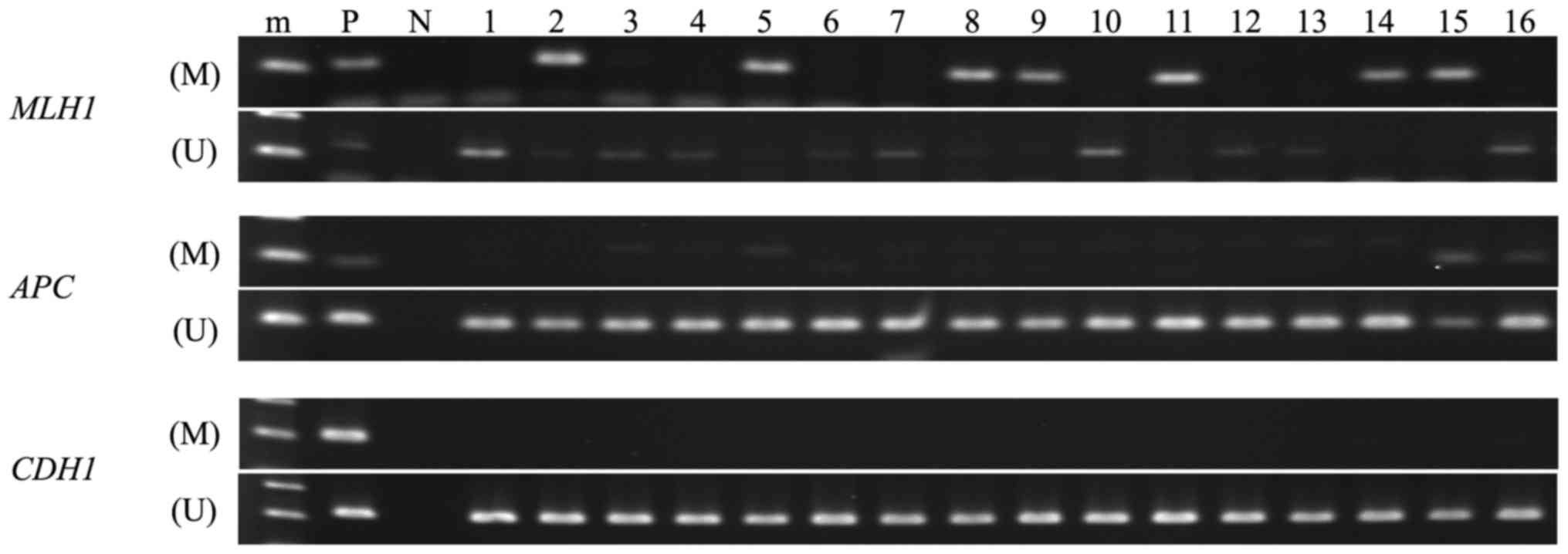
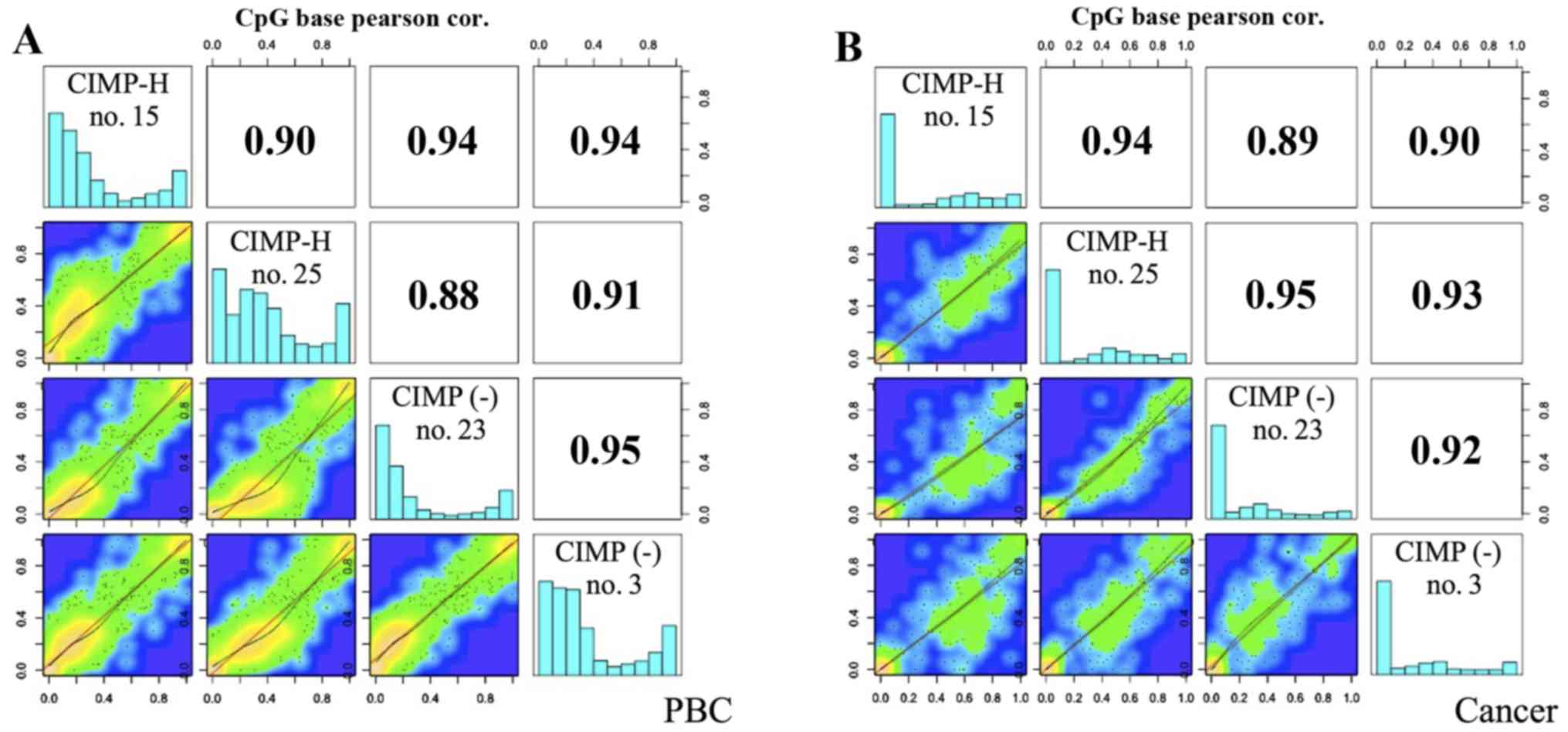
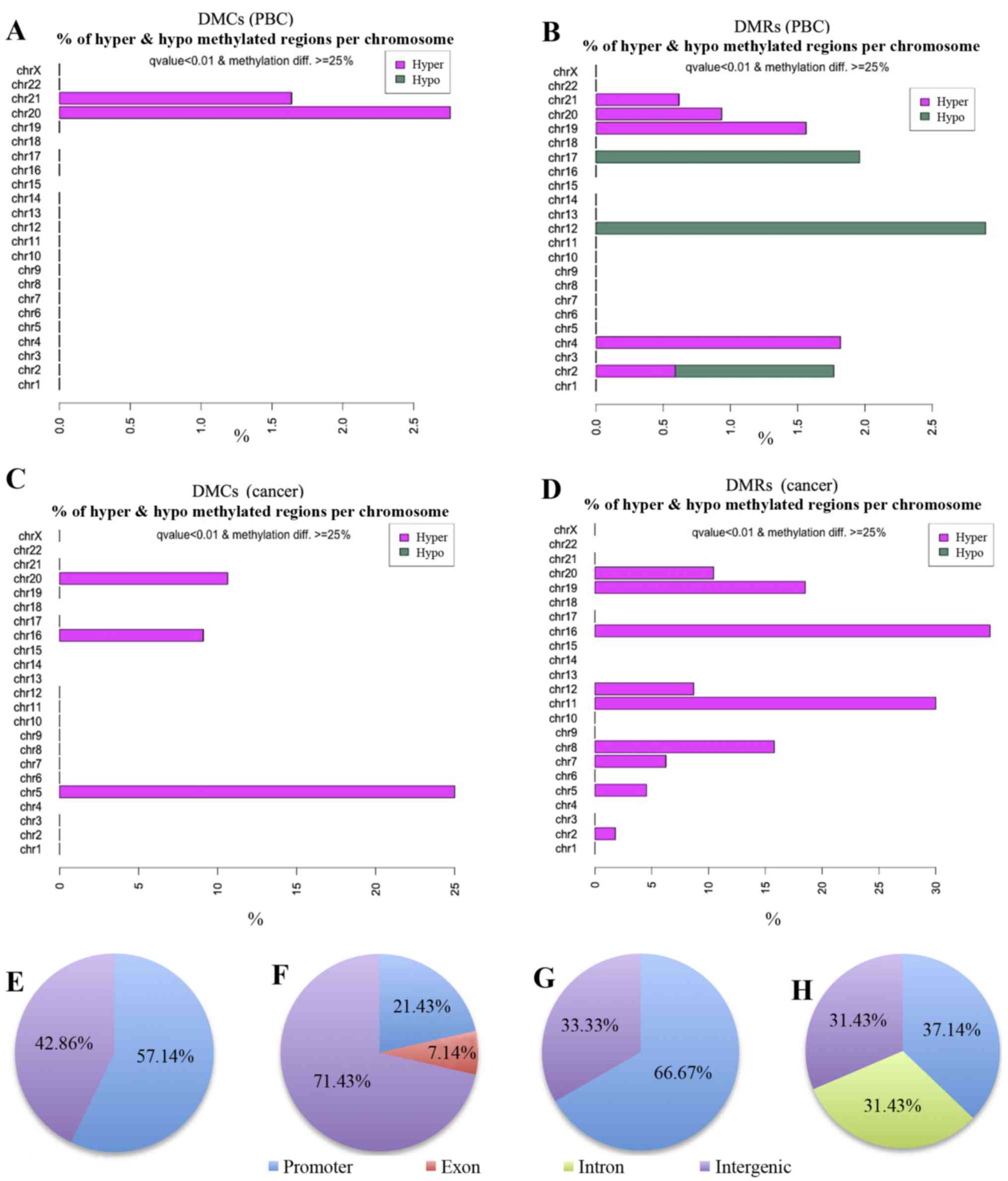
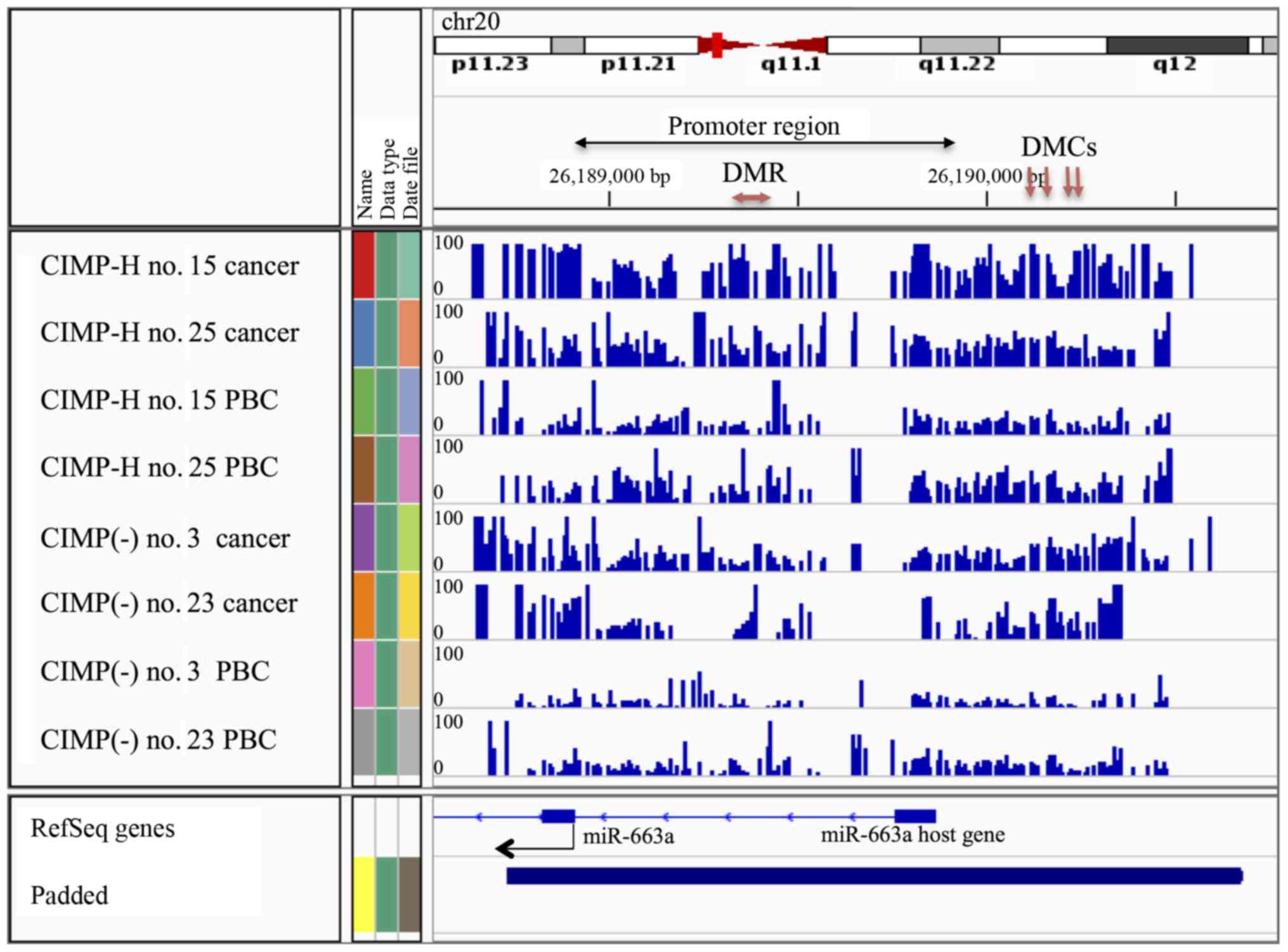
|
1
|
Fiolka R, Zubor P, Janusicova V, Visnovsky
J, Mendelova A, Kajo K, Lasabova Z, Plank L and Danko J: Promoter
hypermethylation of the tumor-suppressor genes RASSF1A, GSTP1 and
CDH1 in endometrial cancer. Oncol Rep. 30:2878–2886.
2013.PubMed/NCBI
|
|
2
|
Guida M, Sanguedolce F, Bufo P, Di Spiezio
Sardo A, Bifulco G, Nappi C and Pannone G: Aberrant DNA
hypermethylation of hMLH-1 and CDKN2A/p16 genes in benign,
premalignant and malignant endometrial lesions. Eur J Gynaecol
Oncol. 30:267–270. 2009.PubMed/NCBI
|
|
3
|
Kanaya T, Kyo S, Maida Y, Yatabe N, Tanaka
M, Nakamura M and Inoue M: Frequent hypermethylation of MLH1
promoter in normal endometrium of patients with endometrial
cancers. Oncogene. 22:2352–2360. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Crépin M, Dieu MC, Lejeune S, Escande F,
Boidin D, Porchet N, Morin G, Manouvrier S, Mathieu M and Buisine
MP: Evidence of constitutional MLH1 epimutation associated to
transgenerational inheritance of cancer susceptibility. Hum Mutat.
33:180–188. 2012. View Article : Google Scholar
|
|
5
|
Ward RL, Dobbins T, Lindor NM, Rapkins RW
and Hitchins MP: Identification of constitutional MLH1 epimutations
and promoter variants in colorectal cancer patients from the Colon
Cancer Family Registry. Genet Med. 15:25–35. 2013. View Article : Google Scholar
|
|
6
|
Cini G, Carnevali I, Quaia M, Chiaravalli
AM, Sala P, Giacomini E, Maestro R, Tibiletti MG and Viel A:
Concomitant mutation and epimutation of the MLH1 gene in a Lynch
syndrome family. Carcinogenesis. 36:452–458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bischoff J, Ignatov A, Semczuk A,
Schwarzenau C, Ignatov T, Krebs T, Küster D, Przadka-Rabaniuk D,
Roessner A, Costa SD, et al: hMLH1 promoter hypermethylation and
MSI status in human endometrial carcinomas with and without
metastases. Clin Exp Metastasis. 29:889–900. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Romero-Pérez L, López-García MÁ,
Díaz-Martín J, Biscuola M, Castilla MÁ, Tafe LJ, Garg K, Oliva E,
Matias-Guiu X, Soslow RA, et al: ZEB1 overexpression associated
with E-cadherin and microRNA-200 downregulation is characteristic
of undifferentiated endometrial carcinoma. Mod Pathol.
26:1514–1524. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park SY, Kook MC, Kim YW, Cho NY, Jung N,
Kwon HJ, Kim TY and Kang GH: CpG island hypermethylator phenotype
in gastric carcinoma and its clinicopathological features. Virchows
Arch. 457:415–422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang QY, Yi DQ, Zhou L, Zhang DH and Zhou
TM: Status and significance of CpG island methylator phenotype in
endometrial cancer. Gynecol Obstet Invest. 72:183–191. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weisenberger DJ, Levine AJ, Long TI,
Buchanan DD, Walters R, Clendenning M, Rosty C, Joshi AD, Stern MC,
Le Marchand L, et al for the Colon Cancer Family Registry:
Association of the colorectal CpG island methylator phenotype with
molecular features, risk factors, and family history. Cancer
Epidemiol Biomarkers Prev. 24:512–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Toyota M, Ahuja N, Ohe-Toyota M, Herman
JG, Baylin SB and Issa JP: CpG island methylator phenotype in
colorectal cancer. Proc Natl Acad Sci USA. 96:8681–8686. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Samowitz WS, Albertsen H, Herrick J, Levin
TR, Sweeney C, Murtaugh MA, Wolff RK and Slattery ML: Evaluation of
a large, population-based sample supports a CpG island methylator
phenotype in colon cancer. Gastroenterology. 129:837–845. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weisenberger DJ, Siegmund KD, Campan M,
Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D,
Buchanan D, et al: CpG island methylator phenotype underlies
sporadic microsatellite instability and is tightly associated with
BRAF mutation in colorectal cancer. Nat Genet. 38:787–793. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Whitcomb BP, Mutch DG, Herzog TJ, Rader
JS, Gibb RK and Goodfellow PJ: Frequent HOXA11 and THBS2 promoter
methylation, and a methylator phenotype in endometrial
adenocarcinoma. Clin Cancer Res. 9:2277–2287. 2003.PubMed/NCBI
|
|
16
|
Kawaguchi M, Yanokura M, Banno K,
Kobayashi Y, Kuwabara Y, Kobayashi M, Nomura H, Hirasawa A, Susumu
N and Aoki D: Analysis of a correlation between the BRAF V600E
mutation and abnormal DNA mismatch repair in patients with sporadic
endometrial cancer. Int J Oncol. 34:1541–1547. 2009.PubMed/NCBI
|
|
17
|
Peterson LM, Kipp BR, Halling KC, Kerr SE,
Smith DI, Distad TJ, Clayton AC and Medeiros F: Molecular
characterization of endometrial cancer: a correlative study
assessing microsatellite instability, MLH1 hypermethylation, DNA
mismatch repair protein expression, and PTEN, PIK3CA, KRAS, and
braf mutation analysis. Int J Gynecol Pathol. 31:195–205. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Banno K, Kisu I, Yanokura M, Masuda K,
Kobayashi Y, Ueki A, Tsuji K, Yamagami W, Nomura H, Susumu N, et
al: Endometrial cancer and hypermethylation: Regulation of DNA and
MicroRNA by epigenetics. Biochem Res Int. 2012:7382742012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Banno K, Yanokura M, Susumu N, Kawaguchi
M, Hirao N, Hirasawa A, Tsukazaki K and Aoki D: Relationship of the
aberrant DNA hypermethylation of cancer-related genes with
carcinogenesis of endometrial cancer. Oncol Rep. 16:1189–1196.
2006.PubMed/NCBI
|
|
20
|
Yanokura M, Banno K, Kawaguchi M, Hirao N,
Hirasawa A, Susumu N, Tsukazaki K and Aoki D: Relationship of
aberrant DNA hypermethylation of CHFR with sensitivity to taxanes
in endometrial cancer. Oncol Rep. 17:41–48. 2007.
|
|
21
|
Yanokura M, Banno K, Susumu N, Kawaguchi
M, Kuwabara Y, Tsukazaki K and Aoki D: Hypermethylation in the p16
promoter region in the carcinogenesis of endometrial cancer in
Japanese patients. Anticancer Res. 26(2A): 851–856. 2006.PubMed/NCBI
|
|
22
|
Akalin A, Kormaksson M, Li S,
Garrett-Bakelman FE, Figueroa ME, Melnick A and Mason CE:
methylKit: A comprehensive R package for the analysis of
genome-wide DNA methylation profiles. Genome Biol. 13:R872012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Avraham A, Cho SS, Uhlmann R, Polak ML,
Sandbank J, Karni T, Pappo I, Halperin R, Vaknin Z, Sella A, et al:
Tissue specific DNA methylation in normal human breast epithelium
and in breast cancer. PLoS One. 9:e918052014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miura F, Enomoto Y, Dairiki R and Ito T:
Amplification-free whole-genome bisulfite sequencing by
post-bisulfite adaptor tagging. Nucleic Acids Res. 40:e1362012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feinberg AP and Vogelstein B:
Hypomethylation distinguishes genes of some human cancers from
their normal counterparts. Nature. 301:89–92. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alvarez H, Opalinska J, Zhou L, Sohal D,
Fazzari MJ, Yu Y, Montagna C, Montgomery EA, Canto M, Dunbar KB, et
al: Widespread hypomethylation occurs early and synergizes with
gene amplification during esophageal carcinogenesis. PLoS Genet.
7:e10013562011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kundaje A, Meuleman W, Ernst J, Bilenky M,
Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ,
et al: Roadmap Epigenomics Consortium: Integrative analysis of 111
reference human epigenomes. Nature. 518:317–330. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yi C, Wang Q, Wang L, Huang Y, Li L, Liu
L, Zhou X, Xie G, Kang T, Wang H, et al: MiR-663, a microRNA
targeting p21WAF1/CIP1, promotes the proliferation and
tumorigenesis of nasopharyngeal carcinoma. Oncogene. 31:4421–4433.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Q, Cheng Q, Chen Z, Peng R, Chen R, Ma
Z, Wan X, Liu J, Meng M, Peng Z, et al: MicroRNA-663 inhibits the
proliferation, migration and invasion of glioblastoma cells via
targeting TGF-β1. Oncol Rep. 35:1125–1134. 2016.PubMed/NCBI
|
|
30
|
Cardenas H, Vieth E, Lee J, Segar M, Liu
Y, Nephew KP and Matei D: TGF-β induces global changes in DNA
methylation during the epithelial-to-mesenchymal transition in
ovarian cancer cells. Epigenetics. 9:1461–1472. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Suga Y, Sugai T, Uesugi N, Kawasaki T,
Fukagawa T, Yamamoto E, Ishida K, Suzuki H and Sugiyama T:
Molecular analysis of isolated tumor glands from endometrial
endometrioid adenocarcinomas. Pathol Int. 65:240–249. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Phipps AI, Limburg PJ, Baron JA,
Burnett-Hartman AN, Weisenberger DJ, Laird PW, Sinicrope FA, Rosty
C, Buchanan DD, Potter JD, et al: Association between molecular
subtypes of colorectal cancer and patient survival.
Gastroenterology. 148:77–87.e2. 2015. View Article : Google Scholar
|
|
33
|
Lister R, Pelizzola M, Dowen RH, Hawkins
RD, Hon G, TontiFilippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al:
Human DNA methylomes at base resolution show widespread epigenomic
differences. Nature. 462:315–322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hitchins MP: Constitutional epimutation as
a mechanism for cancer causality and heritability? Nat Rev Cancer.
15:625–634. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Muraki Y, Banno K, Yanokura M, Kobayashi
Y, Kawaguchi M, Nomura H, Hirasawa A, Susumu N and Aoki D:
Epigenetic DNA hypermethylation: Clinical applications in
endometrial cancer (Review). Oncol Rep. 22:967–972. 2009.PubMed/NCBI
|
|
36
|
Chen QY, Jiao DM, Wang J, Hu H, Tang X,
Chen J, Mou H and Lu W: miR-206 regulates cisplatin resistance and
EMT in human lung adenocarcinoma cells partly by targeting MET.
Oncotarget. 7:24510–24526. 2016.PubMed/NCBI
|
|
37
|
Feng S, Zhu X, Fan B, Xie D, Li T and
Zhang X: miR-p19a-3p targets PMEPA1 and induces prostate cancer
cell proliferation, migration and invasion. Mol Med Rep.
13:4030–4038. 2016.PubMed/NCBI
|
|
38
|
Ge X, Liu X, Lin F, Li P, Liu K, Geng R,
Dai C, Lin Y, Tang W, Wu Z, et al: MicroRNA-421 regulated by HIF-1α
promotes metastasis, inhibits apoptosis, and induces cisplatin
resistance by targeting E-cadherin and caspase-3 in gastric cancer.
Oncotarget. 7:24466–24482. 2016.PubMed/NCBI
|
|
39
|
Ma M, He M, Jiang Q, Yan Y, Guan S, Zhang
J, Yu Z, Chen Q, Sun M, Yao W, et al: MiR-487a promotes
TGF-β1-induced EMT, the migration and invasion of breast bancer
bells by directly targeting MAGI2. Int J Biol Sci. 12:397–408.
2016. View Article : Google Scholar :
|
|
40
|
Zhang L, Wang W, Li X, He S, Yao J, Wang
X, Zhang D and Sun X: MicroRNA-155 promotes tumor growth of human
hepatocellular carcinoma by targeting ARID2. Int J Oncol.
48:2425–2434. 2016.PubMed/NCBI
|
|
41
|
Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L,
Sun L, Liu J, Yang Z and Ran Y: Tumor-suppressive miR-663 gene
induces mitotic catastrophe growth arrest in human gastric cancer
cells. Oncol Rep. 24:105–112. 2010.PubMed/NCBI
|
|
42
|
Lehmann U, Hasemeier B, Christgen M,
Müller M, Römermann D, Länger F and Kreipe H: Epigenetic
inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J
Pathol. 214:17–24. 2008. View Article : Google Scholar
|
|
43
|
Potapova A, Albat C, Hasemeier B,
Haeussler K, Lamprecht S, Suerbaum S, Kreipe H and Lehmann U:
Systematic crossvalidation of 454 sequencing and pyrosequencing for
the exact quantification of DNA methylation patterns with single
CpG resolution. BMC Biotechnol. 11:62011. View Article : Google Scholar
|
|
44
|
Yan-Fang T, Jian N, Jun L, Na W, Pei-Fang
X, Wen-Li Z, Dong W, Li P, Jian W, Xing F, et al: The promoter of
miR-663 is hypermethylated in Chinese pediatric acute myeloid
leukemia (AML). BMC Med Genet. 14:742013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang Y, Wang LL, Li YH, Gao XN, Liu Y and
Yu L: Effect of CpG island methylation on microRNA expression in
the k-562 cell line. Biochem Genet. 50:122–134. 2012. View Article : Google Scholar
|
|
46
|
Hong Q, Yu S, Geng X, Duan L, Zheng W, Fan
M, Chen X and Wu D: High concentrations of uric acid inhibit
endothelial cell migration via miR-663 which regulates phosphatase
and tensin homolog by targeting transforming growth Factor-beta1.
Microcirculation. 22:306–314. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Narkiewicz J, Lapinska-Szumczyk S,
Zurawa-Janicka D, Skorko-Glonek J, Emerich J and Lipinska B:
Expression of human HtrA1, HtrA2, HtrA3 and TGF-beta1 genes in
primary endometrial cancer. Oncol Rep. 21:1529–1537.
2009.PubMed/NCBI
|
|
48
|
Kogure T, Kondo Y, Kakazu E, Ninomiya M,
Kimura O and Shimosegawa T: Involvement of miRNA-29a in epigenetic
regulation of transforming growth factor-beta-induced
epithelialmesenchymal transition in hepatocellular carcinoma.
Hepatol Res. 44:907–919. 2014. View Article : Google Scholar
|
|
49
|
Zhang Q, Chen L, Helfand BT, Jang TL,
Sharma V, Kozlowski J, Kuzel TM, Zhu LJ, Yang XJ, Javonovic B, et
al: TGF-β regulates DNA methyltransferase expression in prostate
cancer, correlates with aggressive capabilities, and predicts
disease recurrence. PLoS One. 6:e251682011. View Article : Google Scholar
|
|
50
|
Kanai Y, Ushijima S, Kondo Y, Nakanishi Y
and Hirohashi S: DNA methyltransferase expression and DNA
méthylation of CPG islands and peri-centromeric satellite regions
in human colorectal and stomach cancers. Int J Cancer. 91:205–212.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Roll JD, Rivenbark AG, Jones WD and
Coleman WB: DNMT3b overexpression contributes to a hypermethylator
phenotype in human breast cancer cell lines. Mol Cancer. 7:152008.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nosho K, Shima K, Irahara N, Kure S, Baba
Y, Kirkner GJ, Chen L, Gokhale S, Hazra A, Spiegelman D, et al:
DNMT3B expression might contribute to CpG island methylator
phenotype in colorectal cancer. Clin Cancer Res. 15:3663–3671.
2009. View Article : Google Scholar : PubMed/NCBI
|