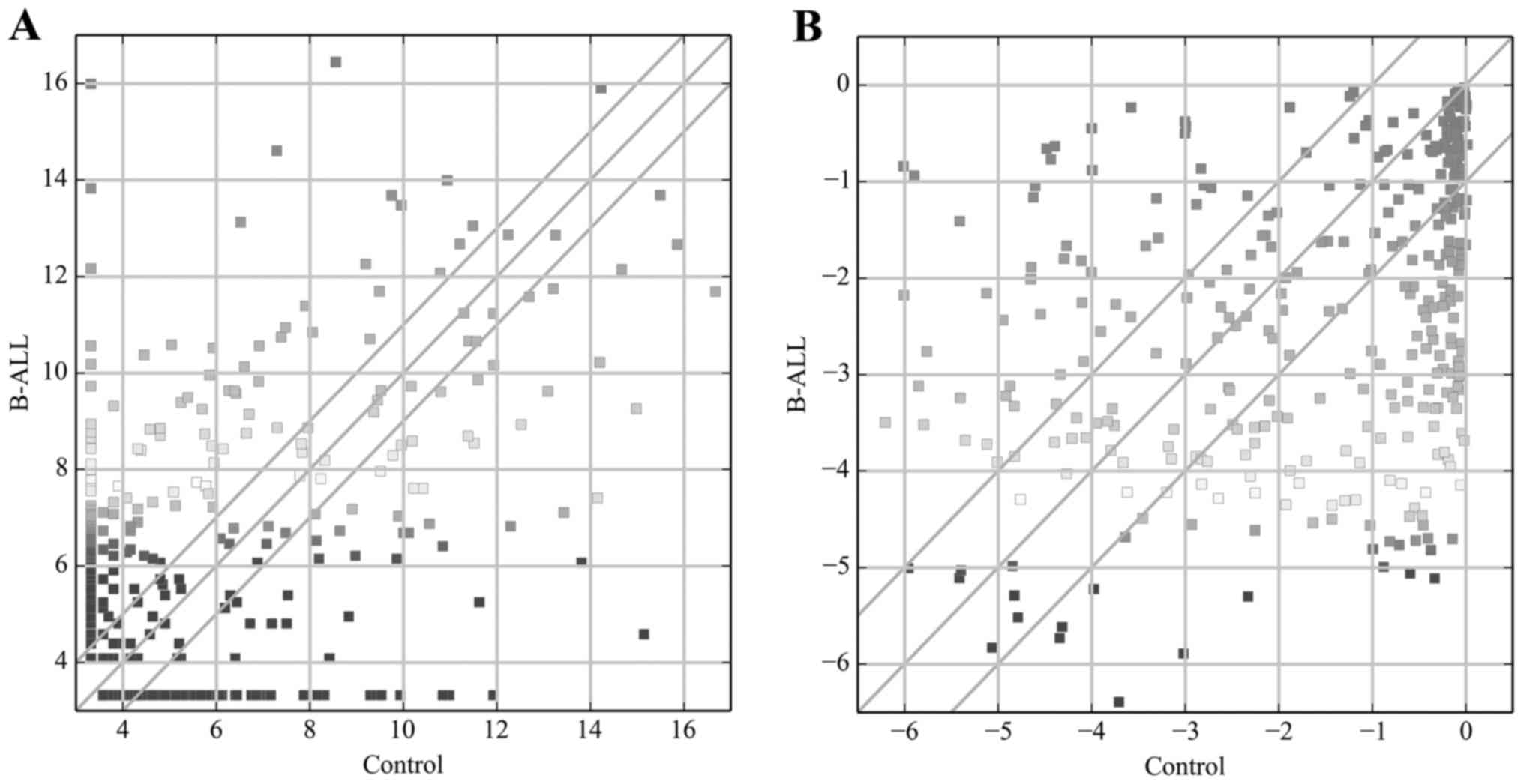
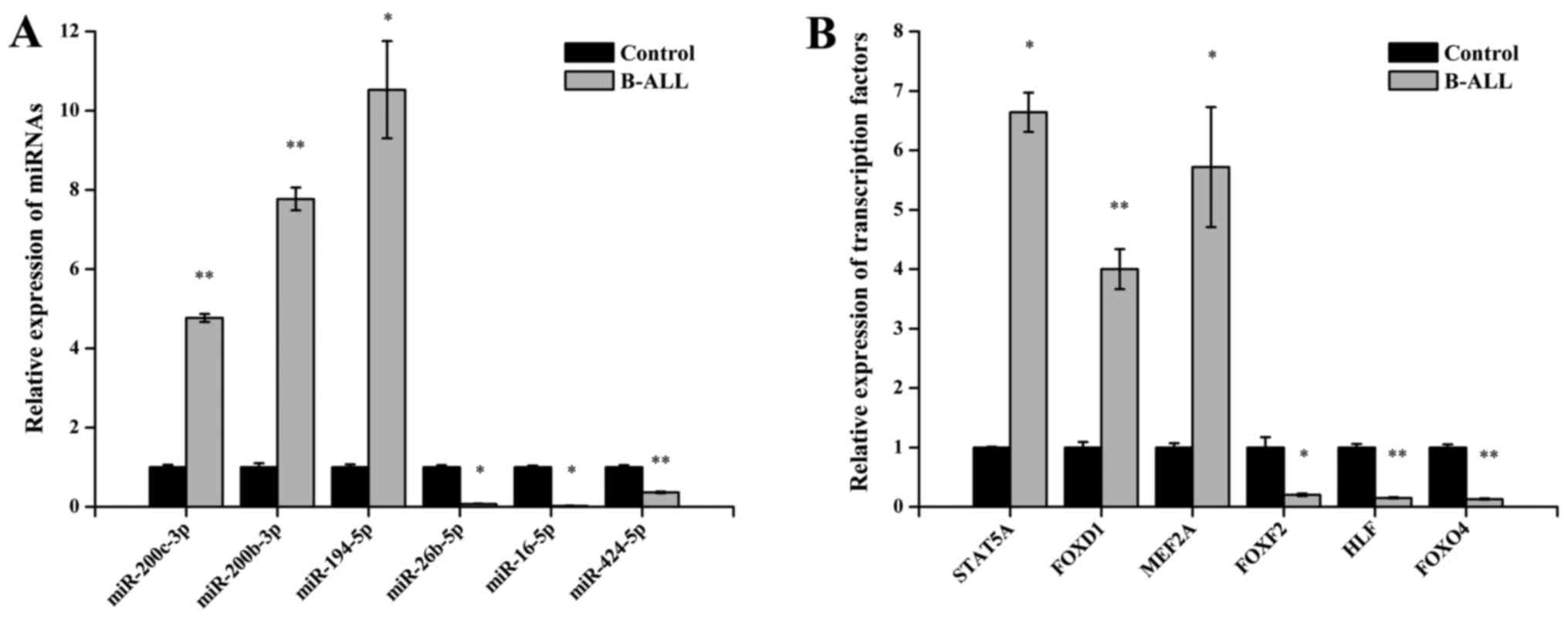
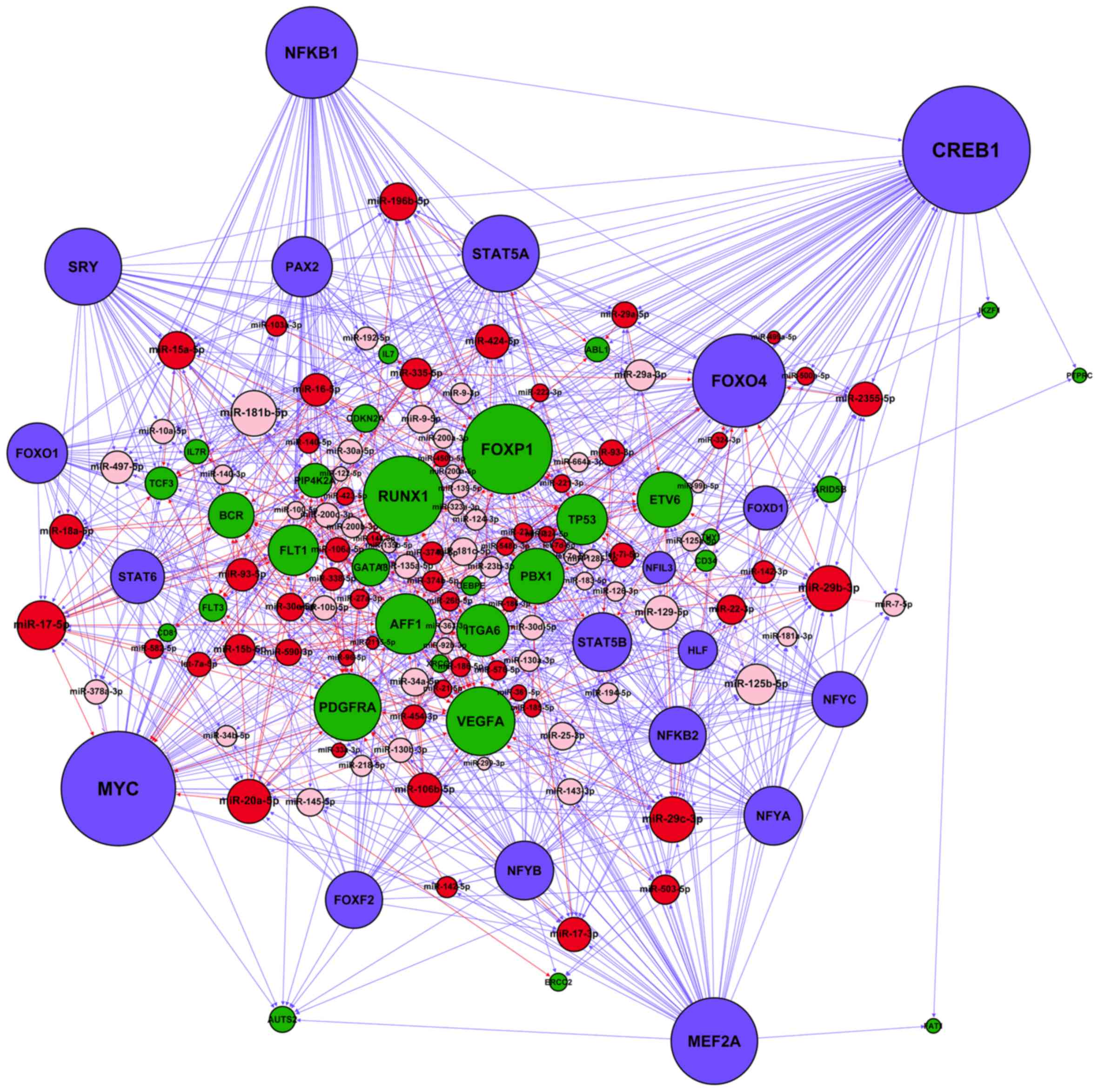
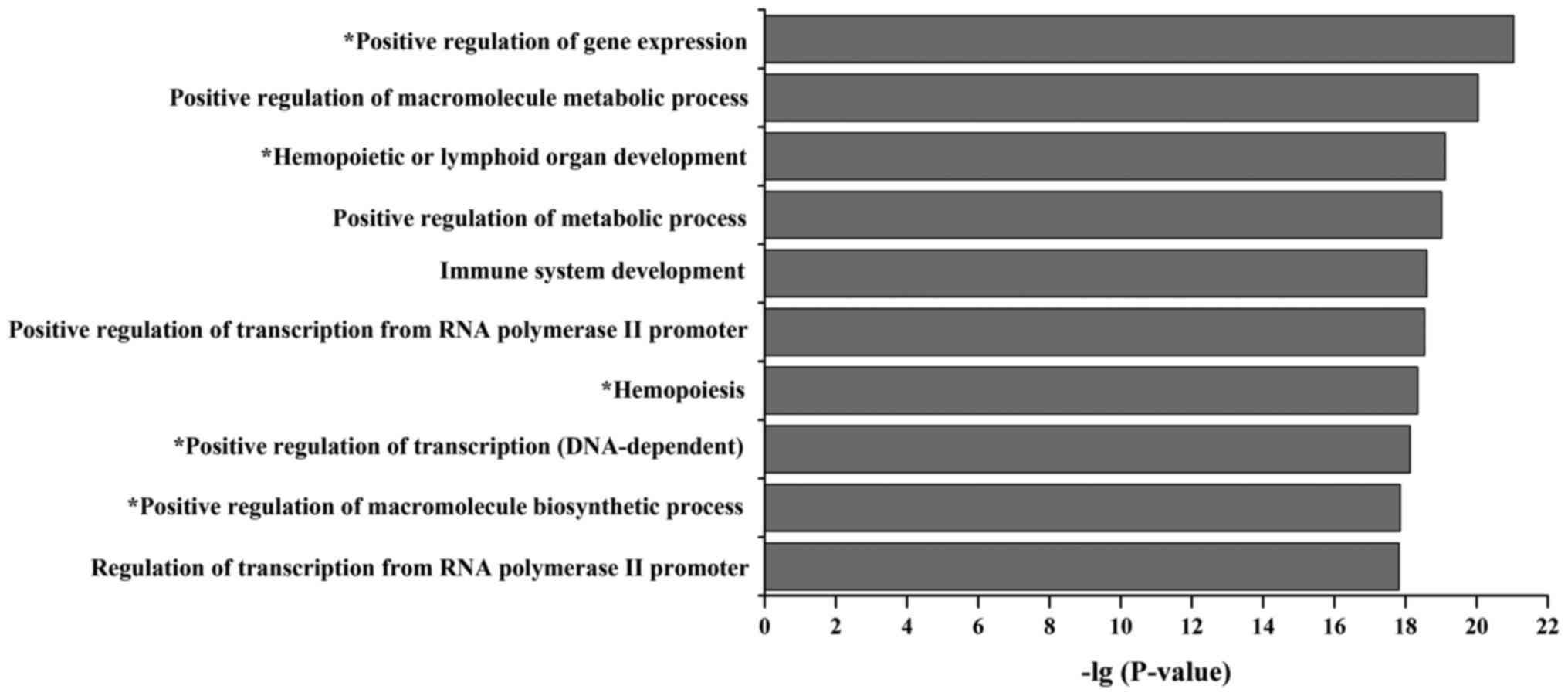
|
1
|
Purizaca J, Meza I and Pelayo R: Early
lymphoid development and microenvironmental cues in B-cell acute
lymphoblastic leukemia. Arch Med Res. 43:89–101. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanjuan-Pla A, Bueno C, Prieto C, Acha P,
Stam RW, Marschalek R and Menéndez P: Revisiting the biology of
infant t(4;11)/MLL-AF4+ B-cell acute lymphoblastic
leukemia. Blood. 126:2676–2685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Korfi K, Smith M, Swan J, Somervaille TC,
Dhomen N and Marais R: BIM mediates synergistic killing of B-cell
acute lymphoblastic leukemia cells by BCL-2 and MEK inhibitors.
Cell Death Dis. 7:e21772016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luna-Aguirre CM, de la Luz Martinez-Fierro
M, Mar-Aguilar F, Garza-Veloz I, Treviño-Alvarado V, Rojas-Martinez
A, Jaime-Perez JC, Malagon-Santiago GI, Gutierrez-Aguirre CH,
Gonzalez-Llano O, et al: Circulating microRNA expression profile in
B-cell acute lymphoblastic leukemia. Cancer Biomark. 15:299–310.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tasian SK and Gardner RA: CD19-redirected
chimeric antigen receptor-modified T cells: A promising
immunotherapy for children and adults with B-cell acute
lymphoblastic leukemia (ALL). Ther Adv Hematol. 6:228–241. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Woo JS, Alberti MO and Tirado CA:
Childhood B-acute lymphoblastic leukemia: A genetic update. Exp
Hematol Oncol. 3:162014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou Y, You MJ, Young KH, Lin P, Lu G,
Medeiros LJ and Bueso-Ramos CE: Advances in the molecular
pathobiology of B-lymphoblastic leukemia. Hum Pathol. 43:1347–1362.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pegram HJ, Smith EL, Rafiq S and Brentjens
RJ: CAR therapy for hematological cancers: Can success seen in the
treatment of B-cell acute lymphoblastic leukemia be applied to
other hematological malignancies? Immunotherapy. 7:545–561. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang XH, Wang CC, Jiang Q, Yang SM, Jiang
H, Lu J, Wang QM, Feng FE, Zhu XL, Zhao T, et al: ADAM28
overex-pression regulated via the PI3K/Akt pathway is associated
with relapse in de novo adult B-cell acute lymphoblastic leukemia.
Leuk Res. 39:1229–1238. 2015. View Article : Google Scholar
|
|
10
|
Sikaria S, Aldoss I and Akhtari M:
Monoclonal antibodies and immune therapies for adult precursor
B-acute lymphoblastic leukemia. Immunol Lett. 172:113–123. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mullighan CG: Molecular genetics of
B-precursor acute lymphoblastic leukemia. J Clin Invest.
122:3407–3415. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Othman MA, Grygalewicz B, Pienkowska-Grela
B, Rygier J, Ejduk A, Rincic M, Melo JB, Carreira IM, Meyer B and
Liehr T: A novel IGH@ gene rearrangement associated with
CDKN2A/B deletion in young adult B-cell acute lymphoblastic
leukemia. Oncol Lett. 11:2117–2122. 2016.PubMed/NCBI
|
|
13
|
Iacobucci I, Iraci N, Messina M, Lonetti
A, Chiaretti S, Valli E, Ferrari A, Papayannidis C, Paoloni F,
Vitale A, et al: IKAROS deletions dictate a unique gene expression
signature in patients with adult B-cell acute lymphoblastic
leukemia. PLoS One. 7:e409342012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moorman AV: The clinical relevance of
chromosomal and genomic abnormalities in B-cell precursor acute
lymphoblastic leukaemia. Blood Rev. 26:123–135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoda A, Yoda Y, Chiaretti S, Bar-Natan M,
Mani K, Rodig SJ, West N, Xiao Y, Brown JR, Mitsiades C, et al:
Functional screening identifies CRLF2 in precursor B-cell acute
lympho-blastic leukemia. Proc Natl Acad Sci USA. 107:252–257. 2010.
View Article : Google Scholar
|
|
16
|
Luan C, Yang Z and Chen B: The functional
role of microRNA in acute lymphoblastic leukemia: Relevance for
diagnosis, differential diagnosis, prognosis, and therapy. Onco
Targets Ther. 8:2903–2914. 2015.PubMed/NCBI
|
|
17
|
Cocco C and Airoldi I: Cytokines and
microRNA in pediatric B-acute lymphoblastic leukemia. Cytokine
Growth Factor Rev. 22:149–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bousquet M, Harris MH, Zhou B and Lodish
HF: MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA.
107:21558–21563. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Scherr M, Elder A, Battmer K, Barzan D,
Bomken S, Ricke-Hoch M, Schröder A, Venturini L, Blair HJ, Vormoor
J, et al: Differential expression of miR-17-92 identifies BCL2 as a
therapeutic target in BCR-ABL-positive B-lineage acute
lymphoblastic leukemia. Leukemia. 28:554–565. 2014. View Article : Google Scholar
|
|
20
|
Piriyapongsa J, Jordan IK, Conley AB,
Ronan T and Smalheiser NR: Transcription factor binding sites are
highly enriched within microRNA precursor sequences. Biol Direct.
6:612011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang HM, Kuang S, Xiong X, Gao T, Liu C
and Guo AY: Transcription factor and microRNA co-regulatory loops:
Important regulatory motifs in biological processes and diseases.
Brief Bioinform. 16:45–58. 2015. View Article : Google Scholar
|
|
22
|
Bhatia S, Kaul D and Varma N: Potential
tumor suppressive function of miR-196b in B-cell lineage acute
lymphoblastic leukemia. Mol Cell Biochem. 340:97–106. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ye H, Liu X, Lv M, Wu Y, Kuang S, Gong J,
Yuan P, Zhong Z, Li Q, Jia H, et al: MicroRNA and transcription
factor co-regulatory network analysis reveals miR-19 inhibits CYLD
in T-cell acute lymphoblastic leukemia. Nucleic Acids Res.
40:5201–5214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A, et al: The 2008 revision of the
World Health Organization (WHO) classification of myeloid neoplasms
and acute leukemia: Rationale and important changes. Blood. 114.
pp. 937–951. 2009, View Article : Google Scholar
|
|
25
|
Alvarnas JC, Brown PA, Aoun P, Ballen KK,
Bellam N, Blum W, Boyer MW, Carraway HE, Coccia PF, Coutre SE, et
al National Comprehensive Cancer Network: Acute lymphoblastic
leukemia. J Natl Compr Canc Netw. 10:858–914. 2012.PubMed/NCBI
|
|
26
|
Allen A, Gill K, Hoehn D, Sulis M, Bhagat
G and Alobeid B: C-myc protein expression in B-cell acute
lymphoblastic leukemia, prognostic significance? Leuk Res.
38:1061–1066. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sui W, Lin H, Peng W, Huang Y, Chen J,
Zhang Y and Dai Y: Molecular dysfunctions in acute rejection after
renal transplantation revealed by integrated analysis of
transcription factor, microRNA and long noncoding RNA. Genomics.
102:310–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dong M, Wang X, Zhao HL, Chen XL, Yuan JH,
Guo JY, Li KQ and Li G: Integrated analysis of transcription
factor, microRNA and LncRNA in an animal model of obliterative
bronchiolitis. Int J Clin Exp Pathol. 8:7050–7058. 2015.PubMed/NCBI
|
|
30
|
Lin Y, Wu J, Chen H, Mao Y, Liu Y, Mao Q,
Yang K, Zheng X and Xie L: Cyclin-dependent kinase 4 is a novel
target in micoRNA-195-mediated cell cycle arrest in bladder cancer
cells. FEBS Lett. 586:442–447. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rappaport N, Nativ N, Stelzer G, Twik M,
Guan-Golan Y, Stein TI, Bahir I, Belinky F, Morrey CP, Safran M, et
al: MalaCards: An integrated compendium for diseases and their
annotation. Database (Oxford). 2013:bat0182013. View Article : Google Scholar
|
|
32
|
Felice B, Cattoglio C, Cittaro D, Testa A,
Miccio A, Ferrari G, Luzi L, Recchia A and Mavilio F: Transcription
factor binding sites are genetic determinants of retroviral
integration in the human genome. PLoS One. 4:e45712009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bang SY, Kim JH, Lee PY, Chi SW, Cho S, Yi
GS, Myung PK, Park BC, Bae KH and Park SG: Candidate target genes
for the Saccharomyces cerevisiae transcription factor, Yap2. Folia
Microbiol (Praha). 58:403–408. 2013. View Article : Google Scholar
|
|
34
|
Lin XC, Xu Y, Sun GP, Wen JL, Li N, Zhang
YM, Yang ZG, Zhang HT and Dai Y: Molecular dysfunctions in acute
myeloid leukemia revealed by integrated analysis of microRNA and
transcription factor. Int J Oncol. 48:2367–2380. 2016.PubMed/NCBI
|
|
35
|
Alexiou P, Vergoulis T, Gleditzsch M,
Prekas G, Dalamagas T, Megraw M, Grosse I, Sellis T and
Hatzigeorgiou AG: miRGen 2.0: A database of microRNA genomic
information and regulation. Nucleic Acids Res. 38:D137–D141. 2010.
View Article : Google Scholar :
|
|
36
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al Gene Ontology Consortium: The Gene Ontology (GO) database and
informatics resource. Nucleic Acids Res. 32:D258–D261. 2004.
View Article : Google Scholar
|
|
37
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T,
et al: KEGG for linking genomes to life and the environment.
Nucleic Acids Res. 36:D480–D484. 2008. View Article : Google Scholar :
|
|
38
|
Arora S, Rana R, Chhabra A, Jaiswal A and
Rani V: miRNA-transcription factor interactions: A combinatorial
regulation of gene expression. Mol Genet Genomics. 288:77–87. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou Y, Kanagal-Shamanna R, Zuo Z, Tang G,
Medeiros LJ and Bueso-Ramos CE: Advances in B-lymphoblastic
leukemia: Cytogenetic and genomic lesions. Ann Diagn Pathol.
23:43–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Brondfield S, Umesh S, Corella A, Zuber J,
Rappaport AR, Gaillard C, Lowe SW, Goga A and Kogan SC: Direct and
indirect targeting of MYC to treat acute myeloid leukemia. Cancer
Chemother Pharmacol. 76:35–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Malin S, McManus S and Busslinger M: STAT5
in B cell development and leukemia. Curr Opin Immunol. 22:168–176.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Berger A, Sexl V, Valent P and Moriggl R:
Inhibition of STAT5: A therapeutic option in BCR-ABL1-driven
leukemia. Oncotarget. 5:9564–9576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chae HD, Mitton B, Lacayo NJ and Sakamoto
KM: Replication factor C3 is a CREB target gene that regulates cell
cycle progression through the modulation of chromatin loading of
PCNA. Leukemia. 29:1379–1389. 2015. View Article : Google Scholar
|
|
44
|
Köhrer S, Havranek O, Seyfried F, Hurtz C,
Coffey GP, Kim E, Ten Hacken E, Jäger U, Vanura K, O'Brien S, et
al: Pre-BCR signaling in precursor B-cell acute lymphoblastic
leukemia regulates PI3K/AKT, FOXO1 and MYC, and can be targeted by
SYK inhibition. Leukemia. 30:1246–1254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Coomans de Brachène A and Demoulin JB:
FOXO transcription factors in cancer development and therapy. Cell
Mol Life Sci. 73:1159–1172. 2016. View Article : Google Scholar
|
|
46
|
Grosjean-Raillard J, Adès L, Boehrer S,
Tailler M, Fabre C, Braun T, De Botton S, Israel A, Fenaux P and
Kroemer G: Flt3 receptor inhibition reduces constitutive NFkappaB
activation in high-risk myelodysplastic syndrome and acute myeloid
leukemia. Apoptosis. 13:1148–1161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vacca A, Felli MP, Palermo R, Di Mario G,
Calce A, Di Giovine M, Frati L, Gulino A and Screpanti I: Notch3
and pre-TCR interaction unveils distinct NF-kappaB pathways in
T-cell development and leukemia. EMBO J. 25:1000–1008. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bruns HA and Kaplan MH: The role of
constitutively active Stat6 in leukemia and lymphoma. Crit Rev
Oncol Hematol. 57:245–253. 2006. View Article : Google Scholar
|
|
49
|
LeBrun DP: E2A basic helix-loop-helix
transcription factors in human leukemia. Front Biosci. 8:s206–s222.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Puissegur MP, Eichner R, Quelen C, Coyaud
E, Mari B, Lebrigand K, Broccardo C, Nguyen-Khac F, Bousquet M and
Brousset P: B-cell regulator of immunoglobulin heavy-chain
transcription (Bright)/ARID3a is a direct target of the oncomir
microRNA-125b in progenitor B-cells. Leukemia. 26:2224–2232. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Strasser A, Harris AW, Bath ML and Cory S:
Novel primitive lymphoid tumours induced in transgenic mice by
cooperation between myc and bcl-2. Nature. 348:331–333. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ott CJ, Kopp N, Bird L, Paranal RM, Qi J,
Bowman T, Rodig SJ, Kung AL, Bradner JE and Weinstock DM: BET
bromodomain inhibition targets both c-Myc and IL7R in high-risk
acute lymphoblastic leukemia. Blood. 120:2843–2852. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Joliot V, Cormier F, Medyouf H, Alcalde H
and Ghysdael J: Constitutive STAT5 activation specifically
cooperates with the loss of p53 function in B-cell lymphomagenesis.
Oncogene. 25:4573–4584. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nakayama J, Yamamoto M, Hayashi K, Satoh
H, Bundo K, Kubo M, Goitsuka R, Farrar MA and Kitamura D: BLNK
suppresses pre-B-cell leukemogenesis through inhibition of JAK3.
Blood. 113:1483–1492. 2009. View Article : Google Scholar :
|
|
55
|
Cho JH, Gelinas R, Wang K, Etheridge A,
Piper MG, Batte K, Dakhallah D, Price J, Bornman D, Zhang S, et al:
Systems biology of interstitial lung diseases: Integration of mRNA
and microRNA expression changes. BMC Med Genomics. 4:82011.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li H: Systems genetics in '-omics' era:
Current and future development. Theory Biosci. 132:1–16. 2013.
View Article : Google Scholar
|
|
57
|
Lin Y, Zhang Q, Zhang HM, Liu W, Liu CJ,
Li Q and Guo AY: Transcription factor and miRNA co-regulatory
network reveals shared and specific regulators in the development
of B cell and T cell. Sci Rep. 5:152152015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jackstadt R and Hermeking H: MicroRNAs as
regulators and mediators of c-MYC function. Biochim Biophys Acta.
1849:544–553. 2015. View Article : Google Scholar
|
|
59
|
Pinz S, Unser S and Rascle A: Signal
transducer and activator of transcription STAT5 is recruited to
c-Myc super-enhancer. BMC Mol Biol. 17:102016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Han SS, Han S and Kamberos NL:
Piperlongumine inhibits the proliferation and survival of B-cell
acute lymphoblastic leukemia cell lines irrespective of
glucocorticoid resistance. Biochem Biophys Res Commun. 452:669–675.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Schepers H, Wierenga AT, Vellenga E and
Schuringa JJ: STAT5-mediated self-renewal of normal hematopoietic
and leukemic stem cells. JAK-STAT. 1:13–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tsuruyama T, Nakamura T, Jin G, Ozeki M,
Yamada Y and Hiai H: Constitutive activation of Stat5a by
retrovirus integration in early pre-B lymphomas of SL/Kh strain
mice. Proc Natl Acad Sci USA. 99:8253–8258. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Gao SM, Yang J, Chen C, Zhang S, Xing CY,
Li H, Wu J and Jiang L: miR-15a/16–1 enhances retinoic
acid-mediated differentiation of leukemic cells and is upregulated
by retinoic acid. Leuk Lymphoma. 52:2365–2371. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huang E, Liu R and Chu Y: miRNA-15a/16: As
tumor suppressors and more. Future Oncol. 11:2351–2363. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Gu TL, Nardone J, Wang Y, Loriaux M,
Villén J, Beausoleil S, Tucker M, Kornhauser J, Ren J, MacNeill J,
et al: Survey of activated FLT3 signaling in leukemia. PLoS One.
6:e191692011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Annesley CE and Brown P: The biology and
targeting of FLT3 in pediatric leukemia. Front Oncol. 4:2632014.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Small D: Targeting FLT3 for the treatment
of leukemia. Semin Hematol. 45(Suppl 2): S17–S21. 2008. View Article : Google Scholar : PubMed/NCBI
|