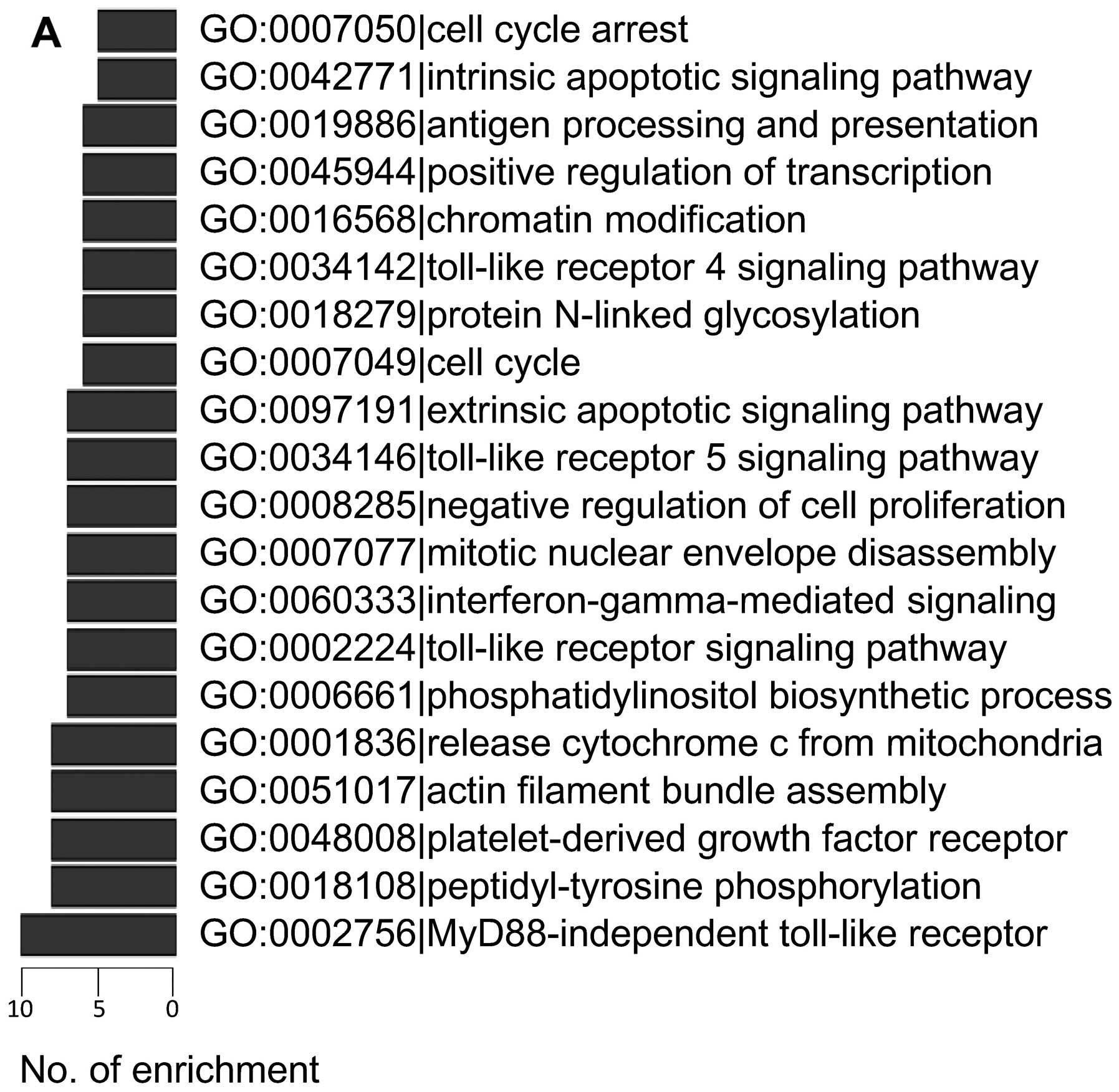
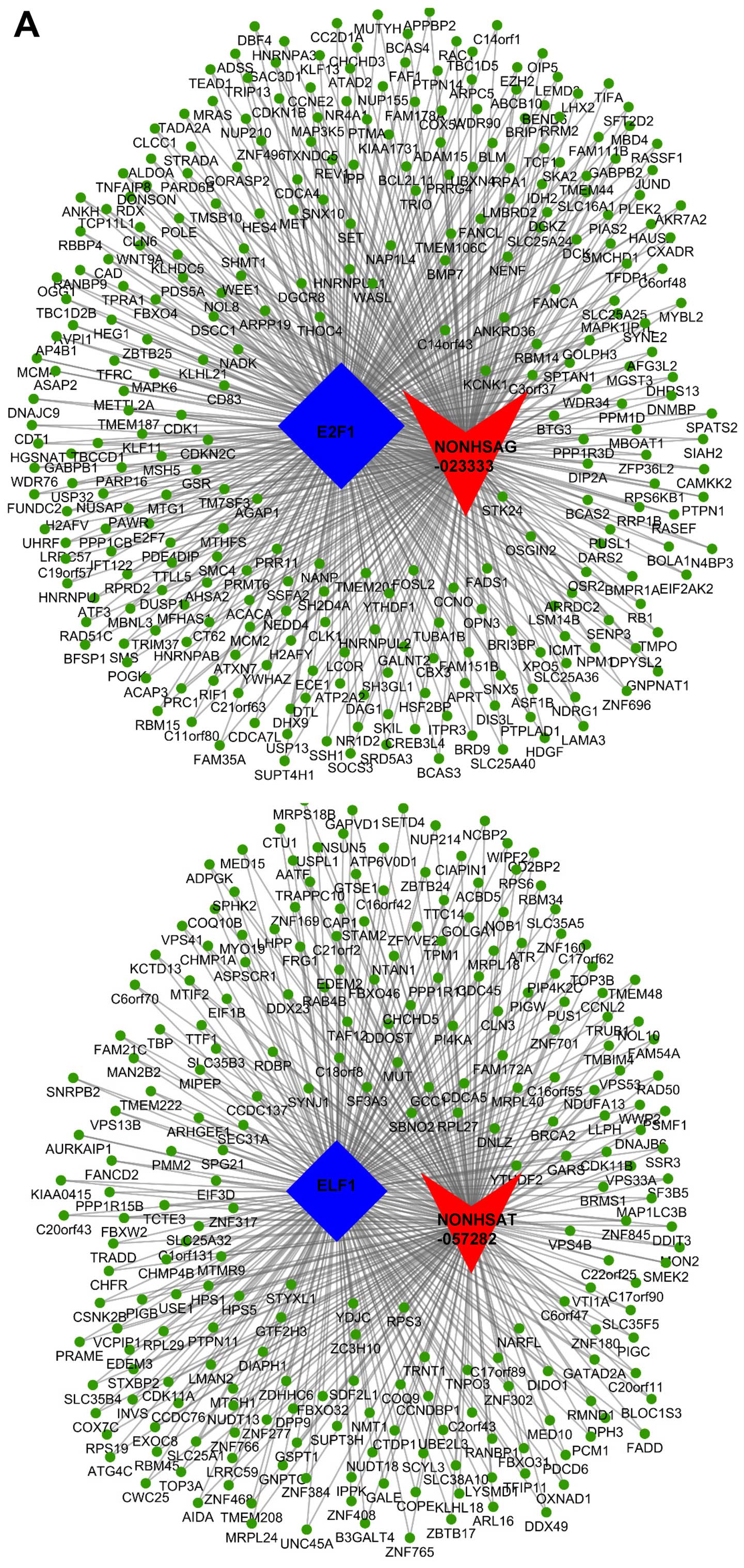
|
1
|
Marques AC, Hughes J, Graham B, Kowalczyk
MS, Higgs DR and Ponting CP: Chromatin signatures at
transcriptional start sites separate two equally populated yet
distinct classes of intergenic long noncoding RNAs. Genome Biol.
14:R1312013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louro R, Smirnova AS and Verjovski-Almeida
S: Long intronic noncoding RNA transcription: Expression noise or
expression choice? Genomics. 93:291–298. 2009. View Article : Google Scholar
|
|
3
|
Lam MT, Cho H, Lesch HP, Gosselin D, Heinz
S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al:
Rev-Erbs repress macrophage gene expression by inhibiting
enhancer-directed transcription. Nature. 498:511–515. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li W, Notani D, Ma Q, Tanasa B, Nunez E,
Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al: Functional
roles of enhancer RNAs for oestrogen-dependent transcriptional
activation. Nature. 498:516–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vance KW and Ponting CP: Transcriptional
regulatory functions of nuclear long noncoding RNAs. Trends Genet.
30:348–355. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiu MT, Hu JW, Yin R and Xu L: Long
noncoding RNA: An emerging paradigm of cancer research. Tumour
Biol. 34:613–620. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He DX, Gu F, Gao F, Hao JJ, Gong D, Gu XT,
Mao AQ, Jin J, Fu L and Ma X: Genome-wide profiles of methylation,
microRNAs, and gene expression in chemoresistant breast cancer. Sci
Rep. 6:247062016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Adam L, Zhong M, Choi W, Qi W, Nicoloso M,
Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al:
miR-200 expression regulates epithelial-to-mesenchymal transition
in bladder cancer cells and reverses resistance to epidermal growth
factor receptor therapy. Clin Cancer Res. 15:5060–5072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chan YC, Khanna S, Roy S and Sen CK:
miR-200b targets Ets-1 and is down-regulated by hypoxia to induce
angiogenic response of endothelial cells. J Biol Chem.
286:2047–2056. 2011. View Article : Google Scholar :
|
|
12
|
Feng X, Wang Z, Fillmore R and Xi Y:
MiR-200, a new star miRNA in human cancer. Cancer Lett.
344:166–173. 2014. View Article : Google Scholar :
|
|
13
|
Neves R, Scheel C, Weinhold S, Honisch E,
Iwaniuk KM, Trompeter HI, Niederacher D, Wernet P, Santourlidis S
and Uhrberg M: Role of DNA methylation in miR-200c/141 cluster
silencing in invasive breast cancer cells. BMC Res Notes.
3:2192010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peter ME: Let-7 and miR-200 microRNAs:
guardians against pluripotency and cancer progression. Cell Cycle.
8:843–852. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma X, Cai Y, He D, Zou C, Zhang P, Lo CY,
Xu Z, Chan FL, Yu S, Chen Y, et al: Transient receptor potential
channel TRPC5 is essential for P-glycoprotein induction in
drug-resistant cancer cells. Proc Natl Acad Sci USA.
109:16282–16287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Wang T, Li Y, Chen D, Yu Z, Jin L,
Ni L, Yang S, Mao X, Gui Y, et al: Identification of long-non
coding RNA UCA1 as an oncogene in renal cell carcinoma. Mol Med
Rep. 13:3326–3334. 2016.PubMed/NCBI
|
|
19
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar
|
|
21
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M, et al: Characterization of HULC, a novel gene with
striking up-regulation in hepatocellular carcinoma, as noncoding
RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Braconi C, Kogure T, Valeri N, Huang N,
Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM and Patel T:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding RNA involved in cancer, neurobiology, and
development. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He D, Gu X, Jiang L, Jin J and Ma X: A
methylation-based regulatory network for microRNA 320a in
chemoresistant breast cancer. Mol Pharmacol. 86:536–547. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He DX, Gu XT, Li YR, Jiang L, Jin J and Ma
X: Methylation-regulated miRNA-149 modulates chemoresistance by
targeting NDST1 in human breast cancer. FEBS J. 281:4718–4730.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marone M, Scambia G, Giannitelli C,
Ferrandina G, Masciullo V, Bellacosa A, Benedetti-Panici P and
Mancuso S: Analysis of cyclin E and CDK2 in ovarian cancer: Gene
amplification and RNA overexpression. Int J Cancer. 75:34–39. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Opyrchal M, Salisbury JL, Iankov I, Goetz
MP, McCubrey J, Gambino MW, Malatino L, Puccia G, Ingle JN, Galanis
E, et al: Inhibition of Cdk2 kinase activity selectively targets
the CD44+/CD24+/Low stem-like subpopulation
and restores chemosensitivity of SUM149PT triple-negative breast
cancer cells. Int J Oncol. 45:1193–1199. 2014.PubMed/NCBI
|
|
30
|
Gerstein MB, Kundaje A, Hariharan M, Landt
SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et
al: Architecture of the human regulatory network derived from
ENCODE data. Nature. 489:91–100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iozzo RV and Sanderson RD: Proteoglycans
in cancer biology, tumour microenvironment and angiogenesis. J Cell
Mol Med. 15:1013–1031. 2011. View Article : Google Scholar
|
|
33
|
Rakoff-Nahoum S and Medzhitov R: Toll-like
receptors and cancer. Nat Rev Cancer. 9:57–63. 2009. View Article : Google Scholar
|
|
34
|
Fu D and Roufogalis BD: Actin disruption
inhibits endosomal traffic of P-glycoprotein-EGFP and resistance to
daunorubicin accumulation. Am J Physiol Cell Physiol.
292:C1543–C1552. 2007. View Article : Google Scholar
|
|
35
|
Gottesman MM: Mechanisms of cancer drug
resistance. Annu Rev Med. 53:615–627. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guil S and Esteller M: Cis-acting
noncoding RNAs: Friends and foes. Nat Struct Mol Biol.
19:1068–1075. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Townsend DM and Tew KD: The role of
glutathione-S-transferase in anti-cancer drug resistance. Oncogene.
22:7369–7375. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Traverso N, Ricciarelli R, Nitti M,
Marengo B, Furfaro AL, Pronzato MA, Marinari UM and Domenicotti C:
Role of glutathione in cancer progression and chemoresistance. Oxid
Med Cell Longev. 2013:9729132013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yu J, Zhang Y, Qi Z, Kurtycz D, Vacano G
and Patterson D: Methylation-mediated downregulation of the B-cell
translocation gene 3 (BTG3) in breast cancer cells. Gene Expr.
14:173–182. 2008.PubMed/NCBI
|
|
40
|
Ru P, Steele R, Hsueh EC and Ray RB:
Anti-miR-203 upregulates SOCS3 expression in breast cancer cells
and enhances cisplatin chemosensitivity. Genes Cancer. 2:720–727.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang W and Figg WD: Secondary BRCA1 and
BRCA2 alterations and acquired chemoresistance. Cancer Biol Ther.
7:1004–1005. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Seth A and Watson DK: ETS transcription
factors and their emerging roles in human cancer. Eur J Cancer.
41:2462–2478. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang X, Brown C, Ni W, Maynard E, Rigby
AC and Oettgen P: Critical role for the Ets transcription factor
ELF-1 in the development of tumor angiogenesis. Blood.
107:3153–3160. 2006. View Article : Google Scholar
|
|
44
|
Li Z, Chao TC, Chang KY, Lin N, Patil VS,
Shimizu C, Head SR, Burns JC and Rana TM: The long noncoding RNA
THRIL regulates TNFα expression through its interaction with
hnRNPL. Proc Natl Acad Sci USA. 111:1002–1007. 2014. View Article : Google Scholar
|
|
45
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: new insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pieraccioli M, Imbastari F, Antonov A,
Melino G and Raschellà G: Activation of miR200 by c-Myb depends on
ZEB1 expression and miR200 promoter methylation. Cell Cycle.
12:2309–2320. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han HB, Gu J, Zuo HJ, Chen ZG, Zhao W, Li
M, Ji DB, Lu YY and Zhang ZQ: Let-7c functions as a metastasis
suppressor by targeting MMP11 and PBX3 in colorectal cancer. J
Pathol. 226:544–555. 2012. View Article : Google Scholar
|
|
48
|
Knoll S, Emmrich S and Pützer BM: The
E2F1-miRNA cancer progression network. Adv Exp Med Biol.
774:135–147. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Frietze S, Lupien M, Silver PA and Brown
M: CARM1 regulates estrogen-stimulated breast cancer growth through
up-regulation of E2F1. Cancer Res. 68:301–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Louie MC, Zou JX, Rabinovich A and Chen
HW: ACTR/AIB1 functions as an E2F1 coactivator to promote breast
cancer cell proliferation and antiestrogen resistance. Mol Cell
Biol. 24:5157–5171. 2004. View Article : Google Scholar : PubMed/NCBI
|