarette smoking in Taiwan - an integrated
molecular and epidemiological study of 58 cases. J Oral Pathol Med.
31:317–322. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Franceschi S, Talamini R, Barra S, Barón
AE, Negri E, Bidoli E, Serraino D and La Vecchia C: Smoking and
drinking in relation to cancers of the oral cavity, pharynx,
larynx, and esophagus in northern Italy. Cancer Res. 50:6502–6507.
1990.PubMed/NCBI
|
|
4
|
Cancela MC, Ramadas K, Fayette JM, Thomas
G, Muwonge R, Chapuis F, Thara S, Sankaranarayanan R and Sauvaget
C: Alcohol intake and oral cavity cancer risk among men in a
prospective study in Kerala, India. Community Dent Oral Epidemiol.
37:342–349. 2009. View Article : Google Scholar
|
|
5
|
Kimple RJ, Smith MA, Blitzer GC, Torres
AD, Martin JA, Yang RZ, Peet CR, Lorenz LD, Nickel KP, Klingelhutz
AJ, et al: Enhanced radiation sensitivity in HPV-positive head and
neck cancer. Cancer Res. 73:4791–4800. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chinn SB and Myers JN: Oral cavity
carcinoma: Current management, controversies, and future
directions. J Clin Oncol. 33:3269–3276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
da Silva SD, Hier M, Mlynarek A, Kowalski
LP and Alaoui-Jamali MA: Recurrent oral cancer: Current and
emerging therapeutic approaches. Front Pharmacol. 3:1492012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Poh CF, Durham JS, Brasher PM, Anderson
DW, Berean KW, MacAulay CE, Lee JJ and Rosin MP: Canadian
Optically-guided approach for Oral Lesions Surgical (COOLS) trial:
Study protocol for a randomized controlled trial. BMC Cancer.
11:4622011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carvalho AL, Kowalski LP, Agra IM, Pontes
E, Campos OD and Pellizzon AC: Treatment results on advanced neck
metastasis (N3) from head and neck squamous carcinoma. Otolaryngol
Head Neck Surg. 132:862–868. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Furness S, Glenny AM, Worthington HV,
Pavitt S, Oliver R, Clarkson JE, Macluskey M, Chan KK and Conway
DI: Interventions for the treatment of oral cavity and
oropharyngeal cancer: Chemotherapy. Cochrane Database Syst Rev.
(4): CD0063862011.PubMed/NCBI
|
|
11
|
National Cancer Institute. Surveillance,
Epidemiology, and End Results (SEER) Program. National Cancer
Institute Surveillance Research Program. based on November 2006
submission of SEER series 9 (1996–2003). 2006
|
|
12
|
Chinn SB, Spector ME, Bellile EL, McHugh
JB, Gernon TJ, Bradford CR, Wolf GT, Eisbruch A and Chepeha DB:
Impact of perineural invasion in the pathologically N0 neck in oral
cavity squamous cell carcinoma. Otolaryngol Head Neck Surg.
149:893–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Daigo Y and Nakamura Y: From cancer
genomics to thoracic oncology: Discovery of new biomarkers and
therapeutic targets for lung and esophageal carcinoma. Gen Thorac
Cardiovasc Surg. 56:43–53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Daigo Y, Takano A, Teramoto K, Chung S and
Nakamura Y: A systematic approach to the development of novel
therapeutics for lung cancer using genomic analyses. Clin Pharmacol
Ther. 94:218–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ishikawa N, Daigo Y, Takano A, Taniwaki M,
Kato T, Hayama S, Murakami H, Takeshima Y, Inai K, Nishimura H, et
al: Increases of amphiregulin and transforming growth factor-alpha
in serum as predictors of poor response to gefitinib among patients
with advanced non-small cell lung cancers. Cancer Res.
65:9176–9184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishikawa N, Daigo Y, Yasui W, Inai K,
Nishimura H, Tsuchiya E, Kohno N and Nakamura Y: ADAM8 as a novel
serological and histochemical marker for lung cancer. Clin Cancer
Res. 10:8363–8370. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kakiuchi S, Daigo Y, Ishikawa N, Furukawa
C, Tsunoda T, Yano S, Nakagawa K, Tsuruo T, Kohno N, Fukuoka M, et
al: Prediction of sensitivity of advanced non-small cell lung
cancers to gefitinib (Iressa, ZD1839). Hum Mol Genet. 13:3029–3043.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kato T, Daigo Y, Hayama S, Ishikawa N,
Yamabuki T, Ito T, Miyamoto M, Kondo S and Nakamura Y: A novel
human tRNA-dihydrouridine synthase involved in pulmonary
carcinogenesis. Cancer Res. 65:5638–5646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kikuchi T, Daigo Y, Katagiri T, Tsunoda T,
Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi
K, et al: Expression profiles of non-small cell lung cancers on
cDNA microarrays: Identification of genes for prediction of
lymph-node metastasis and sensitivity to anti-cancer drugs.
Oncogene. 22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suzuki C, Daigo Y, Ishikawa N, Kato T,
Hayama S, Ito T, Tsuchiya E and Nakamura Y: ANLN plays a critical
role in human lung carcinogenesis through the activation of RHOA
and by involvement in the phosphoinositide 3-kinase/AKT pathway.
Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kakiuchi S, Daigo Y, Tsunoda T, Yano S,
Sone S and Nakamura Y: Genome-wide analysis of organ-preferential
metastasis of human small cell lung cancer in mice. Mol Cancer Res.
1:485–499. 2003.PubMed/NCBI
|
|
22
|
Taniwaki M, Daigo Y, Ishikawa N, Takano A,
Tsunoda T, Yasui W, Inai K, Kohno N and Nakamura Y: Gene expression
profiles of small-cell lung cancers: Molecular signatures of lung
cancer. Int J Oncol. 29:567–575. 2006.PubMed/NCBI
|
|
23
|
Oshita H, Nishino R, Takano A, Fujitomo T,
Aragaki M, Kato T, Akiyama H, Tsuchiya E, Kohno N, Nakamura Y, et
al: RASEF is a novel diagnostic biomarker and a therapeutic target
for lung cancer. Mol Cancer Res. 11:937–951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hayama S, Daigo Y, Yamabuki T, Hirata D,
Kato T, Miyamoto M, Ito T, Tsuchiya E, Kondo S and Nakamura Y:
Phosphorylation and activation of cell division cycle associated 8
by aurora kinase B plays a significant role in human lung
carcinogenesis. Cancer Res. 67:4113–4122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ishikawa N, Daigo Y, Takano A, Taniwaki M,
Kato T, Tanaka S, Yasui W, Takeshima Y, Inai K, Nishimura H, et al:
Characterization of SEZ6L2 cell-surface protein as a novel
prognostic marker for lung cancer. Cancer Sci. 97:737–745. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kato T, Sato N, Hayama S, Yamabuki T, Ito
T, Miyamoto M, Kondo S, Nakamura Y and Daigo Y: Activation of
Holliday junction recognizing protein involved in the chromosomal
stability and immortality of cancer cells. Cancer Res.
67:8544–8553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suzuki C, Takahashi K, Hayama S, Ishikawa
N, Kato T, Ito T, Tsuchiya E, Nakamura Y and Daigo Y:
Identification of Myc-associated protein with JmjC domain as a
novel therapeutic target oncogene for lung cancer. Mol Cancer Ther.
6:542–551. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takahashi K, Furukawa C, Takano A,
Ishikawa N, Kato T, Hayama S, Suzuki C, Yasui W, Inai K, Sone S, et
al: The neuromedin U-growth hormone secretagogue receptor
1b/neurotensin receptor 1 oncogenic signaling pathway as a
therapeutic target for lung cancer. Cancer Res. 66:9408–9419. 2006.
View Article : Google Scholar
|
|
29
|
Taniwaki M, Takano A, Ishikawa N, Yasui W,
Inai K, Nishimura H, Tsuchiya E, Kohno N, Nakamura Y and Daigo Y:
Activation of KIF4A as a prognostic biomarker and therapeutic
target for lung cancer. Clin Cancer Res. 13:6624–6631. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yamabuki T, Takano A, Hayama S, Ishikawa
N, Kato T, Miyamoto M, Ito T, Ito H, Miyagi Y, Nakayama H, et al:
Dikkopf-1 as a novel serologic and prognostic biomarker for lung
and esophageal carcinomas. Cancer Res. 67:2517–2525. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fujitomo T, Daigo Y, Matsuda K, Ueda K and
Nakamura Y: Identification of a nuclear protein, LRRC42, involved
in lung carcinogenesis. Int J Oncol. 45:147–156. 2014.PubMed/NCBI
|
|
32
|
Nguyen MH, Koinuma J, Ueda K, Ito T,
Tsuchiya E, Nakamura Y and Daigo Y: Phosphorylation and activation
of cell division cycle associated 5 by mitogen-activated protein
kinase play a crucial role in human lung carcinogenesis. Cancer
Res. 70:5337–5347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hayama S, Daigo Y, Kato T, Ishikawa N,
Yamabuki T, Miyamoto M, Ito T, Tsuchiya E, Kondo S and Nakamura Y:
Activation of CDCA1-KNTC2, members of centromere protein complex,
involved in pulmonary carcinogenesis. Cancer Res. 66:10339–10348.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tomita Y, Yuno A, Tsukamoto H, Senju S,
Yoshimura S, Osawa R, Kuroda Y, Hirayama M, Irie A, Hamada A, et
al: Identification of CDCA1-derived long peptides bearing both
CD4+ and CD8+ T-cell epitopes: CDCA1-specific
CD4+ T-cell immunity in cancer patients. Int J Cancer.
134:352–366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kobayashi Y, Takano A, Miyagi Y, Tsuchiya
E, Sonoda H, Shimizu T, Okabe H, Tani T, Fujiyama Y and Daigo Y:
Cell division cycle-associated protein 1 overexpression is
essential for the malignant potential of colorectal cancers. Int J
Oncol. 44:69–77. 2014.
|
|
36
|
Harao M, Hirata S, Irie A, Senju S,
Nakatsura T, Komori H, Ikuta Y, Yokomine K, Imai K, Inoue M, et al:
HLA-A2-restricted CTL epitopes of a novel lung cancer-associated
cancer testis antigen, cell division cycle associated 1, can induce
tumor-reactive CTL. Int J Cancer. 123:2616–2625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ciferri C, De Luca J, Monzani S, Ferrari
KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED and
Musacchio A: Architecture of the human ndc80-hec1 complex, a
critical constituent of the outer kinetochore. J Biol Chem.
280:29088–29095. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kaneko N, Miura K, Gu Z, Karasawa H,
Ohnuma S, Sasaki H, Tsukamoto N, Yokoyama S, Yamamura A, Nagase H,
et al: siRNA-mediated knockdown against CDCA1 and KNTC2, both
frequently overexpressed in colorectal and gastric cancers,
suppresses cell proliferation and induces apoptosis. Biochem
Biophys Res Commun. 390:1235–1240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
van Duin M, Broyl A, de Knegt Y,
Goldschmidt H, Richardson PG, Hop WC, van der Holt B,
Joseph-Pietras D, Mulligan G, Neuwirth R, et al: Cancer testis
antigens in newly diagnosed and relapse multiple myeloma:
Prognostic markers and potential targets for immunotherapy.
Haematologica. 96:1662–1669. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wan X, O‘Quinn RP, Pierce HL, Joglekar AP,
Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al:
Protein architecture of the human kinetochore microtubule
attachment site. Cell. 137:672–684. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sundin LJ, Guimaraes GJ and Deluca JG: The
NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to
kinetochore-microtubule attachment in mitosis. Mol Biol Cell.
22:759–768. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
DeLuca JG, Howell BJ, Canman JC, Hickey
JM, Fang G and Salmon ED: Nuf2 and Hec1 are required for retention
of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr
Biol. 13:2103–2109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
DeLuca JG, Moree B, Hickey JM, Kilmartin
JV and Salmon ED: hNuf2 inhibition blocks stable
kinetochore-microtubule attachment and induces mitotic cell death
in HeLa cells. J Cell Biol. 159:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu D, Ding X, Du J, Cai X, Huang Y, Ward
T, Shaw A, Yang Y, Hu R, Jin C, et al: Human NUF2 interacts with
centromere-associated protein E and is essential for a stable
spindle microtubule-kinetochore attachment. J Biol Chem.
282:21415–21424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhai Y, Kronebusch PJ and Borisy GG:
Kinetochore microtubule dynamics and the metaphase-anaphase
transition. J Cell Biol. 131:721–734. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yoshitake Y, Fukuma D, Yuno A, Hirayama M,
Nakayama H, Tanaka T, Nagata M, Takamune Y, Kawahara K, Nakagawa Y,
et al: Phase II clinical trial of multiple peptide vaccination for
advanced head and neck cancer patients revealed induction of immune
responses and improved OS. Clin Cancer Res. 21:312–321. 2015.
View Article : Google Scholar
|