Introduction
Prostate cancer (PCa) has been reported globally as
a type of cancer exclusively diagnosed in males and is defined as
one of the most common malignant tumors in European and American
countries (1). PCa easily and
biochemically relapses along with the development of hormone
refractory cancer, and such condition remains to be a major cause
of poor prognosis (2).
MicroRNAs (miR), as a key regulator, have been
implicated as regulators controlling diverse biological processes
at the post-transcriptional level of repression (3,4).
Previously published studies explained that the excessive
expression of miR-146a influenced the PCa cells in terms of
apoptosis, progression, and viability by directly targeting the
epidermal growth factor receptor (EGFR) (5), Ras-related C3 botulinum toxin
substrate 1 (Rac1) (6), ROCK1
(7), interleukin-1
receptor-associated kinase 1 (IRAK1), and tumor necrosis factor
receptor-associated factor 6 (TRAF6) (8). Although miR-146a acts as a tumor
suppressor in PCa cells, current knowledge on the molecular
mechanisms that contribute to the expression of miR-146a in PCa
cells is still limited. The probable molecular mechanisms that
contribute to the aberrant expression of miRNAs result from gene
mutation, epigenetic modification, and alteration of Dicer
abundance. By means of promoter analysis, miR-146a was predicted to
be transcriptionally regulated by the vital transcriptional factor
YY1. YY1, a uniquely expressed zinc finger transcriptional factor,
played a double-trend regulatory role in a variety of biological
processes, such as embryonic development, cell growth, apoptosis,
differentiation, and oncogenic transformation (9–11).
In this study, we determined that the decreased PCa
cell viability, proliferation and increased apoptosis caused by YY1
depletion accompanied by upregulation of miR-146a. We further
validated the combination of YY1 and its interaction with EZH2 at
the miR-146a promoter prohibited the transcriptional activity of
miR-146a in PCa cells. Together with preceding results concluded by
our team (5–7,12),
our findings provided an in-depth understanding of the correlation
of YY1/EZH2 and miR-146a in PCa cell function disorders, thereby
potentially becoming a therapeutic target for PCa treatment.
Materials and methods
Cell culture
The cell lines were obtained from the Shanghai
Academy of Life Sciences, which is affiliated with the Chinese
Academy of Sciences. PC3 and DU145 human PCa cells were cultured
with DMEM/F12 (1:1) medium supplied with 10% fetal bovine serum and
antibiotics at 37°C in a 5% CO2 incubator.
Tissue samples
A total of 34 tissue samples were obtained between
2012 and 2015 from the radical prostate resection of 34 patients
with PCa admitted to the Zhongda Hospital affiliated with the
Southeast University. The patients were informed about the study
and were asked to sign consents for their tissues used in the
scientific research. Ethical approval for the current study was
previously obtained from the Zhongda Hospital. The diagnoses were
based on the pathological evidence, and the histological features
of the specimens were evaluated by the senior pathologist according
to the classification criteria of the World Health
Organization.
Animal experiments
BALB/c nude male mice (six-week-old) were purchased
from Shanghai SLAC Laboratory Animal Company. Animal experiments
were conducted in accordance with the protocol approved by the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals. PC3 cells were transfected with sh-GFP-YY1 or
sh-GFP-NC lentivirus to establish a stabilized cell line. A 100
µl cell culture mixed with 106 cells was
subcutaneously injected into each flank of the mice. Thereafter,
tumor size was measured every week. All mice were sacrificed on the
29th day. The tumors were removed fresh in preparation for miR-146a
detection.
ONCOMINE database reanalysis
The original YY1 mRNA expression data and the
prognostic data of YY1 in PCa specimens were retrieved from the
ONCOMINE database (www.oncomine.org) and GEO database (Nakagawa prostate,
GSE10645, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE10645.
Grasso prostate, GSE35988, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE35988.
Lapointe prostate, GSE3933, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3933.).
Re-analyses included the relationship among YY1 expression level,
biochemical recurrence within five years, and Gleason scores. These
processes were performed to determine the potential relationship
between PCa prognosis and YY1 expression levels.
RNA isolation and quantitative real-time
polymerase chain reaction (qRT-PCR)
Total RNA was extracted from tissues or cells (after
transfection for 72 h) using TRIzol (Takara, Shiga, Japan)
according to the manufacturer's instruction. RNA concentration was
measured by NanoDrop2000 (Thermo Fisher Scientific, Waltham, MA,
USA). For miR-146a, total RNA (10 ng) was reversed into cDNAs. The
qRT-PCR for miR-146a was performed using the SYBR Green PCR Master
Mix of the Hairpin-it miRNAs RT-PCR Quantitation kit (GenePharma,
Shanghai, China) according to the manufacturer's protocols. The
following PCR conditions were used for detecting miR-146a: 95°C for
3 min, 40 cycles at 95°C for 12 sec, and 62°C for 40 sec (running
in the ABI Step PCR System). The expression of U6 was used as
internal control. The relative expression of miR-146a compared with
U6 was calculated using the 2−ΔΔCt method.
RNA sequencing and bioinformatics
analysis
We depleted YY1 to establish a stabilized sh-YY1-PC3
cell line (targeting sequences are listed in Table I) to assess the role of YY1 in PCa
cells. Stable cell lines were established by puro-mycin. RNA
sequencing was assigned to and performed by Huada Genomics
Technology Co. (Shenzhen, China). RNA samples extracted and
assigned to RNA-sequencing using Illumina-HiSeq2000 system is 500
µg for sh-YY1 and sh-NC respectively. GSEA was used to
identify the pathway or gene sets that are correlated with the YY1
expression profile (http://www.broadinstitute.org/gsea/index.jsp). The
gene sets were acquired from the Molecular Signatures Database on
that website. The normalized enrichment score (NES) and false
discovery rate (FDR) were used to compare the results of the
analysis across gene sets.
 | Table ISequences of si-RNAs, sh-YY1, LNA for
miR-146a and primers of ChIP and Luciferase assay. |
Table I
Sequences of si-RNAs, sh-YY1, LNA for
miR-146a and primers of ChIP and Luciferase assay.
| Item | Sequence
(5′-3′) |
|---|
| si-YY1 |
CGAGGAUCAGAUUCUCAUC |
| si-NC |
UUCUCCGAACGUGUCACGU |
| si-EZH2 |
GAAUGGAAACAGCGAAGGATT |
| sh-YY1 |
CGAGGATCAGATTCTCATC |
| sh-NC |
TTCTCCGAACGTGTCACGT |
| anti-miR-146a |
AACCCAUGGAAUUCAGUUCUCA |
| LNA for ISH |
ACTCTTGACTTAAGGTACCCAA |
| CHIP primer F |
CCTTCAGGGGAGGTCAGGAAAGTGA |
| CHIP primer R |
CTTTGGTGAGGTTAGGGGGACTTGC |
| Luciferase primer
F |
CGAGGTACCTGGGAGCTCAGCAAGATAAGCAAC |
| Luciferase primer
R |
ACGAAGCTTCATCCAGGAAAGGAAGAGCGGTC |
YY1 overexpression and RNA silencing
Human YY1 over-expression pCMV6-YY1 vectors were
obtained from Sangon Biotech Co., Ltd., Shanghai, China. The human
siRNA-YY1 sequence was purchased from GenePharma. Table I lists the detailed sequences of
siRNAs and miR-146a. The transfection process was performed as
previously described (5).
MTT assay
The transfected cells were plated at 2,500
cells/well in 96-well plates. A total of 20 µl of 5 mg/ml
MTT in PBS was added to each well every 24 h after plating. The
cells were lysed after 2 h with the addition of 200 µl
dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO, USA). Absorbance
was measured at 570 nm.
Colony formation assay
After transfection, the cells were seeded in
six-well plates at a density of 800 cells/well and cultured for
10–14 days until colonies appeared. Thereafter, the cells were
stained with crystal violet. The number of colonies was counted
only when they contained more than 50 cells.
Cell apoptosis
Cell apoptosis was detected by a
fluorescence-activated cell sorter. After 72 h of transient
transfections, the cells were harvested by trypsinization, and cell
apoptosis was induced and measured as previously described
(7).
In situ hybridization (ISH) and
immunohistochemical staining (IHC)
ISH and IHC were performed, and subsequent results
were evaluated as previously described (13). ISH was performed using antisense
locked nucleic acid (LNA)-modified probe based on the sequence of
hsa-miR-146a (Boster, Wuhan, China). Table I lists the detailed LNA sequence.
IHC was performed using YY1 antibody (1:250; Proteintech, Chicago,
IL, USA) according to the manufacturer's instructions. Original
magnification, ×200. Evaluation of ISH and IHC was as follows:
Sections with unlabeled or labeled cells <5% were scored 0.
Sections with 5–30% of labeled cells were scored 1, those with
31–70% of labeled cells were scored 2, and those with ≥71% of
labeled cells were scored 3. The staining intensity was scored
similarly using 0 for negative staining, 1 for weakly positive, 2
for moderately positive, and 3 for strongly positive. The scores
for the percentage of positive tumor cells and for the staining
grade were calculated to generate an immune-reactive score for each
specimen. The product of the quantity and intensity scores was
calculated, such that a final score of 0–1 indicates negative
expression (−), 2–3 indicates weak expression (+), 4–5 indicates
moderate expression (++), and 6 indicates strong expression (+++).
Each sample was examined separately and scored by two
researchers.
Luciferase reporter assay
Promoter sequences (14) for miR-146a was reported previously,
and were retrieved from NCBI Genome Bioinformatics (http://www.ncbi.nlm.nih.gov/). Prediction of the
transcription factor binding sites was performed using the JASPAR
analysis tool (http://jaspar.genereg.net/). The promoter segment of
hsa-miR-146a was amplified by PCR (98°C for 2 min, 98°C for 15 sec,
68°C for 2 min with 35 cycles, and 68°C for 5 min) using human
genome DNA. The PCR products were cloned into the KpnI and
HindIII restriction sites (primer sequences are presented in
Table I), downstream of the open
reading frame of luciferase in the pGL3-basic-Vector (Promega,
Madison, WI, USA), to generate the pGL3-basic-miR-146a promoter
reporter. For reporter assays, cells were transfected with
pGL3-miR-146a promoter or mutant reporter plasmid and pRL-TK
plasmid. Luciferase assay activity was measured 48 h after
transfection using the Dual-Luciferase Reporter Assay System
(Promega).
Chromatin immunoprecipitation (ChIP)
Samples were obtained from PC3 cells (after
transfection with si-NC or si-YY1 for 72 h) and processed according
to the manufacturer's protocol (Millipore, Billerica, MA, USA).
Briefly, the chromatin solution was pre-cleared with 50 µl
of protein A-agarose beads (Beyotime, Shanghai, China). ChIP assays
were performed using YY1 antibody (1:50, Abcam, Cambridge, UK),
anti-polymerase II (Beyotime) and IgG (Beyotime), and allowed to
incubate overnight at 4°C. The precipitated samples were analyzed
by PCR (expected segment length, 253 bp), and the sequences of the
primers used for miR-146a promoter region are listed in Table I.
Immunofluorescence (IF)
PC3 cells were cultivated on cover-slips, fixed with
4% formaldehyde for 20 min, permeabilized for 10 min in 0.1% Triton
X-100 in PBS, and incubated for 2 h in blocking buffer (10% normal
goat serum and 0.3% Triton X-100 in PBS) at regular temperature.
Afterward, the cells were incubated with rabbit YY1 antibody
(1:250) and mouse EZH2 antibody (1:250) overnight at 4°C. The
coverslips were washed and incubated with AlexaFluor 488-conjugated
anti-mouse and AlexaFluor 594-conjugated anti-rabbit immunoglobulin
G (Invitrogen, Waltham, MA, USA) and mounted with the mounting
medium (ProLong Gold Antifade Reagent; Invitrogen). Fluorescent
images were acquired using the Olympus FV 1000 microscope.
Western blot analysis and
co-immunoprecipitation (Co-IP)
PC3/DU145 cells were cultured and plated in six-well
plates (5×105 cells/well). After transfection for 72 h
with siRNA-YY1, anti-miR-146a, p-CMV6-YY1, or negative control, the
cells were harvested and homogenized with sodium dodecyl sulfate
(SDS) lysis buffer. The protein concentration was measured by the
BCA method (Beyotime). Total protein was separated by denaturing
with 10% SDS-polyacrylamide gel electrophoresis. Total cell
extracts were pre-cleared with Protein A+G beads (Beyotime) at 4°C
for 2 h. The supernatant was incubated with the anti-EZH2 or
anti-YY1 (EZH2, 1:200; YY1, 1:200; Proteintech, Chicago, IL, USA)
with gentle shacking for 10 h at 4°C followed by addition of 30
µl of Protein A+G beads for another 2 h. The beads were
re-suspended in 100 µl of 2X loading buffer and boiled for
10 min. Then western blot assay was performed according to the
instructions previously described to detect the YY1 and EZH2. The
primary antibodies (EZH2, 1:1000; YY1, 1:1000; Proteintech) and
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1,000,
Proteintech) were purchased.
Statistical analysis
Each experiment was repeated at least thrice.
Numerical data were presented as the mean ± SEM. The difference
among the means was analyzed using Student's t-test. One-way
analysis of variance test was used to compare mean values between
more than two groups. All statistical analyses were performed using
SPSS 19.0 (Chicago, IL, USA). Differences were considered
significant at P<0.05.
Results
ONCOMINE database reanalysis and clinical
significance of YY1 in PCa specimens
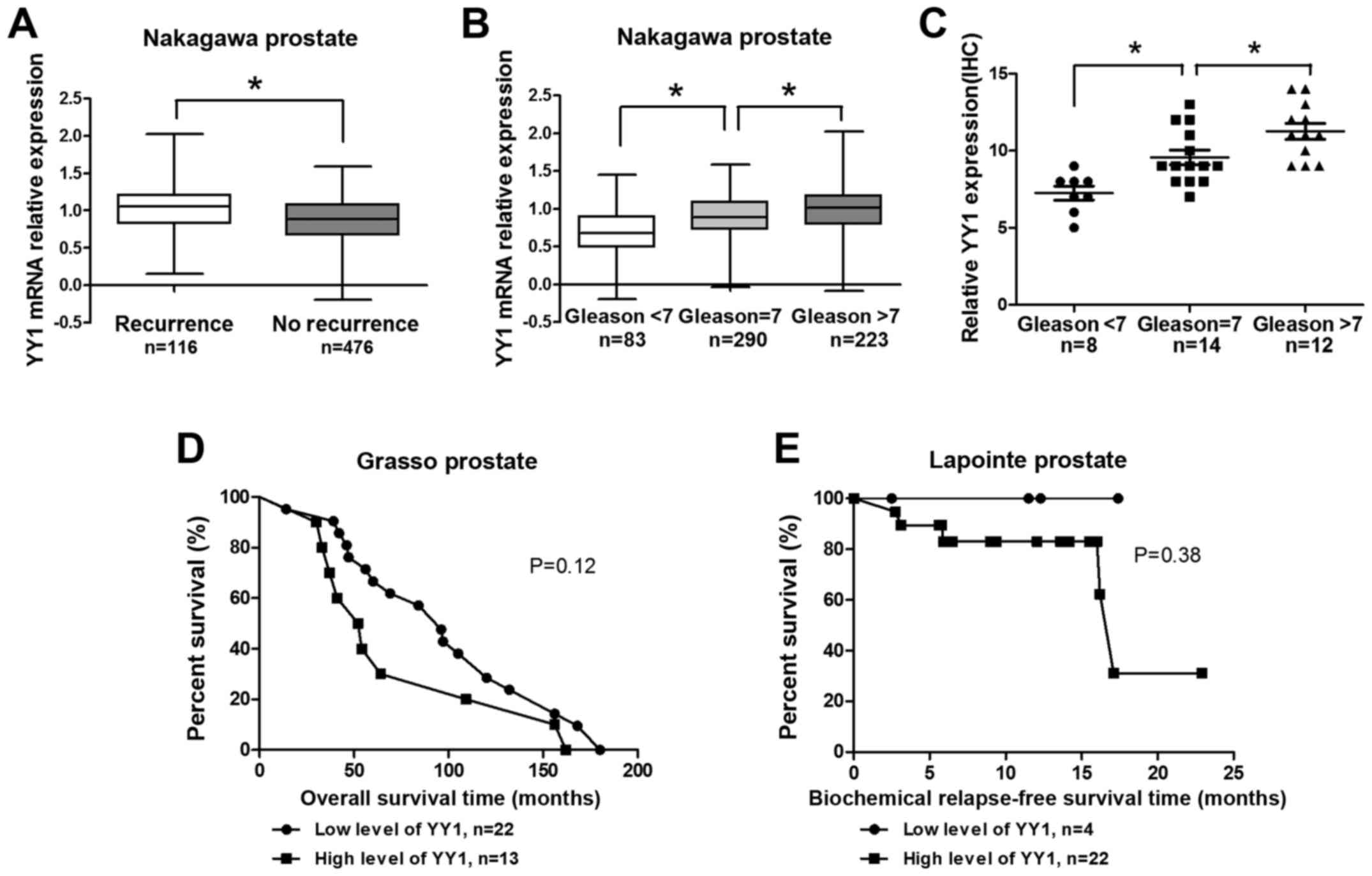
Reanalysis of the Nakagawa prostate indicated that
the YY1 expression level was high in the biochemical recurrence
group (Fig. 1A). In addition, the
YY1 expression levels increased gradually with increasing Gleason
scores (cutline=7) (Fig. 1B). In
the analysis of 34 clinical PCa specimens from Zhongda Hospital,
YY1 scores increased significantly with considerably high Gleason
scores (cutline=7) (Fig. 1C).
Results above were statistically significant. Reanalysis of
correlation of YY1 and overall survival (OS) time in Grasso
prostate project indicated that patients with high level of YY1
showed shorter OS time (P=0.12, Fig.
1D). Moreover, patients with high level of YY1 showed shorter
biochemical relapse-free survival (BRS) (P=0.38, Fig. 1E). The results (Fig. 1D–E) were not statistically
significant due to the small sample limit. However, these results
suggested YY1 might play a critical role in the PCa
progression.
Biological insight into the
co-relationship between YY1 and miR-146a in PCa
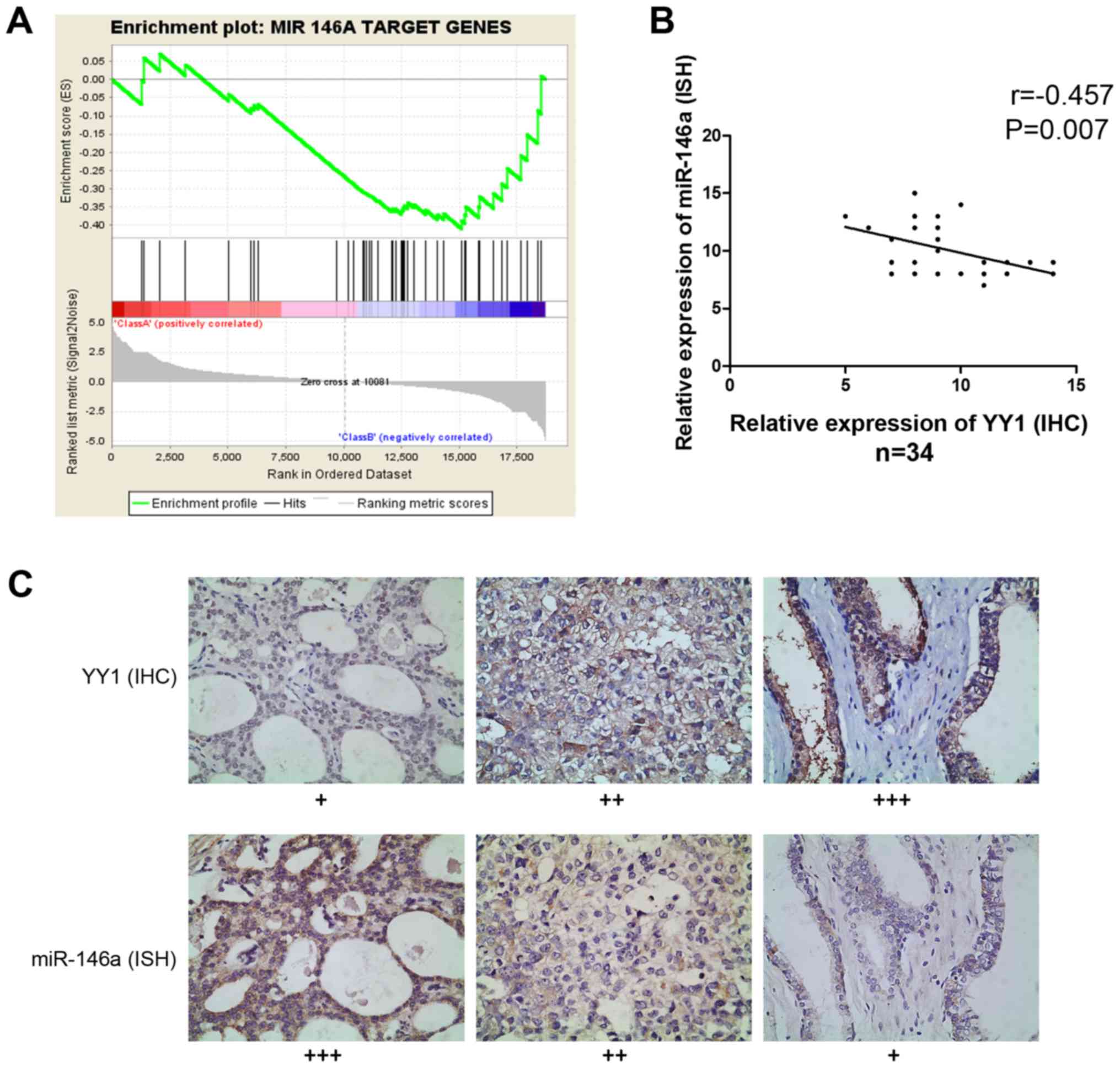
RNA sequencing and GSEA provided information on the
enriched expression of gene sets. When YY1 was knocked down in PC3
cells, the expression of miR-146a target genes demonstrated the
same trend in gene enrichment, indicating the potential
co-relationship between YY1 and miR-146a (NES= −1.039, FDR=0.267
and P=0.15) (Fig. 2A). Then, we
evaluated the miR-146a expression data through ISH and assessed the
YY1 expression data through IHC of 34 PCa specimens to investigate
their co-relationship. Spearman correlation analysis showed a
significantly inverse correlation between miR-146a and YY1
(r= −0.457, P=0.007) (Fig. 2B
and C). Hence YY1 may participate in the regulation of miR-146a
expression process.
Knockdown of YY1 inhibits PCa cell growth
and colony forming in a miR-146a-assisted manner
YY1 was downregulated with si-YY1 to explore the
effect of YY1 on PCa cell lines. In return, YY1 was upregulated
when transfected with pCMV6-YY1. The knockdown or overexpression
efficiency was verified through western blot analysis (Fig. 3A–C). The inhibitory efficiency for
miR-146a after YY1 depletion was verified through qRT-PCR (Fig. 3D and E). The MTT and colony
formation assays were used to detect the importance of miR-146a in
YY1-associated PCa cell growth and proliferation. YY1 depletion
significantly inhibited the growth of PCa cells at 48, 72, and 96 h
(P<0.05) based on the MTT assay. However, no significant
inhibition was observed in the cells transfected with
pCMV6/pCMV6-YY1 (P>0.05). Functional experiments were performed
on the YY1-depleted cells supplemented with anti-miR-146a (miR-146a
inhibitor) to confirm whether miR-146a was required for the
YY1-mediated decrease in cell viability and proliferation.
Significant results were observed when anti-miR-146a was
transfected after YY1 depletion at 48, 72, and 96 h (P<0.05)
(Fig. 3F–K). YY1 depletion
significantly inhibited the proliferation of PCa cells based on
colony formation assay and anti-miR-146a supplementary alleviated
the inhibition effect (Fig. 3L–N).
GSEA showed that a negatively enriched expression of gene sets was
involved in the cell proliferation of YY1 knocked down cells (NES=
−1.17, FDR=0.22 and P=0.11) (Fig.
3O). Collectively, these results indicated that YY1 depletion
inhibited cell viability and proliferation of PCa cells at least in
a miR-146a-assisted manner.
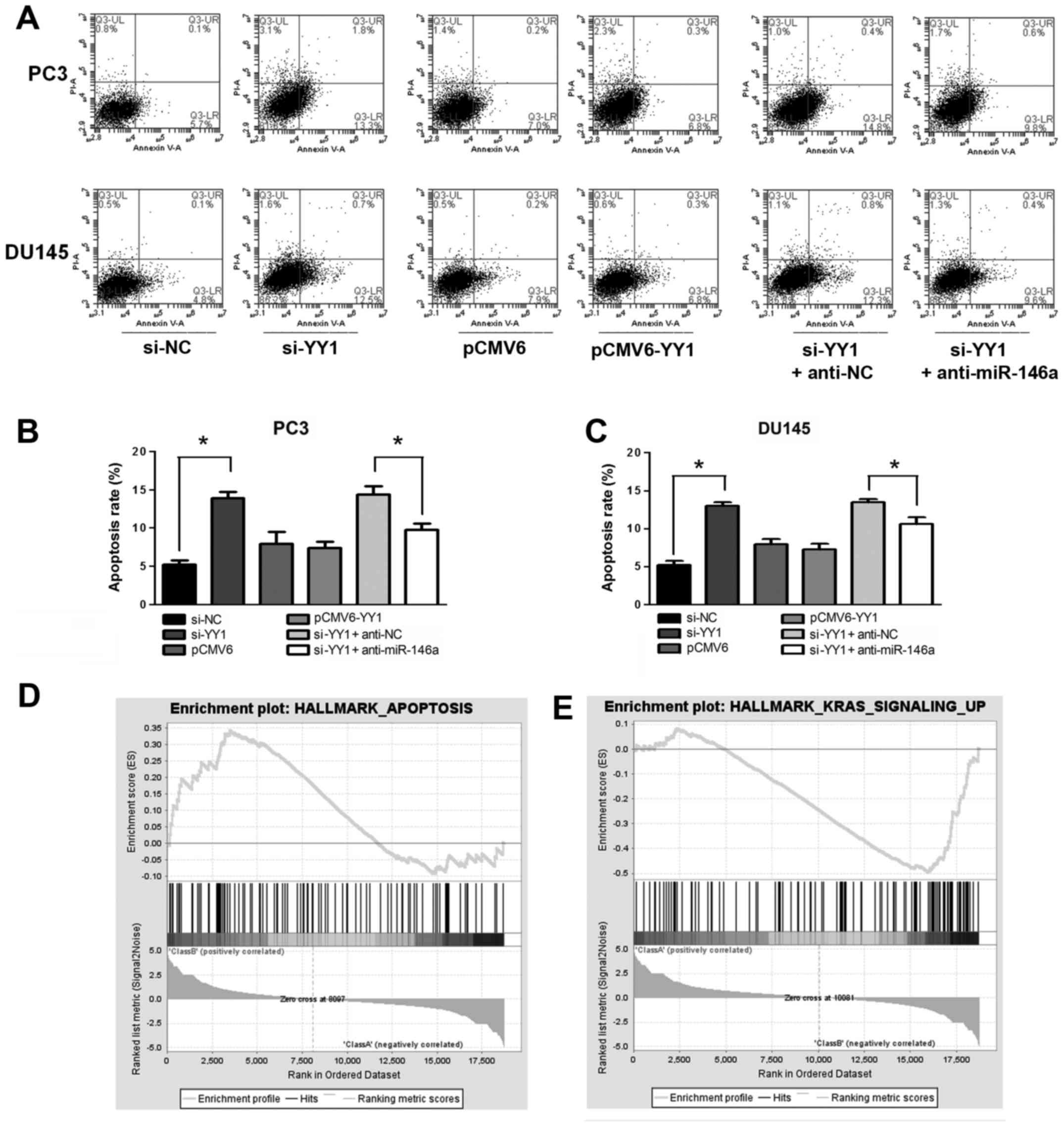
Knockdown of YY1 induces cell apoptosis
in miR-146a-assisted manner
YY1 was downregulated by si-YY1 and upregulated by
transfection with pCMV6-YY1 to assess the role of YY1 in PCa cell
apoptosis. Unobtrusive difference was observed when the cells were
treated with pCMV6-YY1/pCMV6 (P>0.05). YY1 depletion increased
the apoptosis rate of the PCa cells (apoptosis rate: 5.9±0.6% vs.
13.9±0.8% in PC3 and 5.2±0.5% vs. 12.9±0.5% in DU145, P<0.05),
whereas supplementing the cells with miR-146a inhibitor alleviated
the apoptosis effect (apoptosis rate: 14.4±1.0% vs. 9.7±0.8% in PC3
and 13.5±0.5% vs. 10.6±0.9% in DU145, P<0.05) (Fig. 4A–C). GSEA showed that a positively
enriched expression of gene sets was involved in hallmarks of
apoptosis (NES=1.30, FDR=0.24 and P=0.09) (Fig. 4D), and v-Ki-ras2 Kirsten rat
sarcoma viral oncogene homolog (KRAS) (NES= −1.97, FDR= 0.04 and P=
0.01) signal in YY1 knocked down cells (Fig. 4E). KRAS is the downstream molecule
of EFGR signaling pathway, which has been reported be the target of
miR-146a in PCa (5). The negative
correlation of KRAS and YY1 knockdown suggest the potential
regulation between YY1 and miR-146a. Moreover, miR-146a induced
cell apoptosis in PCa cells by activating cleaved caspase-3
(15). Collectively, these results
indicated that YY1 depletion induced cell apoptosis of PCa at least
in a miR-146a-assisted manner.
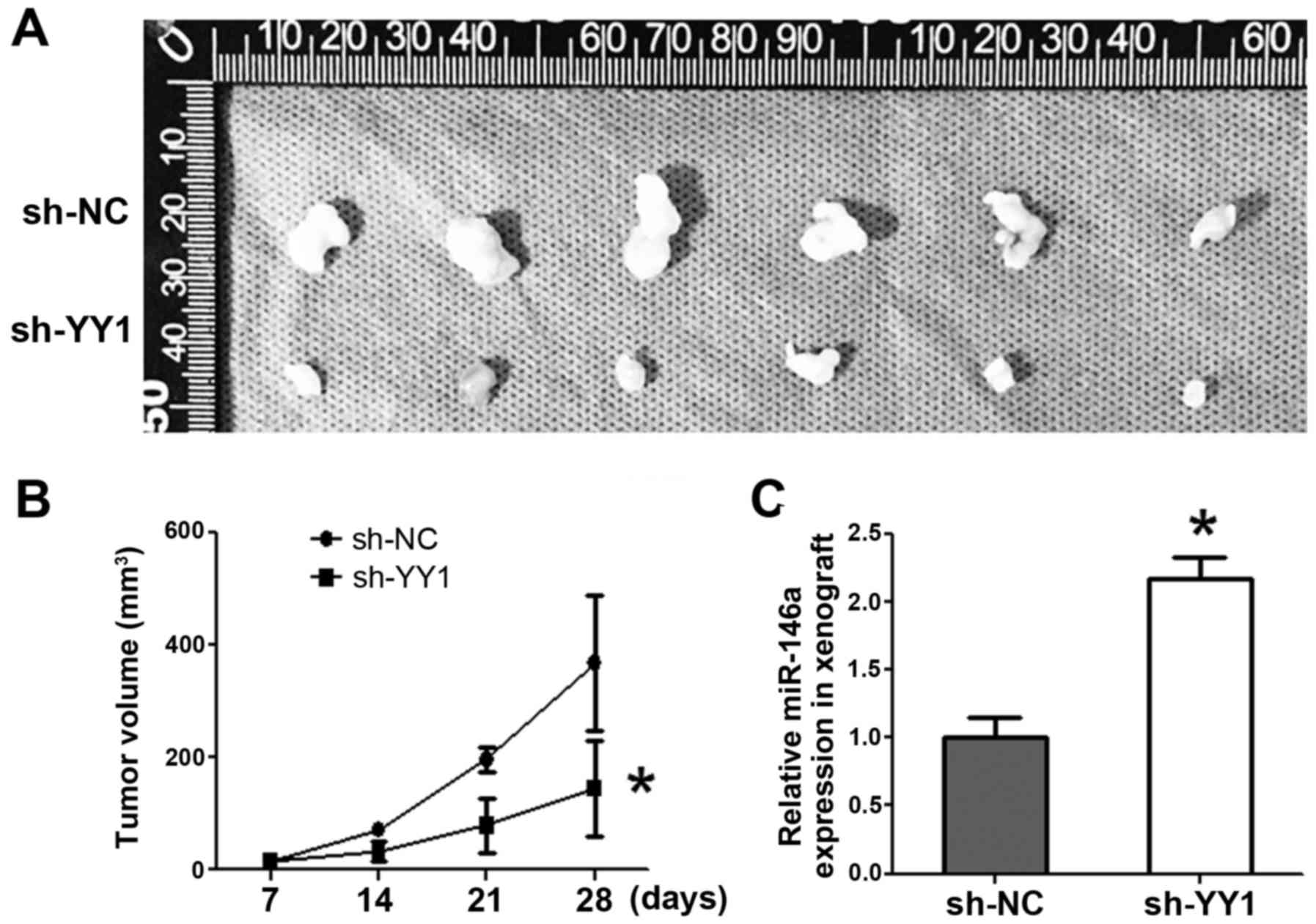
Knockdown of YY1 suppresses xenografts
growth accompanied by miR-146 upregulation
A stabilized sh-YY1-PC3 cell line was established,
and the PCa cells were injected subcutaneously to either flank of
the same nude mice. Tumor volumes were measured every week.
Consequently, sh-YY1 inhibited tumor formation by volume starting
from day 14 (P<0.05) (Fig. 5A and
B). qRT-PCR indicated the significant upregulation of miR-146a
expression in sh-YY1 xenografts (p<0.05) (Fig. 5C).
EZH2 was recruited by YY1 at the promoter
region of miR-146a, thereby repressing the miR-146a expression by
participating in the transcriptional repression
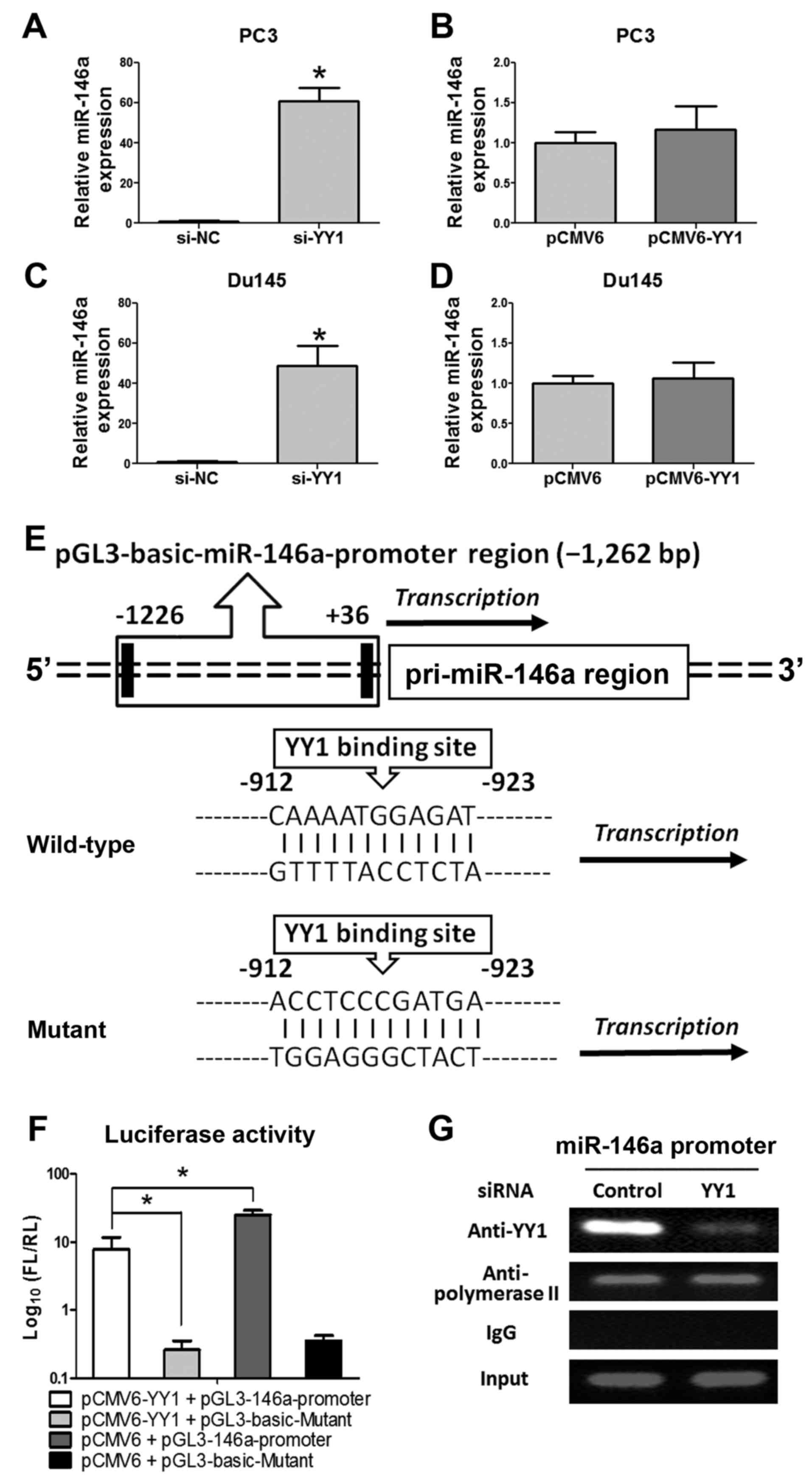
We measured the expression of miR-146a through
qRT-PCR to elucidate the regulatory functions of YY1 and miR-146a
further. Accordingly, miR-146a was significantly upregulated after
si-YY1 treatment, whereas no significant change was observed after
pCMV6-YY1 treatment (Fig. 6A–D).
To elucidate the regulatory mechanism of miR-146a transcription
process, the JARSPA database was consulted to investigate the
concise regulatory mechanism of YY1-miR-146a (−2,000 bp upstream of
miR-146a transcriptional start site, chr5:159895258, GRCH37).
(Fig. 6E) One binding site of YY1
for the pre-miR-146a promoter was predicted (profile score
threshold, 80%). A 1,262 bp fragment upstream of human pri-miR-146a
was constructed and inserted into the luciferase reporter plasmid
pGL3-basic-Vector. The plasmid was co-transfected with pCMV6 or
pCMV6-YY1 into PC-3 cells. Luciferase reporter assay (Fig. 6F) showed that overexpression of YY1
led to a significant decrease in luciferase activity of the
pGL3-miR-146a-promoter plasmid in PC-3 cells. In addition, the ChIP
assay (Fig. 6G) clearly showed YY1
combined at the miR-146a promoter region, thus further determined
that YY1 could directly bind to the miR-146a promoter region
(upstream), which indicated that YY1 could repress miR-146a
expression through transcriptional suppression.
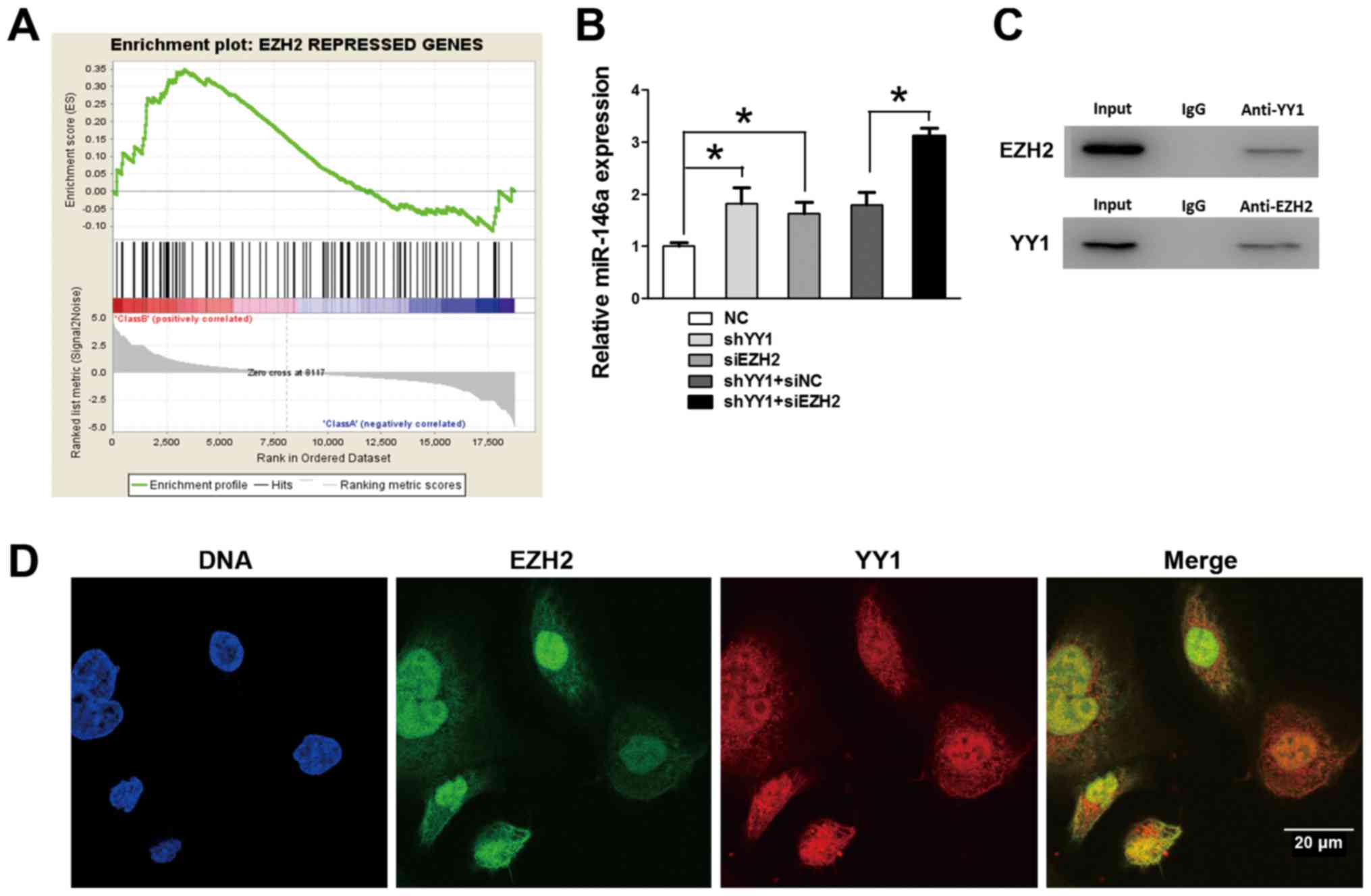
To further study the regulatory mechanism of
miR-146a transcription, we performed GSEA to search for other
co-repressors on the basis of YY1 depleted PC3 cells. Notably, GSEA
showed that the positively enriched expression of gene sets was
repressed by EZH2 in YY1 depleted PC3 cells (NES=1.34, FDR=0.02 and
P=0.02) (Fig. 7A), which suggested
a potential co-relationship between YY1 and EZH2 during the
transcriptional activity of PCa. EZH2 was reported to be positively
co-operating with the tumorigenic transcription factor (i.e., YY1)
(16). To further detect the
influence of EZH2 on miR-146a expression, qRT-PCR was performed and
demonstrated further upregulation of miR-146a after EZH2 knockdown
on the basis of YY1 depletion, suggesting that EZH2 participated in
the YY1-mediated regulatory process (Fig. 7B). IP assays (Fig. 7C) were performed using nuclear
protein from PC3 cells, and validated the directly combined effects
of EZH2 and YY1 in the nucleus. Moreover, the IF provided a clear
view of EZH2 and YY1 located within the cell structure. The general
view by IF (Fig. 7D) of the YY1
and EZH2 conjunction inside the nucleus and cytoplasm was observed
indicated that YY1 and EZH2 might jointly participate in some
biological process. Together with the preceding ChIP and Luciferase
results, we verified that EZH2 was recruited by YY1 at the promoter
region of miR-146a and jointly repressed the miR-146a expression by
participating in the transcriptional repression.
Discussion
miR-146a (miR-146a-5P) is a well-known
anti-carcinoma cell signal modulator in prostate cancer (PCa)
tissues. Our research group have investigated the mechanism of
miR-146a in inhibiting PCa cells (5,7,12).
However, in this study, we focused on whether miR-146a expression
can be regulated by pivotal transcriptional factors. By means of
promoter analysis, YY1 was selected as the transcription factor
with the most potential. We determined that YY1 was upregulated in
PCa tissues with biochemical recurrence, high Gleason score, and
low miR-146a expression. YY1 depletion inhibited cell viability and
proliferation and induced cell apoptosis of PCa cells along with
the upregulation of miR-146a. Through GSEA in YY1 knocked down PC3
cells, we observed the negatively enriched expression of gene sets,
which were in conformity with the miR-146a function of PCa in cell
proliferation (6), cell apoptosis
(7), and KRAS signal (5) as previously reported.
As a transcription factor for miR-146a, YY1 was
first reported by Seligson et al (17) to be overexpressed in PCa cells
compared with normal tissues. A certain level of YY1 inside the
cell membrane is essential in maintaining basic biological
functions, and YY1 also contributes to carcinogenesis (18). YY1 influences gene expression by
directly or indirectly activating or inhibiting gene transcription
(19) and activates a variety of
oncogene factors by interacting with the transforming growth factor
β-3 (20) and v-myc avian
myelocytomatosis viral oncogene homolog (c-Myc) (21).
Notably, in this study, GSEA-represented gene sets
suppressed by EZH2 regulation were positively enriched after YY1
depletion, indicating the potential relationship between essential
transcriptional factors (i.e., EZH2 and YY1) and PCa cells. EZH2, a
crucial component of polycomb repressor complex 2 (PRC2), is
reported to be overexpressed in PCa cells (22). EZH2 represses gene expression by
catalyzing the tri-methylation of histone H3 lysine 27 (H3K27me3)
which regulates gene transcription epigenetically (23). The high expression level of EZH2 in
clinically localized PCa tissues incur poor prognosis (24). Accumulating evidence indicates that
PRC2 exerts strong oncogenic activity. These findings are proven by
investigations on cell proliferation (25), cell invasion, maintenance of
tumor-initiating cells (26), and
tumor xenograft growth in vivo (27). Studies (28,29)
have shown that EZH2 participates in the pre-transcriptional
process of tumor suppressor miRNAs (miR-181a, miR-203, and miR-200
families), thereby implying that a few dominant oncogenes (e.g.,
YY1) and pathways are involved in multiple cellular processes
orchestrated through the regulatory network of EZH2.
As to the incapability of the overexpression of YY1
to enhance the carcinoma ability of PCa cells and to upregulate the
expression of miR-146a, it was speculated to be caused by the
resulting saturation effect on the YY1 regulatory system. Inside
the cytoplasm, special miRNAs cannot be regulated by merely a
single and simplistic regulator (e.g., YY1). For example, in the
regulatory circuitry in renal cell carcinoma cells, the repression
of CCAAT/enhancer-binding protein α (C/EBPα) induced by YY1 leads
to a reduction in miR-34a expression; by contrast, miR-34a exerts a
tumor-suppressive function by directly suppressing the oncogenic
YY1, thereby forming a YY1-C/EBPα-miR-34a positive feedback loop
(30). Nevertheless, YY1 requires
various co-transcriptional factors, such as histone deacetylase 4
(31) and EZH2 (16), to perform its function in the
target genes epigenetic silencing process. As a result,
overexpressed YY1 inside the cytoplasm alone has limited
opportunity or access to the role of tumorigenic transcriptional
factor, thereafter cannot enhance the YY1-associated
transcriptional repression.
In PCa, miR-146a is reported to suppress cell
viability via the nuclear factor κ-light-chain-enhancer of the
activated B cells (NF-κB) signaling pathway (8), whereas the classic NF-κB pathway
activates the transcription of YY1 (32). Together with the preceding results,
a dynamic YY1/EZH2-miR-146a-NF-κB feedback is hypothesized to
modulate the cell viability and proliferation of PCa cell lines. In
addition, as the data shown in the assays, the miR-146a inhibitor
cannot entirely rescue all changes in cell biological functions
after YY1 depletion, suggesting there might be other existing
target pathways affecting the PCa cell biological function.
Considering all of the results, we suggest that the
recruitment of EZH2 by YY1 inhibits the miR-146a transcriptional
activity in PCa cells, and YY1 depletion represses PCa cell
viability and proliferation and induces apoptosis at least in a
miR-146a-assisted manner. This result may provide a promising
strategy for PCa treatment.
Acknowledgments
This work was supported by grants of National
Natural Science Foundation of China (81202034 and 81572517) and the
Scientific Research Innovation Project of University in Jiangsu
Province (KYLX15_0184).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Katzenwadel A and Wolf P: Androgen
deprivation of prostate cancer: Leading to a therapeutic dead end.
Cancer Lett. 367:12–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hua K, Jin J, Zhang H, Zhao B, Wu C, Xu H
and Fang L: MicroRNA-7 inhibits proliferation, migration and
invasion of thyroid papillary cancer cells via targeting CKS2. Int
J Oncol. 49:1531–1540. 2016.PubMed/NCBI
|
|
4
|
Li J, Yang X, Guan H, Mizokami A, Keller
ET, Xu X, Liu X, Tan J, Hu L, Lu Y, et al: Exosome-derived
microRNAs contribute to prostate cancer chemoresistance. Int J
Oncol. 49:838–846. 2016.PubMed/NCBI
|
|
5
|
Xu B, Wang N, Wang X, Tong N, Shao N, Tao
J, Li P, Niu X, Feng N, Zhang L, et al: MiR-146a suppresses tumor
growth and progression by targeting EGFR pathway and in a
p-ERK-dependent manner in castration-resistant prostate cancer.
Prostate. 72:1171–1178. 2012. View Article : Google Scholar
|
|
6
|
Sun Q, Zhao X, Liu X, Wang Y, Huang J,
Jiang B, Chen Q and Yu J: miR-146a functions as a tumor suppressor
in prostate cancer by targeting Rac1. Prostate. 74:1613–1621. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu B, Huang Y, Niu X, Tao T, Jiang L, Tong
N, Chen S, Liu N, Zhu W and Chen M: Hsa-miR-146a-5p modulates
androgen-independent prostate cancer cells apoptosis by targeting
ROCK1. Prostate. 75:1896–1903. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu R, Yi B, Wei S, Yang WH, Hart KM,
Chauhan P, Zhang W, Mao X, Liu X, Liu CG, et al:
FOXP3-miR-146-NF-κB axis and therapy for precancerous lesions in
prostate. Cancer Res. 75:1714–1724. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhalla SS, Robitaille L and Nemer M:
Cooperative activation by GATA-4 and YY1 of the cardiac B-type
natriuretic peptide promoter. J Biol Chem. 276:11439–11445. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee MY, Lu A and Gudas LJ: Transcriptional
regulation of Rex1 (zfp42) in normal prostate epithelial cells and
prostate cancer cells. J Cell Physiol. 224:17–27. 2010.PubMed/NCBI
|
|
11
|
Luo J, Zhou X, Ge X, Liu P, Cao J, Lu X,
Ling Y and Zhang S: Upregulation of Ying Yang 1 (YY1) suppresses
esophageal squamous cell carcinoma development through heme
oxygenase-1. Cancer Sci. 104:1544–1551. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu B, Feng NH, Li PC, Tao J, Wu D, Zhang
ZD, Tong N, Wang JF, Song NH, Zhang W, et al: A functional
polymorphism in Pre-miR-146a gene is associated with prostate
cancer risk and mature miR-146a expression in vivo. Prostate.
70:467–472. 2010. View Article : Google Scholar
|
|
13
|
Tao T, Wang Y, Luo H, Yao L, Wang L, Wang
J, Yan W, Zhang J, Wang H, Shi Y, et al: Involvement of
FOS-mediated miR-181b/miR-21 signalling in the progression of
malignant gliomas. Eur J Cancer. 49:3055–3063. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang B, Wang LL, Ren RJ, Dammer EB, Zhang
YF, Huang Y, Chen SD and Wang G: MicroRNA-146a represses LRP2
translation and leads to cell apoptosis in Alzheimer's disease.
FEBS Lett. 590:2190–2200. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsang DP, Wu WK, Kang W, Lee YY, Wu F, Yu
Z, Xiong L, Chan AW, Tong JH, Yang W, et al: Yin Yang 1-mediated
epigenetic silencing of tumour-suppressive microRNAs activates
nuclear factor-κB in hepatocellular carcinoma. J Pathol.
238:651–664. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seligson D, Horvath S, Huerta-Yepez S,
Hanna S, Garban H, Roberts A, Shi T, Liu X, Chia D, Goodglick L, et
al: Expression of transcription factor Yin Yang 1 in prostate
cancer. Int J Oncol. 27:131–141. 2005.PubMed/NCBI
|
|
18
|
Deng Z, Cao P, Wan MM and Sui G: Yin Yang
1: A multifaceted protein beyond a transcription factor.
Transcription. 1:81–84. 2010. View Article : Google Scholar
|
|
19
|
Thorvaldsen JL, Weaver JR and Bartolomei
MS: A YY1 bridge for X inactivation. Cell. 146:11–13. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Caggia S, Libra M, Malaponte G and Cardile
V: Modulation of YY1 and p53 expression by transforming growth
factor-β3 in prostate cell lines. Cytokine. 56:403–410. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sankar N, Baluchamy S, Kadeppagari RK,
Singhal G, Weitzman S and Thimmapaya B: p300 provides a corepressor
function by cooperating with YY1 and HDAC3 to repress c-Myc.
Oncogene. 27:5717–5728. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aldiri I and Vetter ML: PRC2 during
vertebrate organogenesis: A complex in transition. Dev Biol.
367:91–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiong X, Zhang J, Liang W, Cao W, Qin S,
Dai L, Ye D and Liu Z: Fuse-binding protein 1 is a target of the
EZH2 inhibitor GSK343, in osteosarcoma cells. Int J Oncol.
49:623–628. 2016.PubMed/NCBI
|
|
24
|
Varambally S, Dhanasekaran SM, Zhou M,
Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt
RG, Otte AP, et al: The polycomb group protein EZH2 is involved in
progression of prostate cancer. Nature. 419:624–629. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bracken AP, Pasini D, Capra M, Prosperini
E, Colli E and Helin K: EZH2 is downstream of the pRB-E2F pathway,
essential for proliferation and amplified in cancer. EMBO J.
22:5323–5335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wen S, Tian J, Niu Y, Li L, Yeh S and
Chang C: ASC-J9(®), and not Casodex or Enzalutamide, suppresses
prostate cancer stem/progenitor cell invasion via altering the
EZH2-STAT3 signals. Cancer Lett. 376:377–386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao Q, Mani RS, Ateeq B, Dhanasekaran SM,
Asangani IA, Prensner JR, Kim JH, Brenner JC, Jing X, Cao X, et al:
Coordinated regulation of polycomb group complexes through
microRNAs in cancer. Cancer Cell. 20:187–199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou L, Wang L, Lu L, Jiang P, Sun H and
Wang H: A novel target of microRNA-29, Ring1 and YY1-binding
protein (Rybp), negatively regulates skeletal myogenesis. J Biol
Chem. 287:25255–25265. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weng W, Wang M, Xie S, Long Y, Li F, Sun
F, Yu Y and Li Z: YY1-C/EBPα-miR34a regulatory circuitry is
involved in renal cell carcinoma progression. Oncol Rep.
31:1921–1927. 2014.PubMed/NCBI
|
|
31
|
Ren G, Zhang G, Dong Z, Liu Z, Li L, Feng
Y, Su D, Zhang Y, Huang B and Lu J: Recruitment of HDAC4 by
transcription factor YY1 represses HOXB13 to affect cell growth in
AR-negative prostate cancers. Int J Biochem Cell Biol.
41:1094–1101. 2009. View Article : Google Scholar
|
|
32
|
Wang H, Garzon R, Sun H, Ladner KJ, Singh
R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, et al:
NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis
and rhabdomyosarcoma. Cancer Cell. 14:369–381. 2008. View Article : Google Scholar : PubMed/NCBI
|