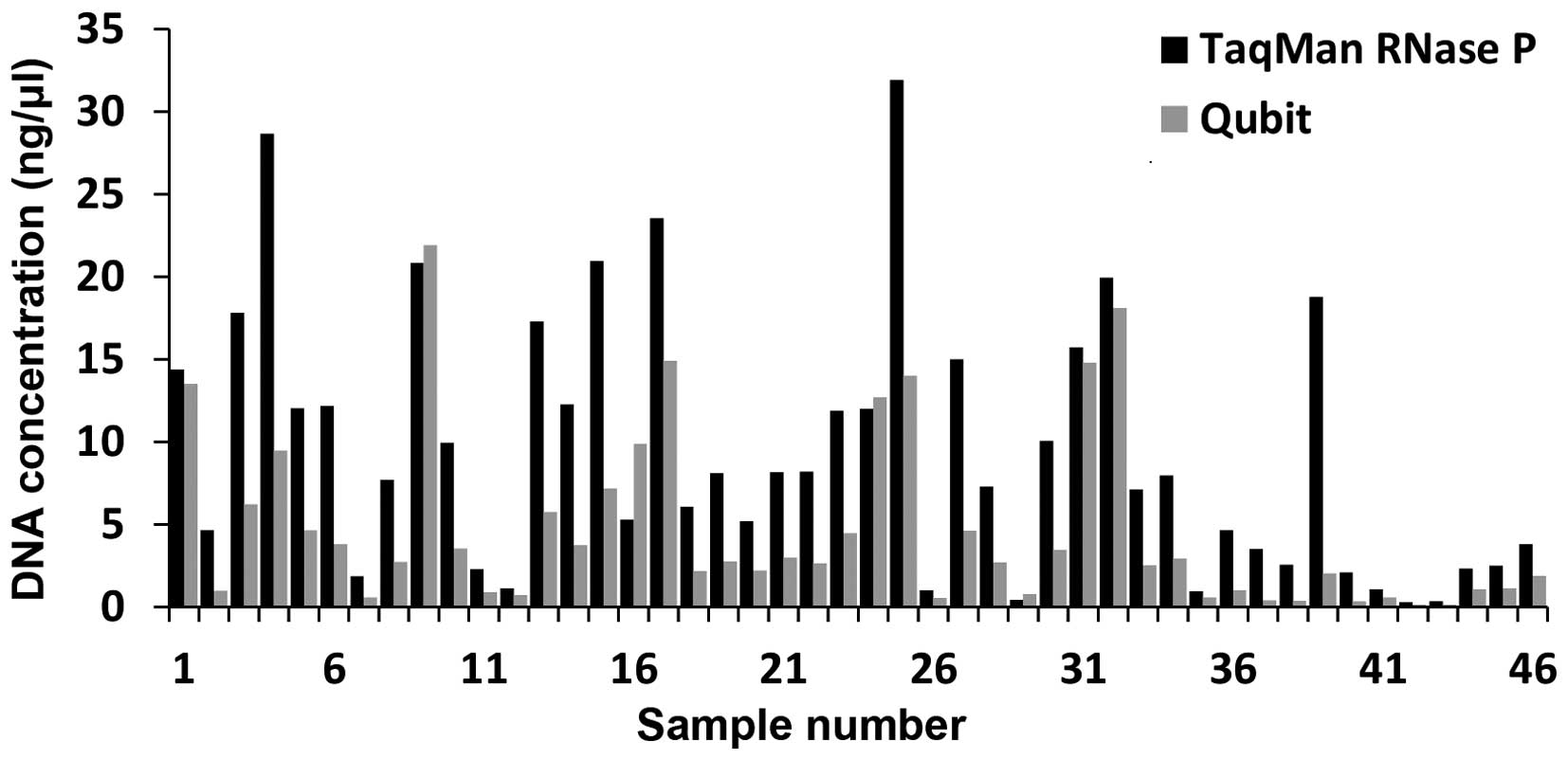
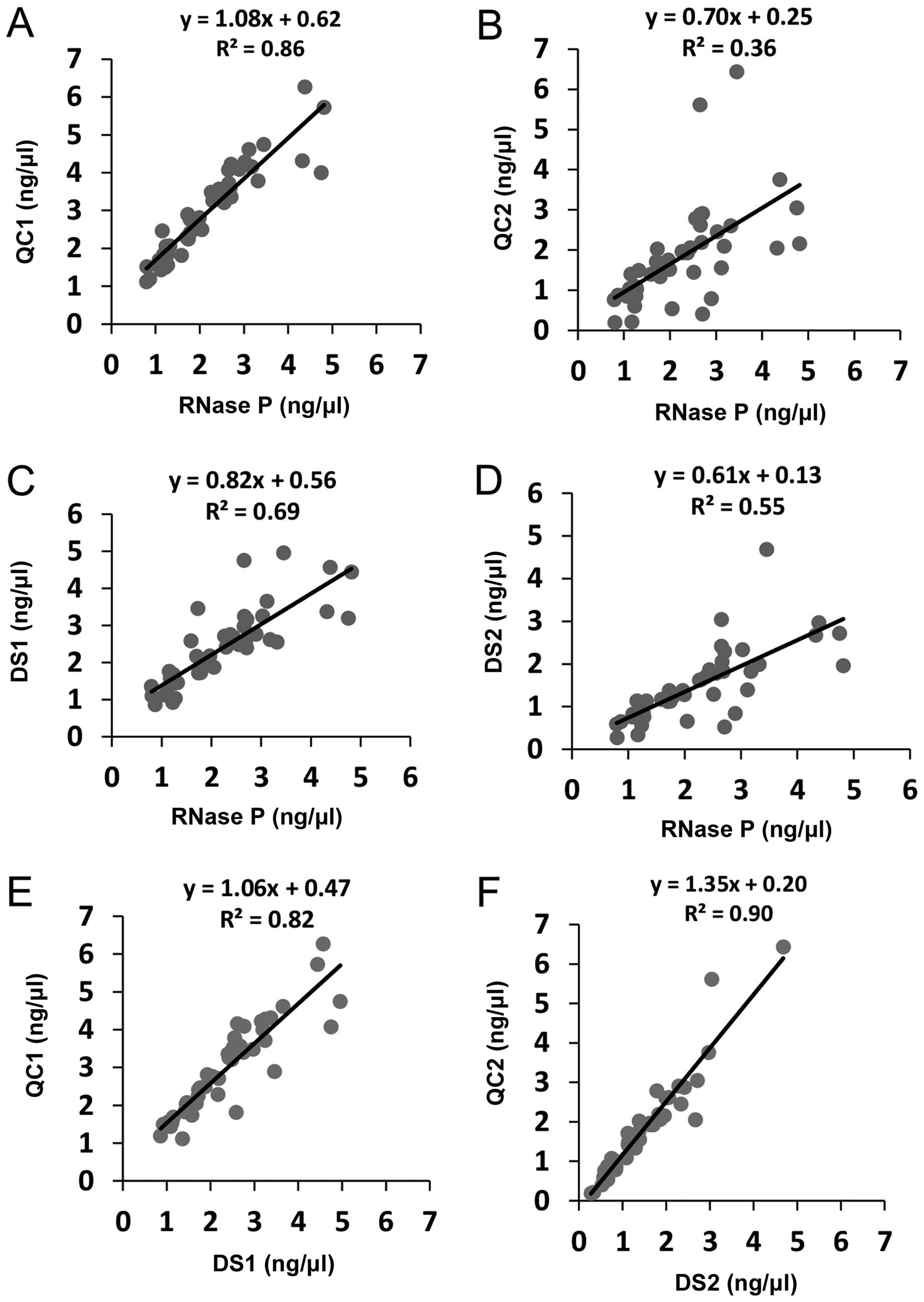
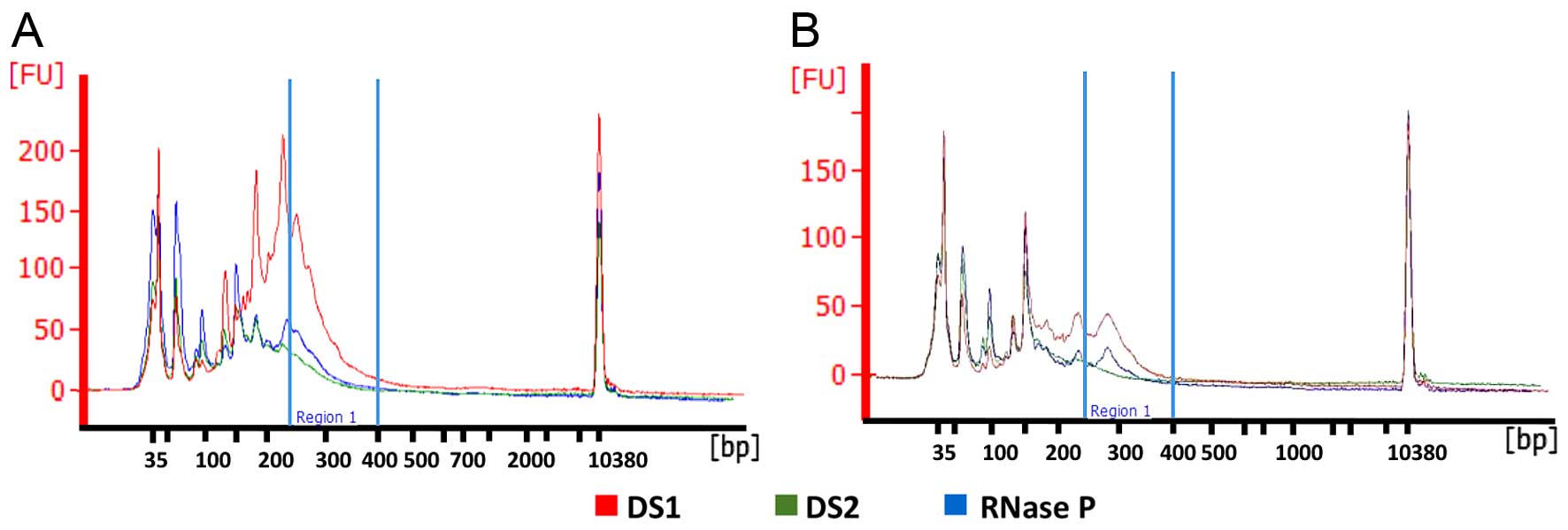
|
1
|
Bennett ST, Barnes C, Cox A, Davies L and
Brown C: Toward the 1,000 dollars human genome. Pharmacogenomics.
6:373–382. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hillier LW, Marth GT, Quinlan AR, Dooling
D, Fewell G, Barnett D, Fox P, Glasscock JI, Hickenbotham M, Huang
W, et al: Whole-genome sequencing and variant discovery in C.
elegans. Nat Methods. 5:183–188. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parsons DW, Jones S, Zhang X, Lin JC,
Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et
al: An integrated genomic analysis of human glioblastoma
multiforme. Science. 321:1807–1812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shah SP, Morin RD, Khattra J, Prentice L,
Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al:
Mutational evolution in a lobular breast tumour profiled at single
nucleotide resolution. Nature. 461:809–813. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fang LT, Lee S, Choi H, Kim HK, Jew G,
Kang HC, Chen L, Jablons D and Kim IJ: Comprehensive genomic
analyses of a metastatic colon cancer to the lung by whole exome
sequencing and gene expression analysis. Int J Oncol. 44:211–221.
2014.
|
|
6
|
Kang HC, Kim HK, Lee S, Mendez P, Kim JW,
Woodard G, Yoon JH, Jen KY, Fang LT, Jones K, et al: Whole exome
and targeted deep sequencing identify genome-wide allelic loss and
frequent SETDB1 mutations in malignant pleural mesotheliomas.
Oncotarget. 7:8321–8331. 2016.PubMed/NCBI
|
|
7
|
Mendez P, Dang J, Kim JW, Lee S, Yoon JH,
Kim T, Sailey CJ, Jablons DM and Kim IJ: Comprehensive evaluation
and validation of targeted next-generation sequencing performance
in two clinical laboratories. Int J Oncol. 49:235–242.
2016.PubMed/NCBI
|
|
8
|
Hadd AG, Houghton J, Choudhary A, Sah S,
Chen L, Marko AC, Sanford T, Buddavarapu K, Krosting J, Garmire L,
et al: Targeted, high-depth, next-generation sequencing of cancer
genes in formalin-fixed, paraffin-embedded and fine-needle
aspiration tumor specimens. J Mol Diagn. 15:234–247. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Han SW, Kim HP, Shin JY, Jeong EG, Lee WC,
Lee KH, Won JK, Kim TY, Oh DY, Im SA, et al: Targeted sequencing of
cancer-related genes in colorectal cancer using next-generation
sequencing. PLoS One. 8:e642712013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cancer Genome Atlas Research Network.
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brennan CW, Verhaak RG, McKenna A, Campos
B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ,
Berman SH, et al; TCGA Research Network. The somatic genomic
landscape of glioblastoma. Cell. 155:462–477. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bass AJ, Thorsson V, Shmulevich I,
Reynolds SM, Miller M, Bernard B, Hinoue T, Laird PW, Curtis C,
Shen H, et al; Cancer Genome Atlas Research Network. Comprehensive
molecular characterization of gastric adenocarcinoma. Nature.
513:202–209. 2014. View Article : Google Scholar :
|
|
14
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, et al: Mapping the hallmarks of lung adenocarcinoma
with massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ng SB, Turner EH, Robertson PD, Flygare
SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler
EE, et al: Targeted capture and massively parallel sequencing of 12
human exomes. Nature. 461:272–276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choi M, Scholl UI, Ji W, Liu T, Tikhonova
IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, et al:
Genetic diagnosis by whole exome capture and massively parallel DNA
sequencing. Proc Natl Acad Sci USA. 106:19096–19101. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ng SB, Buckingham KJ, Lee C, Bigham AW,
Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, et
al: Exome sequencing identifies the cause of a mendelian disorder.
Nat Genet. 42:30–35. 2010. View
Article : Google Scholar :
|
|
18
|
Gawad C, Koh W and Quake SR: Single-cell
genome sequencing: Current state of the science. Nat Rev Genet.
17:175–188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Bourcy CF, De Vlaminck I, Kanbar JN,
Wang J, Gawad C and Quake SR: A quantitative comparison of
single-cell whole genome amplification methods. PLoS One.
9:e1055852014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nielsen K, Mogensen HS, Hedman J,
Niederstätter H, Parson W and Morling N: Comparison of five DNA
quantification methods. Forensic Sci Int Genet. 2:226–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Navarro E, Serrano-Heras G, Castano MJ and
Solera J: Real-time PCR detection chemistry. Clin Chim Acta.
439:231–250. 2015. View Article : Google Scholar
|
|
22
|
Singer VL, Jones LJ, Yue ST and Haugland
RP: Characterization of PicoGreen reagent and development of a
fluorescence-based solution assay for double-stranded DNA
quantitation. Anal Biochem. 249:228–238. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robin JD, Ludlow AT, LaRanger R, Wright WE
and Shay JW: Comparison of DNA quantification methods for next
generation sequencing. Sci Rep. 6:240672016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
White RA III, Blainey PC, Fan HC and Quake
SR: Digital PCR provides sensitive and absolute calibration for
high throughput sequencing. BMC Genomics. 10:1162009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Panaro NJ, Yuen PK, Sakazume T, Fortina P,
Kricka LJ and Wilding P: Evaluation of DNA fragment sizing and
quantification by the agilent 2100 bioanalyzer. Clin Chem.
46:1851–1853. 2000.PubMed/NCBI
|
|
26
|
Arya M, Shergill IS, Williamson M,
Gommersall L, Arya N and Patel HR: Basic principles of real-time
quantitative PCR. Expert Rev Mol Diagn. 5:209–219. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Buh Gasparic M, Tengs T, La Paz JL,
Holst-Jensen A, Pla M, Esteve T, Zel J and Gruden K: Comparison of
nine different real-time PCR chemistries for qualitative and
quantitative applications in GMO detection. Anal Bioanal Chem.
396:2023–2029. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Karro JE, Yan Y, Zheng D, Zhang Z,
Carriero N, Cayting P, Harrrison P and Gerstein M: Pseudogene.org:
A comprehensive database and comparison platform for pseudogene
annotation. Nucleic Acids Res. 35:Database. D55–D60. 2007.
View Article : Google Scholar
|
|
30
|
Bhat S, Curach N, Mostyn T, Bains GS,
Griffiths KR and Emslie KR: Comparison of methods for accurate
quantification of DNA mass concentration with traceability to the
international system of units. Anal Chem. 82:7185–7192. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim IJ, Quigley D, To MD, Pham P, Lin K,
Jo B, Jen KY, Raz D, Kim J, Mao JH, et al: Rewiring of human lung
cell lineage and mitotic networks in lung adenocarcinomas. Nat
Commun. 4:17012013. View Article : Google Scholar : PubMed/NCBI
|