Introduction
Cervical cancer (CC) is the most common cancer in
females and the second most common cause of cancer-related
mortality in China (1). Cervical
cancer is the fourth most common cancer in females in 'developed'
countries (2). The poor prognosis
of cervical cancer is associated with its highly invasive and
diffuse metastatic characteristics (3,4). The
cervical cancer stem cell (CSCC) is recognized as being the 'seed'
for cancer metastasis (5).
However, the CSCC was not identified until recently because of a
shortage of special cell-surface markers. High expression of stem
cell transcription factors (TFs) in cancer is recognized as a
cancer stem cell (CSC) marker and correlates with a poor prognosis
(6). Core stem cell TFs such as
Foxd3 (7), Sox9 (8), Sox2 (9,10),
Nanog (11) and Oct4 (10) have been reported to be highly
expressed in cervical cancer, and to be associated with increased
invasion of cancer cells and carcinogenesis. It has been assumed
that downregulation of expression of these TFs by certain molecules
(e.g., miRNAs) can inhibit invasion and metastasis by cancer cells
(12).
Previously, miR-145 was reported to induce
differentiation of human embryonic stem cells through enzymolysis
of the core stem cell transcription factors Oct4, Sox2, and KLF4
(13). It has also been proposed
as a tumor-suppressor, and its expression is decreased in many
types of cancer (14,15), for example, in colonic
adenocarcinomas where miR-145 expression is lower than in the
mucosa (16). Intravesicular
administration of exogenous miR-145 has been shown to inhibit
growth of mouse orthotopic human bladder cancer xenografts
(17), while miR-145 targets MUC13
to suppress the growth and invasion of pancreatic cancer cells
(14). Overexpression of miR-145
promotes differentiation by inhibiting Oct4 expression in human
endometrial adenocarcinoma cells (18). Based on these data, we investigated
the expression and function of miR-145 in cervical cancer using a
cervical tumorsphere (CT) model.
Materials and methods
Culture of CT
This study protocol was approved by the Medical
Ethics Committees of Xi'an Jiaotong University of Medicine (no.
H34-32-1, Xi'an, China). All investigations were conducted in
accordance with the Declaration of Helsinki. All the patients
involving this study provided a written informed consent. The nude
mice used in this study were treated in accordance with the
institutional guidelines of the Animal Ethics Committee for the
care and use of animals in Xi'an Jiaotong University of Medicine,
China. We confirm that all methods were carried out in accordance
with relevant guidelines and regulations of Xi'an Jiaotong
University of Medicine.
We previously described the method of CT culture
(16). Briefly, 21 samples of
cervical cancer tissue (stage IB, n=14; stage IC, n=4; stage IIa,
n=3; age, 36–69 years) were obtained by resection (Table I). For CT culture, tumors were
washed immediately in phosphate-buffered saline (PBS) containing
500 U/l penicillin G (Gibco; Thermo Fisher Scientific, Waltham, MA,
USA) and 500 mg/l streptomycin (Gibco) <30 min after resection,
and then digested overnight in DMEM/F12 medium supplemented with
0.5 mg/ml collagenase IV (Gibco). Tumors were cultured in stem cell
medium (DMEM/F12), 10 ng/ml basic fibroblast growth factor (bFGF),
10 U/ml leukemia inhibitory factor, 1×105 U/l
penicillin, 100 mg/l streptomycin (all reagents from EMD Millipore,
Merck KGaA, Darmstadt, Germany) at 37°C in a humidified atmosphere
containing 5% CO2. Clones of >50 cells were
recognized as tumorspheres. These were dissociated every 7–10 days
by incubation in a non-enzymatic cell dissociation solution
(Sigma-Aldrich, St. Louis, MO, USA) for 2 min at 37°C and passaged
at 1×103 cells per 100-mm plate. Tumorsphere cells had
differentiated completely by 8 days after switching to stem cell
medium without bFGF.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Sample no. | Age (years) | Diagnosis |
|---|
| 1 | 36 | SCC, Stage IB |
| 2 | 41 | SCC, Stage IB |
| 3 | 49 | SCC, Stage IB |
| 4 | 47 | SCC, Stage IB |
| 5 | 56 | SCC, Stage IB |
| 6 | 54 | SCC, Stage IB |
| 7 | 57 | SCC, Stage IB |
| 8 | 68 | SCC, Stage IB |
| 9 | 56 | SCC, Stage IB |
| 10 | 53 | SCC, Stage IB |
| 11 | 36 | SCC, Stage IB |
| 12 | 39 | SCC, Stage IB |
| 13 | 49 | SCC, Stage IB |
| 14 | 52 | SCC, Stage IB |
| 15 | 61 | SCC, Stage IC |
| 16 | 46 | SCC, Stage IC |
| 17 | 67 | SCC, Stage IC |
| 18 | 38 | SCC, Stage IC |
| 19 | 66 | SCC, Stage IIA |
| 20 | 63 | SCC, Stage IIA |
| 21 | 69 | SCC, Stage IIA |
Transduction of adenovirus vectors
All Ad-vectors had comparable titers of
108–109 transducing units/ml. Virus
suspensions were stored at −80°C until use. Suspensions were
centrifuged briefly and kept on ice immediately before use. For
transduction, 2×104 dissociated tumorsphere cells were
transduced 1 day after initial seeding of cells with a multiplicity
of infection (MOI) of 25. Cells were incubated in stem cell medium
containing adenovirus particles and 4 µg/ml polybrene (Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 18 h at 37°C in
a humidified atmosphere containing 5% CO2. Ad-particles
were removed and the medium replaced with fresh stem cell
medium.
Colony formation assay
A colony formation assay was carried out as
previously described (19).
Briefly, single cervical carcinoma cells or dissociated tumorsphere
cells were cultured in DMEM/F-12 medium supplemented with 10% fetal
calf serum (FCS; Life Technologies, Carlsbad, CA, USA), 2 mmol/l
glutamine (Invitrogen, Carlsbad, CA, USA), 1×105 U/l
penicillin, and 100 mg/l streptomycin. Cells were cultured at
clonal densities of 100–300/cm2 on 2% gelatin
(Sigma-Aldrich)-coated tissue culture dishes (BD Biosciences, San
Jose, CA, USA) at 37°C in 5% CO2 in air. Cloning plates
were monitored every day and medium was changed every 2–3 days.
After 28 days of culture, plates were fixed in 10% formaldehyde/PBS
for 10 min and stained with Harris hematoxylin. Clones (>50
cells) were counted on ≥3 plates per sample and averaged.
Efficiency of colony formation was determined as [(number of
colonies)/(number of cells seeded)] ×100.
Flow cytometric analyses
Dissociated tumorsphere cells or adenovirus
vector-transfected cells were grown in 6-well plates and collected.
Cells were resuspended in PBS containing 2% fetal bovine serum
(FBS) and 0.1% sodium azide, and then analyzed by a FACScalibur
system (BD Biosciences). Acquisition was set for 10,000 events per
sample. Data were analyzed using FACS v4.1.2 (BD Biosciences).
Triplicate samples were analyzed in each experiment.
Real-time polymerase chain reaction
(PCR)
Total RNA was extracted from cells using TRIzol®
(Invitrogen). miRNA levels were assayed using Taqman® probes and
primer sets (Applied Biosystems, Foster City, CA, USA) according to
the manufacturer's instructions. Briefly, first-strand cDNA was
generated using a reverse transcription system kit (Promega,
Madison, WI, USA) with random primers of miR-145. Realtime PCR was
carried out using power SYBR Green PCR master mix (Applied
Biosystems) in a StepOnePlus® system (Applied Biosystems). The
level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was
used as the control for internal normalization. For exact
quantification of gene copies per cell, reverse-transcribed miR-145
cDNA was used as a template to formulate standard curves. Then, the
exact number of copies of miR-145 per cell was calculated according
to their molecular weight and cell counts. Primer sequences are
presented in Table II.
 | Table IIPCR primers and products. |
Table II
PCR primers and products.
| Symbol | Primer | Product (bp) |
|---|
| Nanog | S:
5′-AATGGTGTGACGCAGAAG-3′ | 255 |
| A:
5′-AGATTCCTCTCCACAGTTATAG-3′ | |
| Oct4 | S:
5′-AGCTGGAGAAGGAGAAGC-3′ | 194 |
| A:
5′-AAAGCGGCAGATGGTCGT-3′ | |
| Sox2 | S:
5′-CAATAGCATGGCGAGCGG-3′ | 196 |
| A:
5′-GTCGTAGCGGTGCATGGG-3′ | |
| GAPDH | S:
5′-AAGGCTGAGAATGGGAAAC-3′ | 254 |
| A:
5′-TTCAGGGACTTGTCATACTTC-3′ | |
Implantation of tumorspheres and
tumorsphere-derived cells into nude mice
After dissociation of tumorspheres and
tumorsphere-derived differentiated cells from 21 cervical cancer
patients in a non-enzymatic cell-dissociation solution, cells were
washed in serum-free Hank's balanced salt solution (HBSS). Then,
cells were suspended in a 1:1 (v/v) mixture of serum-free DMEM/F12,
and 1×105 cells were injected (s.c.) into the right
(differentiated cells) or left (tumorsphere cells) mid-abdominal
area of nude mice using a 23-G needle. Animals were subjected to
necropsy 28 days after implantation, and tumor growth assessed by
measuring volume using the formula: V = 1/2 × (L × W2).
In the silenced-miR-145 group, mice were sacrificed at 14 days
after injection to avoid necrosis in transplanted tumors. Sequences
of primary miR-145 or silent miR-145 are shown in Table III.
 | Table IIIPrimers for overexpression of miR-145
or knocked down miR-145. |
Table III
Primers for overexpression of miR-145
or knocked down miR-145.
| Symbol | Primer | Sequence |
|---|
| Pri-miR-145 | Sense |
5′-TGCTGGTCCAGTTTTCCCAGGAATCCCTGTTTTGGCCACTGACTGACAGGGATTCGGGAAAACTGGAC-3′ |
| Antisense |
5′-CCTGGTCCAGTTTTCCCGAATCCCTGTCAGTCAGTGGCCAAAACAGGGATTCCTGGGAAAACTGGACC-3′ |
| Si-miR-145 | Abm | (Catalog number
mh5195) |
| Si-miR-145
control | Abm | (Catalog number
m010) |
Western blotting
Lysates were extracted from tumorspheres or cells.
Proteins (20 µg) were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto nitrocellulose membranes. The latter were blocked
in 5% skimmed milk in Tris-buffered saline-Tween-20, and then
incubated with primary antibodies to core TF proteins or GAPDH
(1:500 dilution; Santa Cruz Biotechnology, Inc.) overnight at 4°C.
After incubation with horseradish peroxidase-labeled secondary
antibodies, membranes were developed using a SuperSignal® West Pico
Trial kit (Pierce, Rockford, IL, USA).
Immunohistochemical analyses
Cervical cancer samples were fixed in
phosphate-buffered 10% formalin (pH 7.2), embedded in paraffin, and
cut into sections (4 µm). Sections were dewaxed in xylene,
dehydrated in alcohol, and incubated in 0.01 M sodium citrate
buffer (pH 6.0) for antigen retrieval. Sections were incubated with
3% H2O2 for 30 min to block endogenous
peroxidase activity and with normal mouse serum at 37°C for 15 min
to block non-specific binding of antibody. Then, sections were
incubated with Sox2, Oct4 or Nanog antibodies (1:100 dilution in
PBS; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature,
followed by incubation with biotinylated secondary antibody (Santa
Cruz Biotechnology, Inc.) and 3,3′-diaminobenzidine.
Mutation of miR-145 binding site within
the sequence of Sox2, Nanog and Oct4
The complementary DNAs of Sox2, Nanog and Oct4 were
purchased from Shanghai Yingji Biological Company (Shanghai,
China). Mutated constructs containing the 6 bp point mutations in
the core TF seed sequence predicted to be the miR-145 target
sequence were synthesized using a QuikChange II site-directed
mutagenesis kit (Stratagene, La Jolla, CA, USA). The primers were
designed by Stratagene using their own software (http://labtools.stratagene.com/QC) and are shown
in Table IV. Ad-vectors
expressing the core TF wild-type or mutation of miR-145 seed
sequence under the control of the U6 promoter were produced by
transient transfection of HEK293T cells, and then transfected into
tumorspheres following the procedures described above.
 | Table IVPrimers for mutation miR-145 target
sequence on Sox2, Nanog and Oct4. |
Table IV
Primers for mutation miR-145 target
sequence on Sox2, Nanog and Oct4.
| Symbol | Primers |
|---|
| Oct4 | S:
GGAGTCGGGGTGGAGAGCAACTCC |
| A:
CCTCAACGAGAGGTGGGGCTGAGG |
| Nanog | S:
CTTTAGTTAATTCATACAATGTC |
| A:
GACATTGTATGAATTAACTAAAG |
| Sox2 | S:
TCTCCCCCCTCGCTGTCCGGCCCT |
| A:
AGGGCCGGACAGCGAGGGGGGAGA |
Injection of Ad-miR-145 into exograft
tumors in null mice
Titers of virus stocks were checked using
TCID50 methods. High-titer stocks were stored at −70°C
until use. Two weeks after injection of tumorspheres into nude
mice, the tumors became palpable and each tumor site was injected
with 5.8×105 pfu of Ad-miR-145 and Ad-Mock with
gadolinium (1:1) (Sigma-Aldrich). Tumor size was measured each
week.
Cell invasion assay
Cell invasion was evaluated using 24-well Transwell®
culture chambers, as previously described (18). Cells were seeded at
5×104 per well and cultured in stem cell medium for 24
h. Then, cells were fixed in methanol and stained with 5% crystal
violet. After examination under a light microscope, cells were
eluted with 33% acetic acid. Optical-density values of the eluate
were read using a Bio-Rad microplate reader at 590 nm.
Patient database bioinformatics
The protocol followed that previously described
(20). Briefly, data on expression
of genes and miRNA were obtained from the Cancer Genome Atlas
(TCGA; https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp)
for patients diagnosed with squamous cervical cancer. Expression of
genes or miR-145 was divided into 'high' and 'low' groups based on
the mean ± 1 SD. Kaplan-Meier survival curves were generated
comparing these two groups via the log-rank test. miR-145
expression combined with expression data for Oct4, Nanog, or Sox2
were subtracted from the miR-145 data for each patient, and the
analysis described above was repeated.
Statistical analyses
Data are shown as the mean ± standard error of the
mean. Comparison between groups were performed using analysis of
variance, Fisher's exact test or two-tailed Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of miR-145 and core stem cell
TFs in undifferentiated cervical cancer stem cells
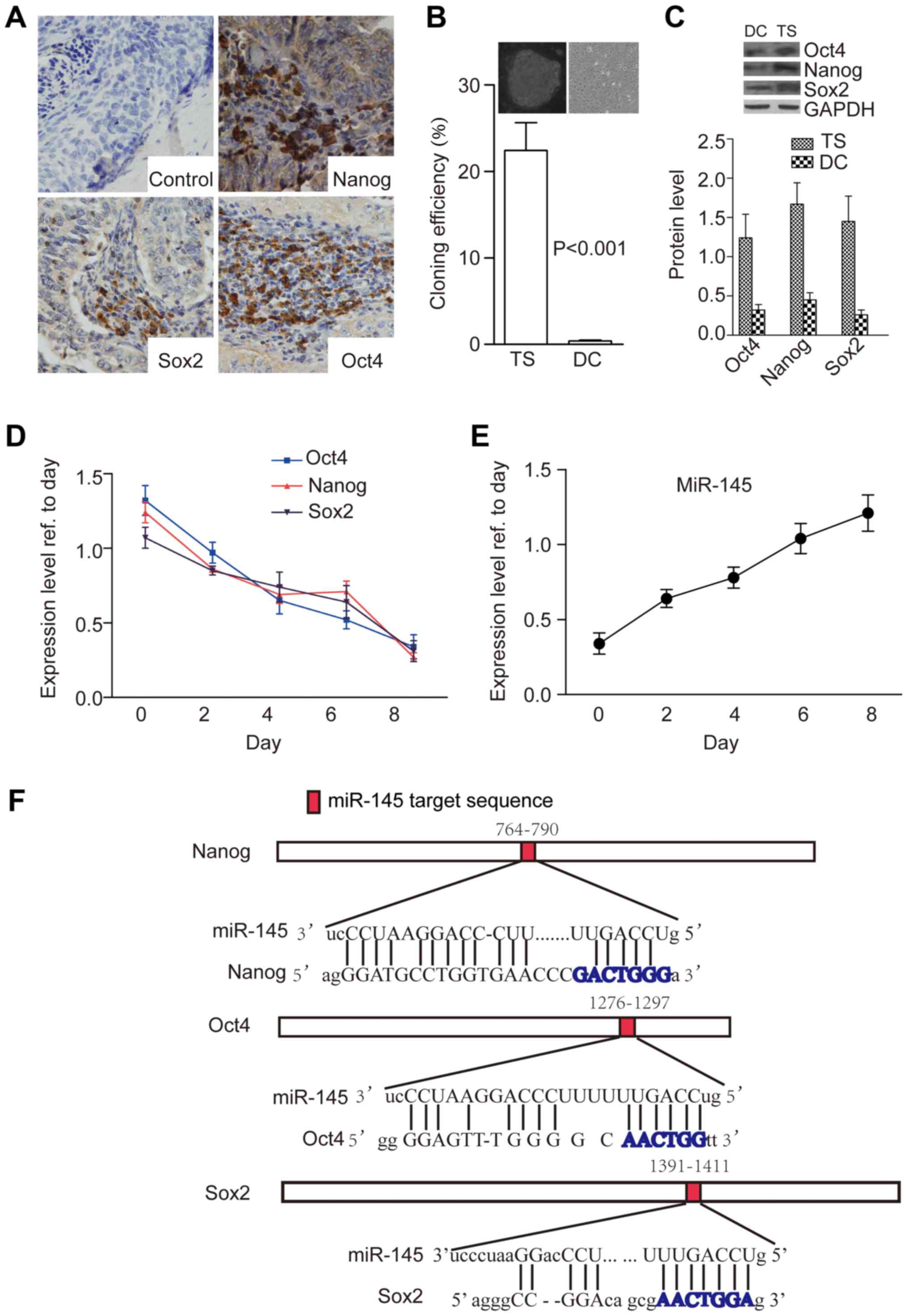
Our results showed that cervical cancer tissues were
immunopositive for Nanog (20/21), Oct4 (19/21) and Sox2 (20/21),
and 19/21 samples were positive for all three TFs. Staining for
these TFs was visible in the cytoplasm and nuclei of a few (but not
all) cancer cells, and we could detect that most cervical cancer
cells expressed these TFs (18/21) (Fig. 1A). After enzymatic dissociation and
culture in stem cell medium, a few colonies (tumorspheres) formed
in all tissues tested (21/21) after 15 days. Colonies were formed
at the rate of 22.41±3.23 per 100 cells dissociated from
tumorspheres, but this rate was reduced to only 0.41±0.07 per 100
cells dissociated from differentiated tumorspheres (P<0.001)
(Fig. 1B) when they detached from
the disks. Furthermore, protein levels of Nanog, Sox2 and Oct4 were
significantly higher in tumorsphere cells than in differentiated
cells (P=0.012, Fig. 1C).
Next, we examined expression of miR-145 and core TFs
according to tumorsphere stages. First, we assessed expression of
miR-145 in cells cultured under tumorsphere conditions or
differentiated conditions [removal of basic fibroblast growth
factor (bFGF) from the stem cell medium]. Quantitative reverse
transcription-polymerase chain reaction (qRT-PCR) showed that the
protein levels of core TFs decreased, whereas miR-145 level
increased, at different time points and underwent dynamic changes
(Fig. 1D and E). Two studies
previously reported potential miR-145 binding sites in the genes of
these TFs (19,21), so we used miRanda software to
predict miR-145 binding sites in the mRNAs of these TFs, the
results confirmed potential miR-145 binding sequences within them
(Fig. 1F).
Overexpression of miR-145 induces
tumorsphere differentiation by decreasing expression of core
TFs
Next, we investigated the role of miR-145 in
cervical cancer. Ad-miR-145-GFP was transduced into tumorspheres or
their differentiated cells to increase miR-145 expression. Flow
cytometric analyses showed that there were 91±1.9% GFP-positive
cells in the overexpressed Ad-miR-145-GFP group and 91±2.3% in the
adenovirus-scrambled sequence-GFP (Ad-scr) group with no homology
to the human genome, which acted as the control group among
transfected tumorsphere cells. The miR-145 level was increased
4.5-fold in the overexpressed Ad-miR-145-GFP group compared with
its control (Fig. 2A).
Accordingly, levels of core TFs were decreased significantly
according to western blotting, and colony formation and cell
invasion was decreased in the overexpressing Ad-miR-145-GFP group
(Fig. 2B–D). Next, dissociated
tumorsphere cells harboring Ad-miR- 145-GFP or Ad-scr-GFP were
injected into nude mice. A total of 21 Ad-scr-GFP-tumorspheres
derived from the 21 patients caused tumor formation in mice after
28 days. In contrast, 21 Ad-miR-145-GFP-tumorspheres from these 21
patients only caused 11 tumors, and those tumors were relatively
small compared with those in the Ad-scr-GFP group (Fig. 2E).
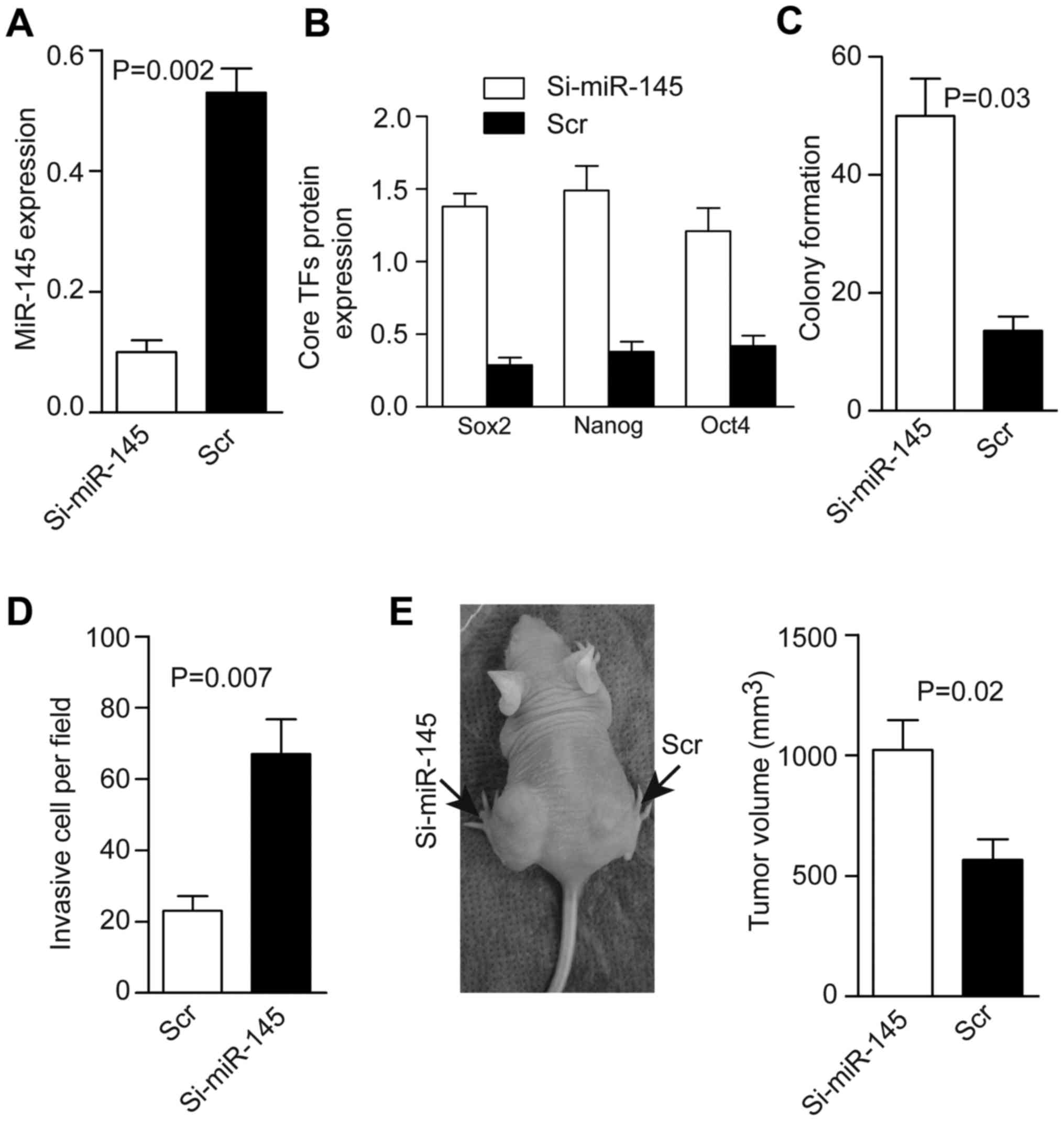
Knockdown of miR-145 promotes tumorsphere
tumorigenicity by maintaining TF expression
Next, we transfected the Ad-silence-miR-145
(Ad-si-miR-145-GFP) vector or its control (Ad-scr-GFP) into CTs.
Flow cytometric analyses showed that there were 90±1.4%
GFP-positive cells in the Ad-si-miR-145-GFP group and 88±3.2% in
the Ad-scr-GFP group. The miR-145 level was decreased fourfold
compared with the control (Fig.
3A), whereas levels of TFs, colony formation, and cell invasion
increased after silencing of miR-145 (Fig. 3B–D). Tumor formation was 21/21 in
the Ad-si-miR-145 group and 20/21 in the scr-GFP group. To prevent
tumor growth leading to necrosis (which would have affected the
measurement of tumor volume), mice were sacrificed 14 days after
injection. Tumors were larger in the si-miR-145 group than in the
scr-GFP group (Fig. 3E).
Mutation of miR-145 binding sites within
the sequences of Sox2, Nanog and Oct4 inhibit their miR-145-induced
enzymolysis
miR-145 seed sequences were predicted within Oct4,
Sox2 3′-untranslated regions (UTR) and Nanog coding sequence (CDS)
by the software miRanda after mutation of miR-145 target sequences
(6 bp deletion of the miR-145 target site) in these core
transcription factors (Fig. 4A),
and the vectors for the mutated structures are shown in Fig. 4B. First, we transfected
Ad-Sox2-EGFP, Ad-Sox2-EGFP-mutation (mut)-miR-145 target sequence
(Ad-Sox2-EGFP-mut) into tumorspheres, a luciferase reporter with no
UTR of Sox2 and Oct4, CDS of Nanog were used as a negative control.
Flow cytometric analyses revealed the percentage of EGFP-positive
cells to be 86-89% (87±2.2%), 88-89% (88±0.6%), and 87-89%
(88±1.9%), respectively. The Sox2 level did not significantly
differ between the wild-type (WT) group and the control (con) group
by 24 or 48 h after transfection (P=0.47 and 0.64, respectively),
but Sox2 expression in the WT group was lower than in the other two
groups (P=0.012 and 0.011, respectively) (Fig. 4C). Similarly, following
transfection of Ad-Oct4-EGFP, Ad-Oct4-EGFP-mut and Ad-control-EGFP,
or Ad-Nanog-EGFP, Ad-Nanog-EGFP-mut and Ad-control-EGFP, flow
cytometric analyses revealed the percentage of GFP-positive cells
to be 83–86% (84±1.9%) and 83–87% (85±1.7%), respectively. The Oct4
and Nanog levels did not differ between the mut group and the
control group (P=0.75 and 0.57, respectively), and their expression
was lower in the WT groups than in the other groups (P=0.002 and
0.005, respectively) (Fig. 4D and
E).
 | Figure 4Endogenous miR-145 represses the
3′-untranslated regions (UTRs) of Oct4, Sox2, and coding sequence
(CDS) of Nanog in CTs. (A) The mutational sequences in the core
TFs. (B) The 3′-UTR or CDS reporters in target validation. Luc,
firefly luciferase; pA, polyadenylation signal; WT, wild-type.
Mutation (mut) has a 6 bp deletion of the miR-145 target site. The
relative luciferase level of 3′-UTR luciferase reporters of Sox2
(C), Oct4 (D), and CDS luciferase reports of Nanog (E) in CTs under
self-renewal conditions at 24 or 48 h after transfection. Mut,
mutant UTR or CDS with a 6 bp deletion of the miR-145 target sites;
Control, the basal luciferase reporter without the UTR of Sox2 and
Oct4, CDS of Nanog (n=3). The difference between 3′-UTR mutant and
wild-type luciferase reporters of Oct4 (F), Sox2 (G), and CDS
mutant and wild-type luciferase reporters of nanog (H) depends on
miR-145 in CTs. On the y-axis, the mutant reporter level is
normalized against the average of the wild-type reporter level to
reflect the magnitude of repression. |
We further addressed the dependence of the reported
repression of Oct4, Sox2 UTR reporter, and Nanog CDS reporter on
the level of miR-145. After depletion of miR-145 by antisense
inhibitor, locked nuclei acid (LNA) in CT completely abolished the
differential regulation between the mutant and WT 3′-UTR reporters
in Oct4 and Sox2 (Fig. 4F–H), and
the mutant and wild-type CDS in Nanog. These results were in
accordance with the reports by Xu et al (13) and Wang et al (21).
Ad-miR-145 inhibits the growth of tumors
derived from cervical tumorspheres
Tumor cells were dissociated from tumorspheres and
injected into null mice. The resulting tumors were visible or
palpable 2 weeks after injection. Next, we injected
5.8×105 pfu of Ad-miR145 or Ad-Mock with gadolinium
combination into tumors three times a week (22). Mice were sacrificed at day 28 and
tumor volume measured. Tumors were smaller in the Ad-miR-145 group
than in the AD-scr group (P=0.009, Fig. 5A), the weak expression of core stem
cell TFs was detected after Ad-pri-miR-145 treatment (Fig. 5B). Moreover, levels of core TFs
were decreased (P=0.03, Fig. 5C),
miR-145 level was increased (P=0.04, Fig. 5D), colony formation was decreased
(P=0.007, Fig. 5E), and cell
invasion was decreased (P=0.012, Fig.
5F) in the Ad-miR-145 group compared with the control
group.
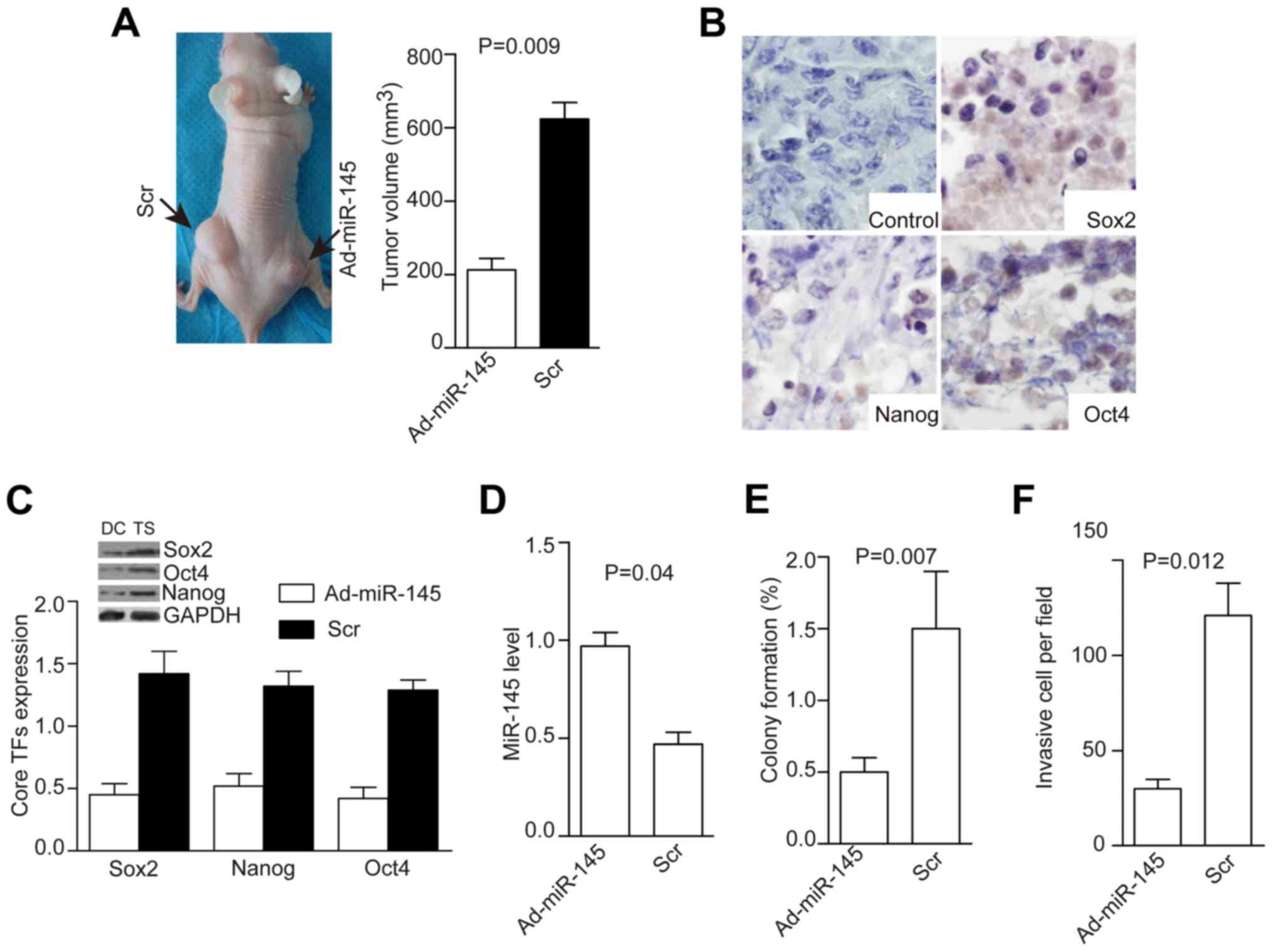 | Figure 5Injection of AD-miR-145 into
transplanted tumors inhibits tumor growth, expression of core TFs,
colony formation, and tumorsphere invasion. Injection of AD-miR-145
(5.8×105 pfu) into transplanted tumors inhibited tumor
growth (A, P=0.09), suppressed the expression of core TFs as shown
by immunohistochemistry (B), decreased levels of core TFs (C,
P=0.02), increased miR-145 level (D, P=0.04), and decreased colony
formation (E, P=0.007) and cell invasion (F, P=0.012). |
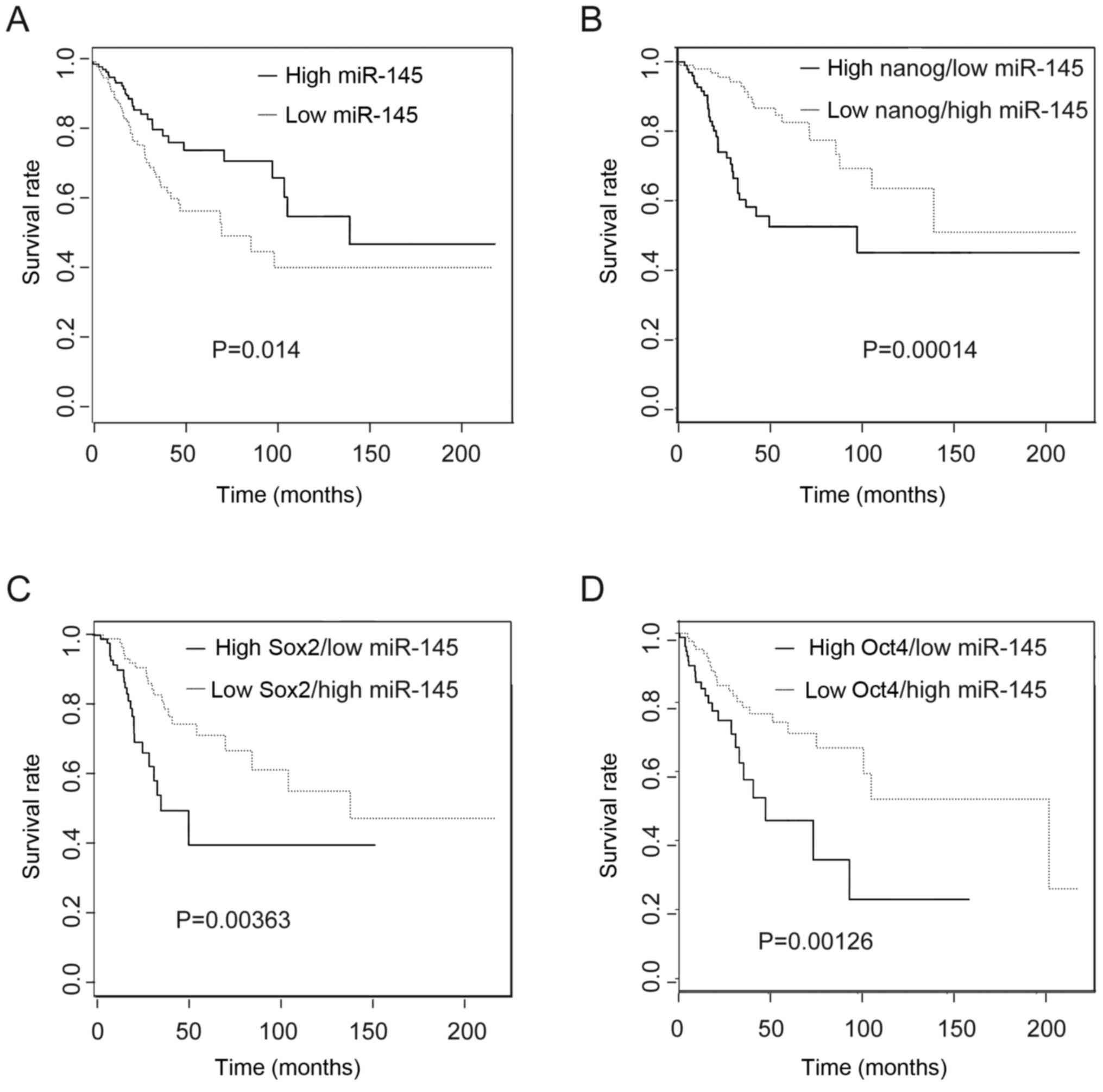
High expression of miR-145 is associated
with increased patient survival
To ascertain the clinical relevance of miR-145 and
these core stem cell transcripts in the prognosis of CC patients,
we evaluated the TCGA dataset. We found that high expression of
miR-145 indicated a better prognosis compared with low expression
of miR-145 (Fig. 6A). When miR-145
levels were combined with Nanog expression from the same patient,
patients in the low Nanog/high miR-145 expression group had a
better prognosis compared with the high Nanog/low miR-145
expression group (Fig. 6B).
Similar results were obtained for expression of low miR-145 with
high Sox2 and Oct4 in these subtypes compared with their opposite
groups. These results implied that these genes could be informative
for survival of CC patients (Fig. 6C
and D).
Discussion
Cervical cancer is caused by human papillomavirus
infection (HPV) (23).
Overexpression of miR-145 in HPV-positive cervical cancer cells
results in reduced genome amplification and late expression of
genes (24). Herein, we focused on
the relationship between miR-145 and mRNAs of core stem cell TFs in
cervical cancer. The core stem cell TFs, Sox2, Nanog and Oct4, are
essential for CSC maintenance. Downregulation of expression of
these TFs induced CSC differentiation and reduced CSC tumorigenesis
(25).
Wang et al reported downregulation of
expression of miR-145 in cervical carcinoma, and suggested miR-145
to be involved in cervical carcinogenesis (26); this speculation was confirmed in
our study. Moreover, we have provided the potential mechanisms by
which miR-145 downregulates expression of core stem cell TFs,
induces CCSC differentiation, and inhibits CCSC tumorigenesis.
First, we examined expression of miR-145 and core stem cell TFs
that maintain the pluripotency of CTs. miR-145 expression was
increased significantly in differentiated cells compared with
tumor-spheres. In addition, we found a negative correlation between
miR-145 levels and core TFs. These results strongly suggest that
overexpression of miR-145 induces tumorsphere differentiation and
inhibits carcinogenesis and invasion of the tumorsphere. When we
employed si-Ad-miR-145 to knockdown miR-145 in tumorspheres, we
found increased levels of core TFs that were involved in the
self-renewal, proliferation and invasion of tumorspheres (27). We also described an association
between miR-145 levels and survival of CC patients based on data
from TGCA. This comparison showed that patients with higher levels
of miR-145 (and in combination with low Sox2, Nanog and Oct4
levels) had higher median survival than patients with lower miR-145
levels. From the TCGA database, it could be concluded miR-145 was
an independent prognosis marker for CC patients. Taken together,
these results suggest that miR-145 downregulates expression of core
TFs to induce CT differentiation. Hence, we demonstrated that
miR-145 is involved in cervical carcinogenesis.
Recently, unmodified miR-145 delivered by
peptide-based vectors applied through a system or location in a
mouse model of colon carcinoma resulted in a 40 or 60% decrease in
tumor growth, with concomitant repression of ERK5 and c-Myc protein
levels compared with negative controls, respectively (28). miR-145 can increase the sensitivity
of tumors to chemotherapy or radiotherapy, and miR-145 treatment
can increase sensitivity to 5-fluorouracil in gastric cancer cells
(29). Similarly, local injection
of Ad-miR-145 results in significant suppression of tumor growth in
orthotopic mouse models of breast cancer (22). In our study, Ad-miR-145 effectively
inhibited the growth and invasion of tumor cells by decreasing
expression of core stem cell TFs in null-mice. These results
suggest that miR-145 could be a potential target of therapy for
cervical carcinoma.
Acknowledgments
This project was supported by the Natural Science
Foundation of Shanxi Province, China (grant no. 2013JM4012) and the
Key projects of Hubei Provincial Department of Education (grant no.
20162103), Natural Science Foundation of Hubei Province, China
(grant no. 2016CFB409).
References
|
1
|
Li J, Kang LN and Qiao YL: Review of the
cervical cancer disease burden in mainland China. Asian Pac J
Cancer Prev. 12:1149–1153. 2011.PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shepherd JH: Cervical cancer. Best Pract
Res Clin Obstet Gynaecol. 26:293–309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shi JF, Canfell K, Lew JB and Qiao YL: The
burden of cervical cancer in China: Synthesis of the evidence. Int
J Cancer. 130:641–652. 2012. View Article : Google Scholar
|
|
5
|
Baccelli I and Trumpp A: The evolving
concept of cancer and metastasis stem cells. J Cell Biol.
198:281–293. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu A, Yu X and Liu S: Pluripotency
transcription factors and cancer stem cells: Small genes make a big
difference. Chin J Cancer. 32:483–487. 2013.PubMed/NCBI
|
|
7
|
Li D, Mei H, Qi M, Yang D, Zhao X, Xiang
X, Pu J, Huang K, Zheng L and Tong Q: FOXD3 is a novel tumor
suppressor that affects growth, invasion, metastasis and
angiogenesis of neuroblastoma. Oncotarget. 4:2021–2044. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang HY, Lian P and Zheng PS: SOX9, a
potential tumor suppressor in cervical cancer, transactivates
p21WAF1/CIP1 and suppresses cervical tumor growth. Oncotarget.
6:20711–20722. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen L, Huang X, Xie X, Su J, Yuan J and
Chen X: High expression of SOX2 and OCT4 indicates radiation
resistance and an independent negative prognosis in cervical
squamous cell carcinoma. J Histochem Cytochem. 62:499–509. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ji J, Wei X and Wang Y: Embryonic stem
cell markers Sox-2 and OCT4 expression and their correlation with
WNT signal pathway in cervical squamous cell carcinoma. Int J Clin
Exp Pathol. 7:2470–2476. 2014.PubMed/NCBI
|
|
11
|
Ding Y, Yu AQ, Li CL, Fang J, Zeng Y and
Li DS: TALEN-mediated Nanog disruption results in less
invasiveness, more chemosensitivity and reversal of EMT in Hela
cells. Oncotarget. 5:8393–8401. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mathieu J and Ruohola-Baker H: Regulation
of stem cell populations by microRNAs. Adv Exp Med Biol.
786:329–351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu N, Papagiannakopoulos T, Pan G, Thomson
JA and Kosik KS: MicroRNA-145 regulates OCT4, SOX2, and KLF4 and
represses pluripotency in human embryonic stem cells. Cell.
137:647–658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khan S, Ebeling MC, Zaman MS, Sikander M,
Yallapu MM, Chauhan N, Yacoubian AM, Behrman SW, Zafar N, Kumar D,
et al: MicroRNA-145 targets MUC13 and suppresses growth and
invasion of pancreatic cancer. Oncotarget. 5:7599–7609. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cui SY, Wang R and Chen LB: MicroRNA-145:
A potent tumour suppressor that regulates multiple cellular
pathways. J Cell Mol Med. 18:1913–1926. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arndt GM, Dossey L, Cullen LM, Lai A,
Druker R, Eisbacher M, Zhang C, Tran N, Fan H, Retzlaff K, et al:
Characterization of global microRNA expression reveals oncogenic
potential of miR-145 in metastatic colorectal cancer. BMC Cancer.
9:3742009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Inamoto T, Taniguchi K, Takahara K,
Iwatsuki A, Takai T, Komura K, Yoshikawa Y, Uchimoto T, Saito K,
Tanda N, et al: Intravesical administration of exogenous
microRNA-145 as a therapy for mouse orthotopic human bladder cancer
xenograft. Oncotarget. 6:21628–21635. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Y, Liu S, Xin H, Jiang J, Younglai E,
Sun S and Wang H: Up-regulation of microRNA-145 promotes
differentiation by repressing OCT4 in human endometrial
adenocarcinoma cells. Cancer. 117:3989–3998. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou X, Gao Q, Wang J, Zhang X, Liu K and
Duan Z: Linc-RNA-RoR acts as a 'sponge' against mediation of the
differentiation of endometrial cancer stem cells by microRNA-145.
Gynecol Oncol. 133:333–339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alvarado AG, Turaga SM, Sathyan P,
Mulkearns-Hubert EE, Otvos B, Silver DJ, Hale JS, Flavahan WA, Zinn
PO, Sinyuk M, et al: Coordination of self-renewal in glioblastoma
by integration of adhesion and microRNA signaling. Neuro-oncol.
18:656–666. 2016. View Article : Google Scholar
|
|
21
|
Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao
L, Wu M, Xiong J, Guo X and Liu H: Endogenous miRNA sponge
lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem
cell self-renewal. Dev Cell. 25:69–80. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim SJ, Oh JS, Shin JY, Lee KD, Sung KW,
Nam SJ and Chun KH: Development of microRNA-145 for therapeutic
application in breast cancer. J Control Release. 155:427–434. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Crosbie EJ, Einstein MH, Franceschi S and
Kitchener HC: Human papillomavirus and cervical cancer. Lancet.
382:889–899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gunasekharan V and Laimins LA: Human
papillomaviruses modulate microRNA 145 expression to directly
control genome amplification. J Virol. 87:6037–6043. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luo W, Li S, Peng B, Ye Y, Deng X and Yao
K: Embryonic stem cells markers SOX2, OCT4 and Nanog expression and
their correlations with epithelial-mesenchymal transition in
nasopharyngeal carcinoma. PLoS One. 8:e563242013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chhabra R and Saini N: MicroRNAs in cancer
stem cells: Current status and future directions. Tumour Biol.
35:8395–8405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ibrahim AF, Weirauch U, Thomas M,
Grünweller A, Hartmann RK and Aigner A: MicroRNA replacement
therapy for miR-145 and miR-33a is efficacious in a model of colon
carcinoma. Cancer Res. 71:5214–5224. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
-145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|