Introduction
Long non-coding RNAs (lncRNAs) are encoded similarly
to coding genes but do not contain a protein-coding sequence. The
transcripts of lncRNA are >200 nucleotides long and expressed at
lower levels than protein-coding transcripts. Some lncRNAs directly
modulate gene expression in a cis manner; these lncRNAs may
be expressed from the promoter (1), intron (2) and enhancer regions (3) of certain genes and then regulate
neighboring protein-coding genes on the same chromosome.
Furthermore, some lncRNAs distally regulate gene expression across
multiple chromosomes in a trans manner. These lncRNAs can
facilitate enhancer function via long-range DNA looping
interactions (4). Also, some
lncRNAs appear to modulate the DNA-binding of certain transcription
factors (TFs) and non-DNA-binding cofactors at several target
sites, thus affecting gene expression in trans (5).
In recent years, several lncRNAs have been
implicated in cancer development and progression (6–8).
Chemoresistance is an important feature of cancer progression,
since chemoresistant cells are insensitive to the apoptotic signals
delivered by cytotoxic chemotherapeutic agents. They also show a
strong ability to proliferate. A large degree of chemoresistance is
acquired during the response of cancer cells to unfavorable niches,
and their rapid response depends on the effective and heritable
epigenetic regulation of gene expression, including DNA methylation
and microRNA regulation (9). For
example, methylation of microRNA-200c inhibits microRNA expression,
so the targets of the microRNA, including modulators of epithelial
mesenchymal transition pathways that essentially promote cell
proliferation and diminish apoptosis, are activated (10–14).
In the current decade, emerging evidence indicates that 70–90% of
the mammalian genome produces lncRNAs, which are another important
component of epigenetic regulation (15,16).
However, although >10,000 mammalian lncRNAs have been catalogued
(5), functional studies are as yet
limited in number, and how the network of lncRNAs is involved in
cancer chemoresistance still requires elucidation.
In the present study, we set out to analyze lncRNA
expression in adriamycin-resistant breast cancer cells using
microarrays, and compare their lncRNA expression profile with that
of parental chemosensitive cells in order to identify and
characterize dysregulated lncRNAs that may be involved in breast
cancer chemoresistance (Fig.
1A).
Materials and methods
Cell culture
MCF-7/WT human breast cancer cells were obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). Adriamycin (ADM)-resistant cells (MCF-7/ADM) and paclitaxel
(PTX)-resistant cells (MCF-7/PTX) were derived by treating MCF-7
cells with stepwise increasing concentrations of ADM or PTX over 8
months (17). The cells were
cultured in RPMI-1640 supplemented with 10% FBS, 100 μg/ml
penicillin and 100 U/ml streptomycin.
Microarray
The microarray profiling was conducted in the
laboratory of OE Biotechnology Co. (Shanghai, China). RNA from
MCF-7/ADM and MCF-7/WT cells was separately extracted using the
acid-phenol and chloroform method. Cyanine-3-CTP-labeled cRNA was
obtained using a Quick Amp Labeling kit (Agilent Technologies,
Santa Clara, CA, USA) and then purified with an RNeasy Mini kit
(Qiagen, Valencia, CA, USA). The labeled cRNAs were then hybridized
onto Agilent-062918 OE Human lncRNA Microarray V4.0 028004 (Agilent
Technologies), which is a Custom Gene Expression Array for OE
Biotechnology Co. and detects 46,506 lncRNAs. After washing, the
arrays were scanned with an Agilent scanner (G2505C).
Quality control of the microarray
data
Total RNA was quantified by the NanoDrop 2000
(Thermo Fisher Scientific, Waltham, MA, USA) and the RNA integrity
was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies).
RNA with RNA integrity number (RIN) value >7 and 28S/18S >0.7
was used for microarray analysis. After the raw data extraction,
the median CV (%) for each probe set was reported as the array
reproducibility. To analyze the biological repeatability of the
microarrays between MCF-7/ADM (n=3) or MCF-7/WT (n=3) cells,
two-dimensional principal component analysis was performed.
Data deposition
The microarray data have been submitted to GEO
(GSE81971).
Data analysis
Raw data of microarray was generated using Agilent
Feature Extraction software (Agilent Technologies) and then
normalized using GeneSpring’s quantile normalization (version 12.5;
Agilent Technologies). Differentially-expressed lncRNAs were
identified with a fold change ≥2.0 and a P-value <0.05.
WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/) was used
for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analysis.
Real-time PCR
LncRNA expression was analyzed using qRT-PCR.
Briefly, total RNA was extracted from MCF-7/WT and /ADM cells with
TRIzol (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from
total RNA (3 μg) using the SuperScript First Strand Synthesis
system (Invitrogen) with Oligo (dT) primers. Primers used for
real-time PCR were as previously described for UCA1 (18,19),
HOTAIR (6), GAS5 (20), HULC (21), MEG3 (22), SPRY4-IT1 (23) and CRNDE (24). Primer for NONHSAT028712 was:
forward, 5′-AAATACCTCACCCTCATCTATACCAAC-3′ and reverse,
5′-TTTCCCGTTGCCATTGAT-3′; for NONHSAT057282 was forward,
5′-AGCCGGAGGTGAGGAAGTT-3′ and reverse,
5′-AAGATTTTATTAGATTTTGGAACCTGAG-3′; for NONHSAG023333 was forward,
5′-GTTGGGAAATCAAG CATCGT-3′ and reverse, 5′-TTTAGCAAAAATGCAACTA
CATCC-3′.
The RT-PCR values were normalized to GAPDH and
calculated using the 2−ΔΔCT method.
LncRNA inhibition and functional
studies
LncRNA Smart Silencer was synthesized by Guangzhou
RiboBio Co., Ltd., (Guangzhou, China) and used to inhibit lncRNA.
The inhibitor was then transfected into MCF-7 cells using
Lipofectamine (Invitrogen).
MCF-7/ADM or MCF-7/PTX cells transfected with lncRNA
inhibitor were seeded onto 96-well plates and exposed to different
concentrations of ADM or PTX for 48 h. Then cell viability and
IC50 were assessed as previously described (25,26).
In experiments analyzing the cell cycle, MCF-7/ADM
cells transfected with lncRNA inhibitor were fixed and stained with
100 μg/ml propidium iodide (PI; Sigma Life Science) containing
RNase. PI fluorescence was detected using a FACSCalibur flow
cytometer on the PE-Texas Red channel for DNA content.
Results
Expression of lncRNAs in chemoresistant
breast cancer cells
To gain insights into the role of lncRNAs in
chemoresistance, we used microarray-based profiling to analyze the
lncRNAs and mRNAs in adriamycin-resistant MCF-7/ADM cells and their
chemosensitive parental control MCF-7/WT cells (17). In MCF-7/ADM cells, 4030 lncRNAs and
unannoted transcripts were upregulated and 3708 were downregulated
(Fig. 1B; Submitted online as
GSE81971), while 3423 mRNAs were upregulated and 2950 were
downregulated (Fig. 1C;
fold-change ≥2, P<0.05), suggesting that lncRNAs may be
dysregulated and participate in the development of
chemoresistance.
We then validated several cancer-related lncRNAs
with RT-PCR (6,7,18–24).
Among these, SPRY4-IT1 was upregulated in chemoresistant MCF-7/ADM
cells by both microarray analysis and RT-PCR, suggesting that it
may play a role in chemoresistance; other lncRNAs were not
significantly changed either in microarray analysis or RT-PCR
(Fig. 1D). These data, thus,
confirm the accuracy of microarrays, while prompted us to search
for new lncRNAs that may mediate chemoresistance.
In order to identify new candidate lncRNAs in
chemoresistance, we then chose the top 200 most significantly
changed lncRNAs for further analysis.
LncRNAs correlate with mRNA
expression
LncRNAs influence gene expression by regulating
chromatin remodeling, transcription and post-transcriptional
processing (5). To identify
potential lncRNA targets, we calculated the Pearson correlation of
each significantly changed lncRNA with each significantly changed
mRNA. An lncRNA and an mRNA were considered to be correlated when
the coefficient was >0.7 (P<0.05), so such mRNAs might be
regulated by their correlated lncRNAs. The most correlated mRNAs
for top 10 changed lncRNAs are exemplified in Table I.
 | Table ITop 10 changed lncRNAs and their
correlated mRNAs. |
Table I
Top 10 changed lncRNAs and their
correlated mRNAs.
| LncRNAa | FC (abs)a | Up/downa | Location | Top 3 correlated
mRNA (P-value; coefficience) |
|---|
| NONHSAT082326 | 16783 | Down |
chr21:43782390-6644 | SLC30A1
(0.00010002310686108;0.991822939253643)
FKBP1A (0.00010007371243696;−0.991820868137818)
PRDX2 (0.000100126235187625;0.991818719110682) |
|
ENST00000455354 | 3060 | Down |
chr21:41755010-7285 | LAMB2
(0.000100025125846499;−0.991822856613367)
DENND2C (0.000100084970897196;0.991820407437975)
USP2 (0.000100086690841299;−0.991820337059573) |
|
ENST00000422749 | 2703 | Down |
chr21:41755010-7285 | NEBL
(0.000100168472873419;0.991816991316235)
ABLIM1 (0.000100266459849063;0.991812984413778)
C17orf51 (0.000100447057119302;−0.99180560450735) |
|
ENST00000444046 | 2522 | Down |
chr21:41755010-7285 | ANPEP
(0.000100005971417187;−0.991823640668079)
KIF23 (0.000100364967970128;−0.991808958168178)
ARL6IP5 (0.000100426675196719;0.991806437060535) |
| NONHSAT128425 | 1756 | Down |
chr8:120221107-55888 | GDA
(0.00010028234061338;−0.991812335198366)
GHITM (0.000100331837803225;0.991810312051648)
MRPL13 (0.000100359825255418;−0.991809168313582) |
| NONHSAT023895 | 2501 | Up |
chr11:102667774-8070 | UBXN8
(0.000100363702178807;0.991809009891383)
SSH1 (0.000100433168514824;0.991806171814755)
NUP188 (0.000100566546204432;−0.991800725355188) |
| NONHSAT022443 | 894 | Up |
chr11:67353574-910 | HSCB
(0.000100041635865034;−0.991822180863287)
INPPL1 (0.000100075383041166;−0.991820799774519)
SNTB2 (0.000100118418071562;−0.991819038921128) |
| NONHSAT091446 | 621 | Up |
chr3:120123741-30173 | PTP4A1
(0.000100232420515941;−0.991814376135381)
DPY19L3 (0.000100276937161676;−0.991812556089039)
LOC100129846 (0.000100350020328222;−0.9918095689) |
| NONHSAT023896 | 574 | Up |
chr11:102668127-877 | ACSS3
(0.000100063079112198;−0.991821303280653)
RYBP (0.000100092305234907;−0.991820107328388)
ZNF57 (0.000100124371169228;−0.991818795369462) |
| NONHSAT005455 | 531 | Up |
chr1:117282602-5231 | BACE1
(0.000100119326125993;−0.991819001770562)
RBMS1 (0.000100213122112613;0.991815165268592)
NEXN (0.000100296526373889;0.991811755319101) |
The 500 lncRNA-mRNAs with the highest Pearson
correlation coefficient values were chosen for functional analysis
in GO and KEGG using the method described by Guttman et al
(27). The GO and KEGG functions
of each lncRNA-correlated mRNA were analyzed, then a hypergeometric
cumulative distribution function was applied to calculate the
enrichment of functional terms in the annotation of these mRNAs.
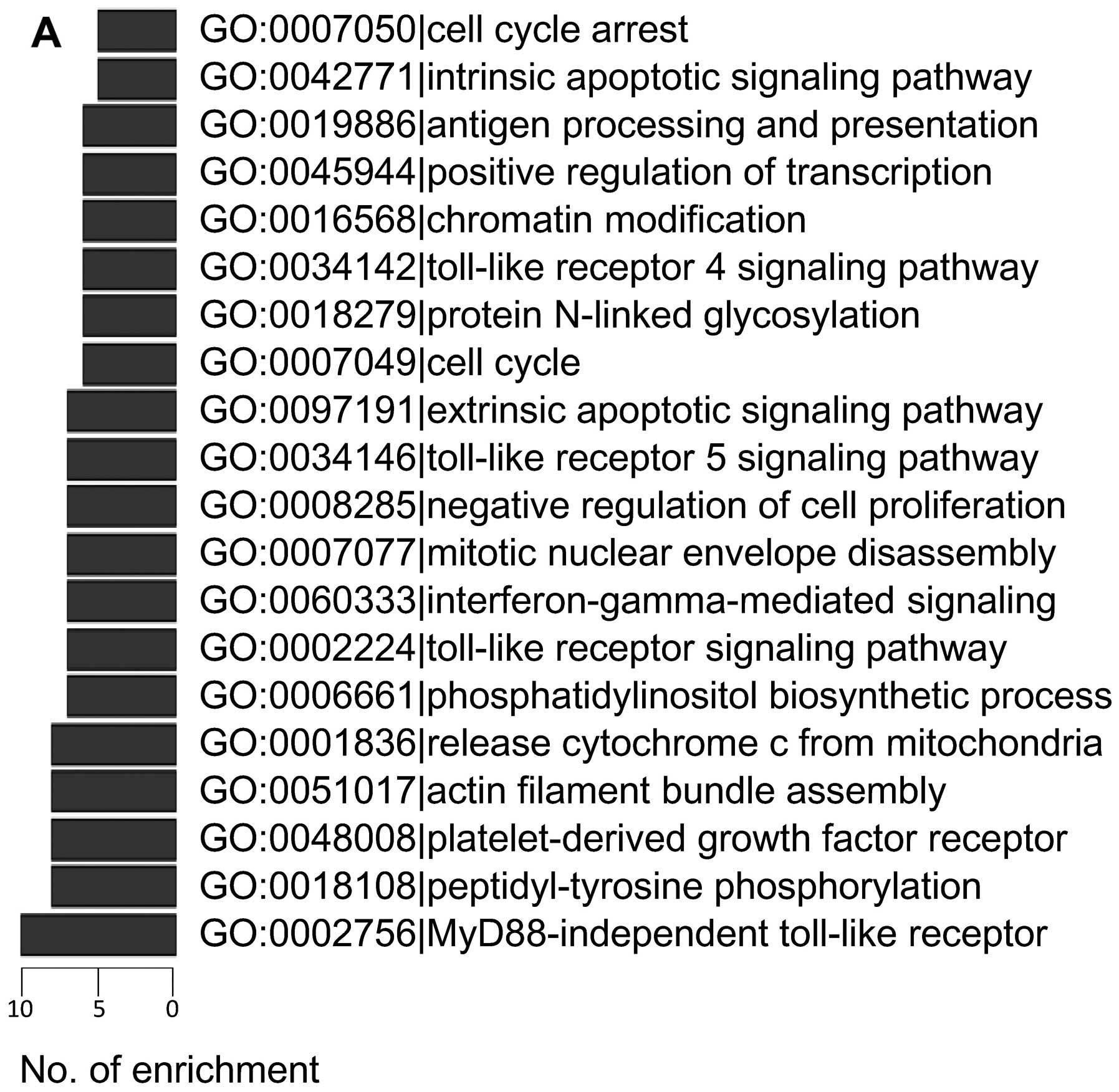
The most enriched GO processes and KEGG pathways are shown in
Fig. 2. Both analyses showed that
pathways directly associated with apoptosis and cell proliferation
were frequently regulated by lncRNAs, including release of
cytochrome c from mitochondria (GO:0001836), cell cycle
(GO:0007049), cell cycle arrest (GO:0007050), apoptosis (KEGG
04210), negative regulation of cell proliferation (GO:0008285),
MAPK signaling pathway (KEGG 04010) and p53 signaling pathway (KEGG
04115).
Cis-regulation of lnRNAs
Cis-regulation by lncRNAs of their correlated
mRNAs was then analyzed in chemoresistant MCF-7/ADM cells vs.
MCF-7/WT cells. Several lncRNAs were located 100K windows upstream
or downstream of the given mRNA, and the mRNA expression was
significantly correlated with the lncRNA. Possible
cis-regulation of their correlated mRNAs is exemplified in
Table II. Among these, we found
that NONHSAT028712 was significantly increased in MCF-7/ADM cells
(Table II and Fig. 3A). We also analyzed the expression
of NONHSAT028712 in another chemoresistant breast cancer cell line
MCF-7/PTX, which is paclitaxel-resistant (9). This lncRNA also significantly
increased in MCF-7/PTX cells (Fig.
3A). These data suggest a possible role of NONHSAT028712 in
mediating chemoresistance.
 | Table IICis-regulation of top 10
changed lncRNAs on their correlated mRNAs. |
Table II
Cis-regulation of top 10
changed lncRNAs on their correlated mRNAs.
| LncRNAs
(downregulated) | Possible targets of
cis-regulation (coefficient) | LncRNAs
(upregulated) | Possible targets of
CIS-regulation (coefficient) |
|---|
| NONHSAT128425 | NOV (−0.983) | NONHSAT022443 | SSH3 (−0.985);
RPS6KB2 (−0.975); RAD9A (−0.971) |
| NONHSAT006799 | PMF1 (0.958); LMNA
(−0.9878); SEMA4A (0.962) | NONHSAT028712 | ZC3H10 (−0.917);
RAB5B (0.985002614214423); RPS26 (0.954); OR10P1 (0.972); METTL7B
(0.975); DGKA (0.991); CDK2 (0.986) |
| NONHSAT042185 | CTDSPL2 (−0.821);
CASC4 (0.970) | NONHSAT098174 | FGF2 (0.943) |
| NONHSAT143304 | CDH3 (0.991) | NONHSAT057176 | RAB12 (0.961) |
| NONHSAT012940 | CSGALNACT2
(−0.983); RET (0.966); BMS1 (−0.977) | NONHSAT022441 | RPS6KB2 (−0.968);
RAD9A (−0.972); SSH3 (−0.983); ADRBK1 (−0.989) |
The cis-regulation of NONHSAT028712 is shown
in Fig. 3B. Genes of several
significantly-changed mRNAs was found to locate near the coding
sequence of NONHSAT028712. Among these mRNAs, the cell cycle kinase
CDK2 showed a high mRNA level in MCF-7/ADM and MCF-7/PTX cells
(Fig. 3A), and this has been
associated with cancer progression and chemoresistance (28,29).
We therefore inhibited the expression of NONHSAT028712 with a
synthesized inhibitor (Fig. 3C),
then analyzed the chemoresistance of the treated MCF-7/ADM cells.
The IC50 significantly decreased in MCF-7/ADM cells when
NONHSAT028712 was inhibited (Fig.
3D), along with a lower CDK2 mRNA level (Fig. 3E) and a higher rate of cell cycle
arrest in G1 (Fig. 3F). These data
strongly suggest that NONHSAT028712 regulates chemoresistance via a
CDK2-related pathway, while the mechanism requires further
exploration.
Interaction of transcription factors with
lncRNAs
To identify the possible role of lncRNA-TF
interactions in regulating gene expression, we first predicted the
TFs of lncRNA-correlated mRNAs using data from Gerstein et
al (30) that showed the
genomic binding information of different TFs. Then the
intersections of lncRNA-mRNA and mRNA-TF were calculated with a
hypergeometric cumulative distribution. Each lncRNA was
significantly associated with several TFs (data not shown). For
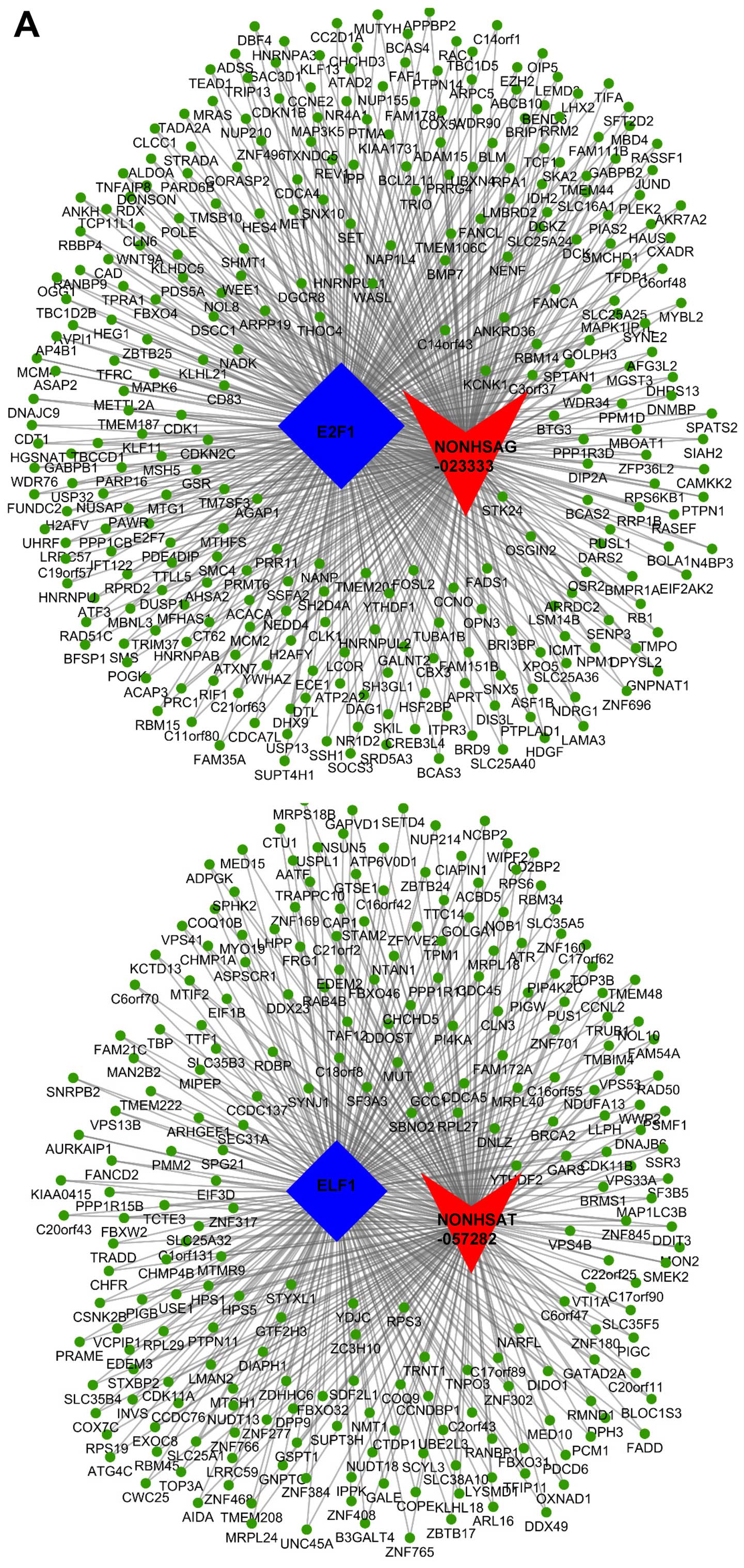
instance, NONHSAT057282 and NONHSAG023333 were significantly
correlated with ELF1 and E2F1, and enriched the most mRNAs of all
lncRNA-TF interactions. NONHSAT057282-ELF1 co-regulated 241 genes
and NONHSAG023333-E2F1 co-regulated 308 genes. We then drew a
ternary relation graph of the two interactions with Cytoscape 3.01
software (Agilent) (Fig. 4A).
Furthermore, NONHSAT05728 was upregulated in chemoresistant
MCF-7/ADM and MCF-7/PTX cells vs. chemosensitive MCF-7/WT cells,
while NONHSAG023333 was upregulated in MCF-7/WT vs. MCF-7/ADM and
MCF-7/PTX cells (Fig. 4B),
suggesting that NONHSAT05728 may enhance chemoresistance, but
NONHSAG023333 may negatively regulate chemoresistance. Indeed, when
we knocked down NONHSAT05728, chemoresistance decreased in both
MCF-7/ADM and MCF-7/PTX cells. On the other hand, knockdown of
NONHSAG023333 increased the chemoresistance in MCF-7/WT cells
(Fig. 4C). Therefore, these
results suggest that both NONHSAT05728 and NONHSAG023333 are
involved in chemoresistance, and their activity may be facilitated
by TFs.
The top 15 TFs with the highest enrichment of
lncRNAs are summarized in Fig. 4D,
indicating the potential involvement of certain TFs in regulating
chemoresistance via lncRNAs. In order to visualize these most
significantly-related lncRNA-TF interactions, the top 100 with the
lowest Q-values were then used to draw a two-element relation graph
(Fig. 4E). ELF1 was still most
frequently associated with several lncRNAs, and PBX3 and ZEB1 were
also intensively associated with lncRNAs.
Discussion
In the present study, for the first time we assessed
the genome-wide lncRNA expression patterns in adriamycin resistant
breast cancer cells using microarrays and explored their possible
functions by analyzing their cis-regulated mRNAs, as well as
TF-regulated mRNAs.
We first identified dysregulated lncRNAs in
adriamycin-resistant MCF-7/ADM cells; these lncRNAs correlated with
a list of dysregulated mRNAs. Because most of the lncRNAs in
current databases have not yet been functionally annotated, we
predicted their functions based on their correlated mRNAs.
Chemoresistance is an important feature of cancer progression, so
the lncRNAs dysregulated in chemoresistant MCF-7/ADM cells also
showed functions associated with hallmarks of cancer progression
(31). For instance, proteoglycans
in cancer (KEGG 05205) are responsible for increased cancerous
angiogenesis and provide a favorable microenvironment for cancer
cells (32); and toll-like
receptor signaling pathways (GO:0002224 and 0002756) provide cancer
cells with sustained proliferative signals (33). Furthermore, the key feature of
chemoresistant cancer cells is insensitivity to the cytotoxicity of
chemotherapeutic agents. Such insensitivity could be achieved by a
low efficacy of cellular drug transport, which may be associated
with actin filament bundle assembly (GO:0051017) (34) and endocytosis (KEGG 04144)
(35). Importantly, apoptosis
inactivation and cell proliferation enhancement, whose pathways
were frequently enriched in both the GO and KEGG pathways, not only
support cancer growth and metastasis but also contribute to the
insensitivity of cancer cells to chemotherapeutic agents.
Therefore, being the ‘mission critical’ of cancer progression and
chemoresistance regulated by various genetic and epigenetic
mechanisms, we suggest apoptosis and cell proliferation may be
still the main targets of regulation by lncRNAs.
Cis-action on target genes located at or near
the same locus is one of the main mechanism by which lncRNAs
regulate gene expression (36). We
therefore identified genes whose expression was correlated with
that of nearby lncRNAs. This analytical method greatly facilitated
the identification of lncRNAs critical for chemoresistance. Based
on the roles of lncRNAs in apoptosis and cell proliferation during
chemoresistance, we then explored the possible mechanism of action
of NONHSAT028712 in MCF-7/ADM and MCF-7/PTX cells because it may
cis-regulate CDK2. The preliminary results strongly suggest
an interaction between NONHSAT028712, CDK2, the cell cycle, and
chemoresistance, and further studies are needed to clarify how
NONHSAT028712 modulates expression of the nearby CDK2 gene; it may
directly interact with the gene, facilitate the 3D folding of
chromatin, or interact with other genetic (e.g. TFs) and epigenetic
(e.g. microRNAs) regulators of the CDK2 gene (36).
LncRNAs frequently physically interact with TFs to
regulate gene expression. We found that NONHSAT057282 and
NONHSAG023333 were involved in chemoresistance. Then the lncRNA-TF
interaction analysis suggests that these two lncRNAs may interact
with ELF1 and E2F1 respectively, and subsequently modulated a group
of chemoresistance-related genes such as GSTP1 (37,38),
BTG3 (39), SOCS3 (40) and BRAC2 (41). Furthermore, among the identified
lncRNA-TF interactions, ELF1 showed the highest enrichment
frequency; 50 lncRNAs were significantly associated with this TF.
ELF1 belong to the ETS transcription factor family, which is
important for cancer progression (42) and breast cancer chemoresistance, as
we demonstrated previously (25).
ELF1 is associated with tumor angiogenesis (43), but its role in cancer
chemoresistance has not been identified. Our results, thus, suggest
ELF1 as a new participant in chemoresistance by potentially
interacting with different lncRNAs. In future studies, mass
spectrometry could be applied to confirm the lncRNA-TF interactions
(44), and chromatin
immunoprecipitation-based sequencing might be needed to verify the
ELF1-related target genes (6). The
TF ZEB1 also frequently interacted with lncRNAs. ZEB1 modulates the
epithelial-mesenchymal transition pathway, which is essential for
chemoresistance (45). Previous
studies have shown that the epigenetic regulation of ZEB1 by
microRNAs and DNA methylation effectively generates chemoresistance
(10,46), but few studies have explored the
ZEB1-lncRNA interaction in chemoresistance. Therefore, considering
the importance of ZEB1, it would be worthwhile to investigate the
molecular mechanism by which lncRNAs mediate chemoresistance via
ZEB1; here, we have provided several target lncRNAs that are likely
to interact with ZEB1. Furthermore, other significantly enriched
TFs, such as PBX3 (47) and E2F1
(48–50) are also involved in breast cancer
progression; their interactions with lncRNAs may be important
mechanisms by which they control gene expression and enhance
chemoresistance.
In summary, we provide an overview of lncRNA
regulation at the combined levels of lncRNA and gene expression in
breast cancer chemoresistance, and systematically identify novel
dysregulated targets in chemoresistant breast cancer cells.
Experimental validation of specific interactions between lncRNAs
and genes, and lncRNAs and TFs allowed us to identify the key
players in chemoresistance, and decipher the underlining molecular
mechanism of action of lncRNAs in cancer progression. Based on this
analytical approach, we have shown the relationship and possible
mechanism of action of several lncRNAs in the development of
chemoresistance, suggesting that the analysis is precise and
valuable for the support of future studies.
Acknowledgements
The present study was supported by the China
National Natural Science Foundation (81572940 and 91439131 to X.M.;
31550006 to D.X.H.), the Natural Science Foundation for
Distinguished Young Scholars of Jiangsu Province (BK20140004 to
X.M.), the National High Technology Research and Development
Program (863 Program) of China (SQ2015AA020948 to X.M.) and the
Fundamental Research Funds for the Central Universities
(JUSRP51311A and JUSRP51615B to X.M. and D.X.H.). We thank
Professor I.C. Bruce for critical reading of the manuscript.
References
|
1
|
Marques AC, Hughes J, Graham B, Kowalczyk
MS, Higgs DR and Ponting CP: Chromatin signatures at
transcriptional start sites separate two equally populated yet
distinct classes of intergenic long noncoding RNAs. Genome Biol.
14:R1312013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louro R, Smirnova AS and Verjovski-Almeida
S: Long intronic noncoding RNA transcription: Expression noise or
expression choice? Genomics. 93:291–298. 2009. View Article : Google Scholar
|
|
3
|
Lam MT, Cho H, Lesch HP, Gosselin D, Heinz
S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al:
Rev-Erbs repress macrophage gene expression by inhibiting
enhancer-directed transcription. Nature. 498:511–515. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li W, Notani D, Ma Q, Tanasa B, Nunez E,
Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al: Functional
roles of enhancer RNAs for oestrogen-dependent transcriptional
activation. Nature. 498:516–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vance KW and Ponting CP: Transcriptional
regulatory functions of nuclear long noncoding RNAs. Trends Genet.
30:348–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiu MT, Hu JW, Yin R and Xu L: Long
noncoding RNA: An emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He DX, Gu F, Gao F, Hao JJ, Gong D, Gu XT,
Mao AQ, Jin J, Fu L and Ma X: Genome-wide profiles of methylation,
microRNAs, and gene expression in chemoresistant breast cancer. Sci
Rep. 6:247062016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Adam L, Zhong M, Choi W, Qi W, Nicoloso M,
Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al:
miR-200 expression regulates epithelial-to-mesenchymal transition
in bladder cancer cells and reverses resistance to epidermal growth
factor receptor therapy. Clin Cancer Res. 15:5060–5072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chan YC, Khanna S, Roy S and Sen CK:
miR-200b targets Ets-1 and is down-regulated by hypoxia to induce
angiogenic response of endothelial cells. J Biol Chem.
286:2047–2056. 2011. View Article : Google Scholar :
|
|
12
|
Feng X, Wang Z, Fillmore R and Xi Y:
MiR-200, a new star miRNA in human cancer. Cancer Lett.
344:166–173. 2014. View Article : Google Scholar :
|
|
13
|
Neves R, Scheel C, Weinhold S, Honisch E,
Iwaniuk KM, Trompeter HI, Niederacher D, Wernet P, Santourlidis S
and Uhrberg M: Role of DNA methylation in miR-200c/141 cluster
silencing in invasive breast cancer cells. BMC Res Notes.
3:2192010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peter ME: Let-7 and miR-200 microRNAs:
guardians against pluripotency and cancer progression. Cell Cycle.
8:843–852. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma X, Cai Y, He D, Zou C, Zhang P, Lo CY,
Xu Z, Chan FL, Yu S, Chen Y, et al: Transient receptor potential
channel TRPC5 is essential for P-glycoprotein induction in
drug-resistant cancer cells. Proc Natl Acad Sci USA.
109:16282–16287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Wang T, Li Y, Chen D, Yu Z, Jin L,
Ni L, Yang S, Mao X, Gui Y, et al: Identification of long-non
coding RNA UCA1 as an oncogene in renal cell carcinoma. Mol Med
Rep. 13:3326–3334. 2016.PubMed/NCBI
|
|
19
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar
|
|
21
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M, et al: Characterization of HULC, a novel gene with
striking up-regulation in hepatocellular carcinoma, as noncoding
RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Braconi C, Kogure T, Valeri N, Huang N,
Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM and Patel T:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding RNA involved in cancer, neurobiology, and
development. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He D, Gu X, Jiang L, Jin J and Ma X: A
methylation-based regulatory network for microRNA 320a in
chemoresistant breast cancer. Mol Pharmacol. 86:536–547. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He DX, Gu XT, Li YR, Jiang L, Jin J and Ma
X: Methylation-regulated miRNA-149 modulates chemoresistance by
targeting NDST1 in human breast cancer. FEBS J. 281:4718–4730.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marone M, Scambia G, Giannitelli C,
Ferrandina G, Masciullo V, Bellacosa A, Benedetti-Panici P and
Mancuso S: Analysis of cyclin E and CDK2 in ovarian cancer: Gene
amplification and RNA overexpression. Int J Cancer. 75:34–39. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Opyrchal M, Salisbury JL, Iankov I, Goetz
MP, McCubrey J, Gambino MW, Malatino L, Puccia G, Ingle JN, Galanis
E, et al: Inhibition of Cdk2 kinase activity selectively targets
the CD44+/CD24+/Low stem-like subpopulation
and restores chemosensitivity of SUM149PT triple-negative breast
cancer cells. Int J Oncol. 45:1193–1199. 2014.PubMed/NCBI
|
|
30
|
Gerstein MB, Kundaje A, Hariharan M, Landt
SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et
al: Architecture of the human regulatory network derived from
ENCODE data. Nature. 489:91–100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iozzo RV and Sanderson RD: Proteoglycans
in cancer biology, tumour microenvironment and angiogenesis. J Cell
Mol Med. 15:1013–1031. 2011. View Article : Google Scholar
|
|
33
|
Rakoff-Nahoum S and Medzhitov R: Toll-like
receptors and cancer. Nat Rev Cancer. 9:57–63. 2009. View Article : Google Scholar
|
|
34
|
Fu D and Roufogalis BD: Actin disruption
inhibits endosomal traffic of P-glycoprotein-EGFP and resistance to
daunorubicin accumulation. Am J Physiol Cell Physiol.
292:C1543–C1552. 2007. View Article : Google Scholar
|
|
35
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guil S and Esteller M: Cis-acting
noncoding RNAs: Friends and foes. Nat Struct Mol Biol.
19:1068–1075. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Townsend DM and Tew KD: The role of
glutathione-S-transferase in anti-cancer drug resistance. Oncogene.
22:7369–7375. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Traverso N, Ricciarelli R, Nitti M,
Marengo B, Furfaro AL, Pronzato MA, Marinari UM and Domenicotti C:
Role of glutathione in cancer progression and chemoresistance. Oxid
Med Cell Longev. 2013:9729132013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu J, Zhang Y, Qi Z, Kurtycz D, Vacano G
and Patterson D: Methylation-mediated downregulation of the B-cell
translocation gene 3 (BTG3) in breast cancer cells. Gene Expr.
14:173–182. 2008.PubMed/NCBI
|
|
40
|
Ru P, Steele R, Hsueh EC and Ray RB:
Anti-miR-203 upregulates SOCS3 expression in breast cancer cells
and enhances cisplatin chemosensitivity. Genes Cancer. 2:720–727.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang W and Figg WD: Secondary BRCA1 and
BRCA2 alterations and acquired chemoresistance. Cancer Biol Ther.
7:1004–1005. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Seth A and Watson DK: ETS transcription
factors and their emerging roles in human cancer. Eur J Cancer.
41:2462–2478. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang X, Brown C, Ni W, Maynard E, Rigby
AC and Oettgen P: Critical role for the Ets transcription factor
ELF-1 in the development of tumor angiogenesis. Blood.
107:3153–3160. 2006. View Article : Google Scholar
|
|
44
|
Li Z, Chao TC, Chang KY, Lin N, Patil VS,
Shimizu C, Head SR, Burns JC and Rana TM: The long noncoding RNA
THRIL regulates TNFα expression through its interaction with
hnRNPL. Proc Natl Acad Sci USA. 111:1002–1007. 2014. View Article : Google Scholar
|
|
45
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: new insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pieraccioli M, Imbastari F, Antonov A,
Melino G and Raschellà G: Activation of miR200 by c-Myb depends on
ZEB1 expression and miR200 promoter methylation. Cell Cycle.
12:2309–2320. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han HB, Gu J, Zuo HJ, Chen ZG, Zhao W, Li
M, Ji DB, Lu YY and Zhang ZQ: Let-7c functions as a metastasis
suppressor by targeting MMP11 and PBX3 in colorectal cancer. J
Pathol. 226:544–555. 2012. View Article : Google Scholar
|
|
48
|
Knoll S, Emmrich S and Pützer BM: The
E2F1-miRNA cancer progression network. Adv Exp Med Biol.
774:135–147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Frietze S, Lupien M, Silver PA and Brown
M: CARM1 regulates estrogen-stimulated breast cancer growth through
up-regulation of E2F1. Cancer Res. 68:301–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Louie MC, Zou JX, Rabinovich A and Chen
HW: ACTR/AIB1 functions as an E2F1 coactivator to promote breast
cancer cell proliferation and antiestrogen resistance. Mol Cell
Biol. 24:5157–5171. 2004. View Article : Google Scholar : PubMed/NCBI
|