Introduction
Squamous cell carcinoma (SCC) is the most frequent
tumour entity within head and neck cancers occurring in 90% of
patients (1, 2) and the 6th most frequent cancer
worldwide (3, 4). Despite advances in therapy, the
prognosis for HNSCC patients remains poor, with a 5-year survival
of 46–50% (5, 6). As exogenous factors, alcohol
consumption and smoking are assumed to cause lesions and HNSCC in a
dose-dependent manner (7–9). An infection of human papilloma virus
(HPV) is present in ~25% of HNSCC and is associated with an
improved prognosis (10, 11). Also, endogenous risk factors, such
as genetic predisposition may promote hereditary HNSCC (12, 13).
The acquisition of somatic mutations during HNSCC
development is well understood for the tumour suppressor gene TP53,
which is mutated frequently in 30–78% of HNSCCs (14–19).
TP53 mutations are associated with a reduced radiosensitivity of
HNSCC tumours and with poor prognosis (20). Other potentially important genes in
HNSCC are the genes known to be mutated in Fanconi anemia (FA).
Patients with this rare autosomal-recessive disorder have a 500- to
700-fold increased risk to develop HNSCC (21–23)
and suffer from chromosomal instability, predisposition to
congenital anomalies, bone marrow failure and cancer.
FA genes are candidate genes for
HNSCC
Sixteen FA genes are currently known, the gene
products are involved in the surveillance and repair of DNA
crosslinks (24). A recent study
found germline mutations of FA genes in 27 analysed FA families
(25). All FA genes are active in
the S-phase of the cell cycle, where DNA damage recognized during
DNA replication triggers the accumulation of FA proteins and their
interaction with other repair mechanisms. In the present study, we
focused on three FA genes that have not been studied extensively,
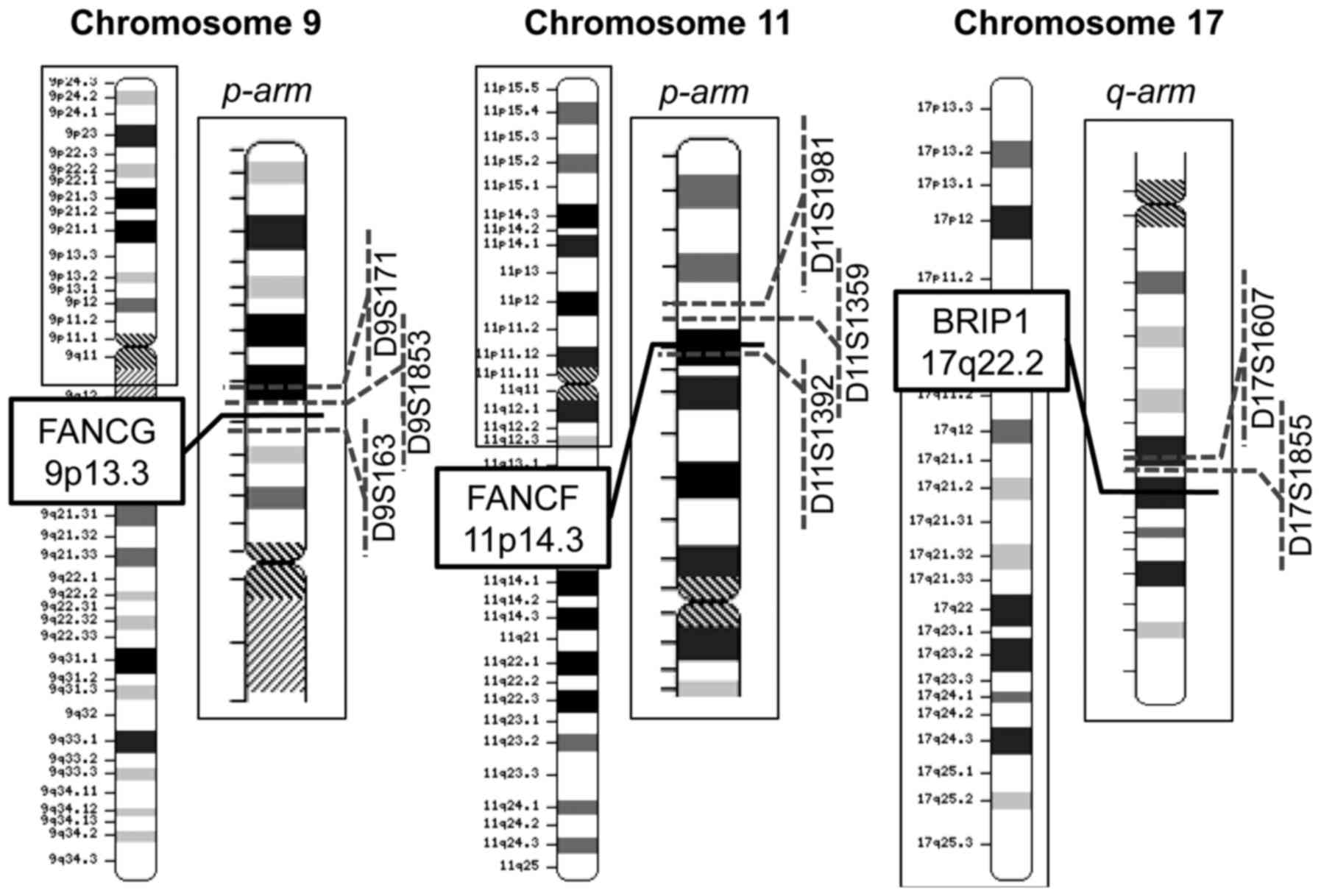
the FANCF, FANCG and BRIP1.
Expression analyses using quantitative PCR showed
reduced expression of all three genes, FANCF, FANCG
and BRIP1 in HNSCC, with a reduction of gene expression by
up to 40-fold (26). A possible
cause for the reduced expression of FA genes is an allelic gene
loss that can be detected as a loss of heterozygosity (LOH).
Measurements of LOH have previously indicated FA genes frequently
altered in oral HNSCC (27–31).
A frequent LOH on chromosome 9 for instance, is potentially
overlapping with many genes such as tumour suppressors p16 and
FANCG (32).
Clinical relevance
LOH in FA genes may impact the prognosis for HNSCC
patients under different treatment regimens. It was shown that
poly(ADP-ribose) polymerase (PARP) inhibitors can be used to kill
specifically FANCD1 (BRCA2)-deficient tumour cells
(33). Recently, the assessment of
FA genes, such as FANCD2 was proposed to aid patient
stratification for treatment with DNA inter-strand crosslinking
agents and PARP inhibitors (34).
Thus, FA genes may be used as predictive biomarkers in cancer
therapy.
The three FA genes studied here
Fanconi anemia, complementation group F -
FANCF is localized on chromosome 11p14.3 and involved in
stabilization of multimeric FA protein complexes (35, 36). The protein, Fancf interacts with
Fanca, Fancc and Fancg to stabilize the dimers of Fanca and Fancc
as well as Fanca and Fancg which are core components of the FA DNA
repair pathway (24, 37). Methylation of FANCF is
associated with a decreased expression of the gene and with ovary,
breast, lung, cervix and testis cancer (38–42).
Fanconi anemia, complementation group G -
FANCG (XRCC9) is localized on chromosome 9p13.3 (43). Fancg interacts with Fanca and Fancf
and supports the formation of the DNA repair core complex (44). Mutations in FANCG lead to
chromosomal instability in a number of different cancers (45).
The BRCA1 interacting protein C-terminal
helicase 1 -BRIP1 (FANCJ) is localized on chromosome 17q22.2
(46–48). BRIP1 associates with
BRCA1 and thus, contributes to genomic stability through its
role in cellular DNA repair (48,
49). Mutations in BRIP1
are associated with hereditary breast cancer and Fanconi anemia
(50).
In this study, we established microsatellite markers
for the assessment of LOH in three genetic loci comprising FA
genes. To identify a potential effect on patient survival in oral
HNSCC, we evaluated LOH in FA genes and clinical parameters in
survival analyses. We then discuss an observed weak association of
LOH in one of the three FA genes and considerable frequencies of
LOH in all three genes in light of potential therapeutic
relevance.
Materials and methods
Samples
All patients were enrolled for observation and
treatment of HNSCC between 1993 and 2007 at the Department of Oral
and Maxillofacial Surgery, University Hospital Carl Gustav Carus,
Technische Universität Dresden (Dresden, Germany). Fifty-four
patients, 40 male and 14 female, with a median age of 60 years were
included in the study (Table I).
We collected clinical information about the tumour-, node- and
metastatic stage as well the radiotherapy and chemotherapy used. To
study the effects of lifestyle factors we obtained data on smoking
and alcohol consumption for each patient.
 | Table IClinical data for patients evaluated
for loss of heterozygosity (LOH) in FA gene regions. |
Table I
Clinical data for patients evaluated
for loss of heterozygosity (LOH) in FA gene regions.
| Variables | No LOH (n=23) | LOH FANCF
region (n=9) | LOH FANCG
region (n=22) | LOH BRIP1
region (n=6) | Any LOH (n=30) | All (n=54) |
|---|
| Age (median, 1st
and 3rd quartile) | 59 (46;66.50) | 55 (53;68) | 59.5
(52.25;66) | 57
(51.25;62.75) | 60 (53.5;66) | 60 (52;66.75) |
| Gender | | | | | | |
| 0-Female | 6 (26.09%) | 2 (22.22%) | 4 (18.18%) | 2 (33.33%) | 7 (23.33%) | 14 (25.93%) |
| 1-Male | 17 (73.91%) | 7 (77.78%) | 18 (81.81%) | 4 (66, 67%) | 23 (76.67%) | 40 (74.07%) |
| Tumour stage | | | | | | |
| 2 | 2 (8.7%) | 1 (11.11%) | 2 (9.09%) | 0 (0%) | 2 (10%) | 5 (9.25%) |
| 3 | 2 (8.7%) | 2 (22.22%) | 6 (27.27%) | 1 (16.67%) | 8 (26.67%) | 10 (18.52%) |
| 4 | 19 (82.61%) | 6 (66.67%) | 14 (63.63%) | 5 (83.33%) | 19 (63.33%) | 39 (72.22%) |
| Node stage | | | | | | |
| 0 | 12 (52.17%) | 1 (11.11%) | 5 (22.72%) | 2 (33.33%) | 6 (20%) | 19 (35.19%) |
| 1 | 1 (4.35%) | 2 (22.22%) | 5 (22.72%) | 2 (33.33%) | 9 (30%) | 10 (18.52%) |
| 2 | 8 (34.78%) | 6 (66.67%) | 11 (50%) | 2 (33.33%) | 14 (46.67%) | 22 (40.74%) |
| 3 | 0 (0%) | 0 (0%) | 1 (4.55%) | 0 (0%) | 1 (3.33%) | 1 (1.85%) |
| Metastasis
stage | | | | | | |
| 0 | 19 (82.61%) | 8 (88.89%) | 22 (100%) | 6 (100%) | 29 (96.67%) | 49 (90.74%) |
| 1 | 1 (4.35%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1.85%) |
| Tumour grading | | | | | | |
| 1 | 1 (4.35%) | 0 (0%) | 2 (9.09%) | 0 (0%) | 2 (6.67%) | 3 (5.56%) |
| 2 | 13 (56.52%) | 4 (44.44%) | 14 (63.63%) | 5 (83.33%) | 20 (66.67%) | 34 (62.96%) |
| 3 | 3 (13.04%) | 3 (33.33%) | 4 (18.18%) | 0 (0%) | 5 (16.67%) | 8 (14.81%) |
| Recurrence | | | | | | |
| 0 | 15 (65.22%) | 5 (55.56%) | 14 (63.64%) | 4 (66.67%) | 20 (66.67%) | 35 (64.81%) |
| 1 | 8 (34.78%) | 4 (44.44%) | 8 (36.36%) | 2 (33.33%) | 10 (33.33%) | 19 (35.19%) |
| Surgery | 21 (91.30%) | 7 (77.78%) | 18 (81.82%) | 6 (100%) | 26 (86.67%) | 48 (88.89%) |
| Radiotherapy | 21 (91.30%) | 9 (100%) | 20 (90.90%) | 6 (100%) | 28 (93.33%) | 49 (90.74%) |
| Chemotherapy | 8 (34.78%) | 4 (44.44%) | 5 (22.73%) | 1 (16.67%) | 7 (23.33%) | 15 (27.78%) |
|
Nicotin/Alcohol | 18 (78.26%) | 7 (77.78%) | 19 (86.36%) | 3 (50%) | 25 (83.33%) | 43 (79.63%) |
| Survival (months,
median, 1st and 3rd Qu.) | 21.33 (13.61;
73.77) | 10.30 (7.53;
22.13) | 20.40 (8.30;
98.21) | 40.73 (16.10;
73.72) | 19.48 (7.80;
78.31) | 20.60 (11.30;
78.80) |
Tumour specimens were initially chosen to represent
primary HNSCC tumours of the locally advanced stages T3 and T4
(51) and were collected in
surgeries and biopsies. Five tumour samples were later re-assigned
to stage T2. Tumour and corresponding blood samples were collected
during the time of surgery or biopsy, snap-frozen in liquid
nitrogen and stored at −80°C. Written informed consent was obtained
from all the patients.
Ethics statement
The study was approved by the ethics committee of
the Faculty for Medicine at the Technische Universität Dresden,
Germany (study ID EK37022001).
DNA extraction, microsatellite
amplification and detection of LOH
DNA extraction from tumour samples and corresponding
blood was performed with the QIAamp DNA Mini kit (Qiagen, Hilden,
Germany) following standard procedures. Eight microsatellite
markers were used to determine heterozygosity and microsatellite
instability (MSI) using their polymorphic repeat length. We used
eight pairs of oligonucleotide primers (Applied Biosystems,
Darmstadt, Germany) which were previously published (Table II) in polymerase chain reaction
(PCR) to amplify loci in proximity to three FA genes (Fig. 1) (43, 44,
52–59). Six loci were repeats of
dinucleotides (D9S1853, D9S171, D9S163, D11S1359, D17S1855 and
D17S1607) and two were repeats of tetranucleotides (D11S1392 and
D11S1981). We established three multiplex PCR reactions and one
singleplex PCR. The reactions contained primers, MgCl2
(concentrations given in Table
III), and 100 ng of template DNA in the Qiagen Multiplex PCR
kit (Qiagen) and were run in a total reaction volume of 25
µl on a PCR cycler (Bio-Rad Laboratories, München, Germany).
After an initial heating step at 95°C for 15 min, PCR cycling was
carried out at 95°C for 35 sec, 56°C for 90 sec and 72°C for 90 sec
for 27 cycles. Fluorescent labelling of one primer each per pair
was utilized to detect the amplification products in capillary gel
electrophoresis using a 3130xl Genetic Analyzer, the GeneScan 500
LIZ size standard and software GeneMapper version 4.0 (all Applied
Biosystems).
 | Table IIPrimers used for amplification of
microsatellite markers. |
Table II
Primers used for amplification of
microsatellite markers.
| Primer | Sequence | Label | Product size
(bp) | Refs. |
|---|
| D9S171 s |
AGCTAAGTGAACCTCATCTCTGTCT | VTC | 158–177 | (52–54) |
| D9S171 as |
ACCCTAGCACTGATGGTATAGTCT | | | |
| D9S1853 s |
GATCCAGCCTCACTGAA | 6-FAM | 247–265 | (44, 52) |
| D9S1853 as |
TTGGGCATAGAATTTTTACTTT | | | |
| D9S163 s |
TGCTGCACATCTTAGGGAGT | NED | 270–271 | (52, 55) |
| D9S163 as |
ACAGCGCTCAGAAATCATATAA | | | |
| D11S1359 s |
TTGGAAGACACATGCACAAA | NED | 148 | (43) |
| D11S1359 as |
ATTTTCCAGCCTCCATAATC | | | |
| D11S1981 s |
AATTCCTTTACTCCAGAAAGG | VTC | 134–178 | (56) |
| D11S1981 as |
CAGATTTCTGCTTTCCCAGA | | | |
| D11S1392 s |
TTGCATCCATACGGAAAGTC | 6-FAM | 200–220 | (57) |
| D11S1392 as |
ACATCTGAGACTTGTAGTAGAAGGC | | | |
| D17S1607 s |
CAGATAAAAAACACAAGTTTCTGAC | NED | 103–123 | (58) |
| D17S1607 as |
GCTCCACCCCAGACCTA | | | |
| D17S1855 s |
GGGGACCNCTAGAAACC | PET | 219–225 | (52, 58) |
| D17S1855 as |
GAGAATACATTGTAACAACTCCAGT | | | |
 | Table IIICombinations of primers and
concentrations for multiplex PCR. |
Table III
Combinations of primers and
concentrations for multiplex PCR.
| Combination | Primer name | Primer
concentration | MgCl2
concentration |
|---|
| 1 | D9S171 | 0.20 µM | 1.8 mM |
| D9S163 | 0.15 µM |
| D17S1607 | 0.30 µM |
| 2 | D11S1392 | 0.20 µM | 1.5 mM |
| D11S1359 | 0.20 µM |
| 3 | D17S1855 | 0.20 µM | 1.5 mM |
| D11S1981 | 0.20 µM |
| 4 | D9S1853 | 0.20 µM | 1.5 mM |
Larger microsatellite markers may show LOH more
frequently than shorter markers, because they amplify less well in
PCR. To avoid this problem we used a high amount of input DNA (100
ng). Consequently, the larger microsatellite markers showed a lower
frequency of LOH for FANCF and FANCG and a higher
frequency of LOH for BRIP1 when compared to the smaller
markers in the same locus (Table
IV). When comparing the two alleles of each microsatellite
marker, the LOH affected the larger allele in 33% of the markers
(standard deviation 16.6). Thus, a preferential amplification of
the smaller microsatellite markers is not apparent in our
samples.
 | Table IVLoss of heterozygosity (LOH) in FA
gene regions. |
Table IV
Loss of heterozygosity (LOH) in FA
gene regions.
| Tumour no. | FANCG
| FANCF
| BRIP1
|
|---|
D9S171
158–177 bp | D9S1853
247–265 bp | D9S163
270–271 bp | D11S1981
134–178 bp | D11S1359
148 bp | D11S1392
200–220 bp | D17S1607
103–123 bp | D17S1855
219–225 bp |
|---|
| 2 | LOH | LOH | n.i. | i. | i. | i. | i. | i. |
| 3 | n.i. | i. | i. | i. | i. | i. | i. | i. |
| 7 | LOH | n.i. | LOH | i. | i. | n.i. | i. | i. |
| 14 | i. | i. | i. | LOH | LOH | i. | LOH | n.i. |
| 20 | i. | n.i. | i. | i. | i. | i. | i. | i. |
| 23 | i. | LOH | i. | i. | i. | i. | n.i. | i. |
| 26 | LOH | n.i. | n.i. | LOH | LOH | LOH | i. | i. |
| 27 | i. | i. | i. | n.i. | i. | n.i. | n.i. | i. |
| 30 | i. | n.i. | i. | i. | i. | i. | i. | i. |
| 37 | i. | i. | n.i. | n.i. | n.i. | n.i. | i. | i. |
| 40 | i. | i. | i. | i. | n.i. | i. | n.i. | i. |
| 48 | n.i. | LOH | i. | i. | i. | i. | i. | i. |
| 53 | LOH | LOH | LOH | i. | i. | i. | n.i. | i. |
| 60 | i. | i. | n.i. | i. | i. | i. | n.i. | LOH |
| 101 | i. | i. | i. | i. | i. | i. | i. | i. |
| 107 | i. | i. | i. | i. | LOH | LOH | i. | n.i. |
| 114 | i. | n.i. | n.i. | i. | i. | i. | i. | n.i. |
| 116 | LOH | n.i. | LOH | n.i. | LOH | n.i. | LOH | i. |
| 117 | i. | i. | n.i. | i. | n.i. | i. | n.i. | i. |
| 118 | LOH | n.i. | n.i. | i. | n.i. | n.i. | n.i. | i. |
| 120 | i. | i. | n.i. | i. | i. | i. | i. | i. |
| 121 | n.i. | i. | i. | n.i. | n.i | i. | i. | i. |
| 123 | LOH | LOH | i. | n.i. | i. | i. | i. | i. |
| 144 | LOH | LOH | i. | i. | i. | i. | i. | i. |
| 145 | i. | n.i. | LOH | i. | n.i. | i. | i. | n.i. |
| 150 | LOH | LOH | LOH | i. | i. | MSI | i. | n.i. |
| 152 | i. | i. | i. | i. | i. | i. | i. | n.i. |
| 154 | LOH | LOH | LOH | i. | i. | i. | i. | i. |
| 155 | n.i. | i. | n.i. | i. | i. | n.i. | n.i. | LOH |
| 157 | n.i. | i. | i. | i. | i. | n.i. | i. | i. |
| 171 | i. | n.i. | i. | LOH | LOH | LOH | i. | i. |
| 179 | i. | n.i. | i. | i. | n.i. | i. | i. | i. |
| 180 | n.i. | i. | n.i. | i. | i. | i. | n.i. | LOH |
| 181 | i. | n.i. | n.i. | i. | i. | n.i. | i. | i. |
| 185 | i. | i. | n.i. | i. | i. | i. | i. | i. |
| 193 | n.i. | i. | i | n.i. | n.i. | LOH | i. | i. |
| 196 | i. | i. | n.i. | i. | i. | i. | i. | i. |
| 206 | i. | i. | i. | n.i. | i. | i. | n.i. | i. |
| 213 | i. | i. | n.i. | i. | i. | i. | i. | n.i. |
| 325 | LOH | LOH | LOH | i. | n.i. | i. | i. | i. |
| 326 | LOH | n.i. | n.i. | i. | i. | i. | i. | i. |
| 328 | LOH | n.i. | LOH | i. | n.i. | n.i. | i. | i. |
| 336 | i. | i. | i. | i. | n.i. | i. | i. | i. |
| 385 | LOH | i. | LOH | i. | i. | i. | i. | i. |
| 386 | n.i. | LOH | LOH | i. | n.i. | i. | i. | i. |
| 393 | n.i. | n.i. | i. | i. | i. | LOH | i. | i. |
| 401 | n.i. | i. | i. | i. | i. | i. | i. | i. |
| 409 | i. | i. | i. | i. | i. | i. | i. | i. |
| 457 | i. | n.i. | i. | i. | i. | i. | i. | i. |
| 458 | n.i. | LOH | n.i. | n.i. | n.i. | LOH | n.i. | i. |
| 474 | LOH | LOH | n.i. | i. | i. | i. | i. | i. |
| 477 | n.i. | n.i. | i. | i. | i. | i. | n.i. | i. |
| 478 | LOH | n.i. | LOH | i. | i. | i. | LOH | n.i. |
| 479 | LOH | LOH | LOH | LOH | LOH | n.i. | i. | MSI |
| ∑ i. | 42/54 | 37/54 | 37/54 | 46/54% | 41/54 | 44/54 | 42/54 | 46/54 |
| 77,78% | 68,52% | 68,52% | 85,19 | 75,93% | 81,48% | 77,78% | 85,19% |
| ∑ LOH | 17/42 | 13/37 | 12/37 | 4/46 | 6/41 | 6/44 | 3/42 | 3/46 |
| 40,48% | 35,14% | 32,43% | 8,69% | 14,63% | 13,64% | 7,14% | 6,52% |
To detect LOH all informative microsatellite markers
were analysed for their peak area (60–62).
The peak area of the higher peak was divided by the area of the
lower peak, and a quotient of the values for tumour and blood was
calculated. When above 1.5, we called LOH in the tumour tissue as
previously described (62).
Non-informative and unstable (MSI) markers were excluded.
Statistical analysis of clinical
data
Clinical data were obtained from the regional clinic
cancer registry Dresden (Table I).
To investigate if LOH was more frequent in subgroups of the
patients we tested for an association of LOH with age (above vs.
below 60 years), gender, tumour stages (T2, T3 and T4), smoking,
alcohol consumption and recurrent disease (Table I). For this we used Chi-square
tests and Fisher's exact tests if expected frequencies were <5
using IBM SPSS Statistics version 19 (IBM, Ehningen, Germany).
We further investigated the potential impact of LOH
on patient survival R v. 3.0. (63). Survival time was obtained and right
censored for alive subjects and also if death occurred not due to
HNSCC. First, we tested for an association between LOH in regions
around the FA genes, the mentioned clinical parameters and overall
patient survival using Kaplan-Meier curves and log-rank tests
(Table V). We reported raw
P-values using a significance level alpha of 0.05 as well as
corrected significance levels (alpha') depending on the number of
tests carried out according to the Bonferroni method (64).
 | Table VP-values from log-rank tests
comparing survival in HNSCC patients related to clinical variables
and LOH at FA gene regions. |
Table V
P-values from log-rank tests
comparing survival in HNSCC patients related to clinical variables
and LOH at FA gene regions.
| Test no. | Variables | All patients
| Patients with
tumour stage T4
|
|---|
| P-value | n | P-value | n |
|---|
| 1 | Gender | 0.495 | 53 | 0.819 | 39 |
| 2 | Tumour
stage | 0.636 | 53 | | |
| 3 | Node
stage |
2.69e-12 | 51 |
2.58e-09 | 37 |
| 4 | Metastasis
stage | 0.662 | 49 | 0.868 | 36 |
| 5 | Tumour
grading | 0.0418 | 44 | 0.108 | 32 |
| 6 | Surgery |
0.000466 | 53 | 0.00368 | 39 |
| 7 |
Radiotherapy | 0.246 | 53 | 0.197 | 39 |
| 8 |
Chemotherapy | 0.0742 | 53 | 0.133 | 39 |
| 9 |
Nicotin/Alcohol | 0.24 | 53 | 0.434 | 39 |
| 10 | D11S1981 | 0.19 | 45 | 0.286 | 34 |
| 11 | D11S1359 | 0.0248 | 40 | 0.0617 | 32 |
| 12 | D11S1392 | 0.0326 | 42 | 0.00254 | 31 |
| 13 | FANCF
region | 0.00617 | 52 | 0.00595 | 38 |
| 14 | D9S171 | 0.344 | 41 | 0.902 | 31 |
| 15 | D9S1853 | 0.901 | 36 | 0.309 | 27 |
| 16 | D9S163 | 0.438 | 35 | 0.573 | 26 |
| 17 | FANCG
region | 0.682 | 53 | 0.452 | 39 |
| 18 | D17S1607 | 0.983 | 41 | 0.876 | 31 |
| 19 | D17S1855 | 0.768 | 44 | 0.586 | 30 |
| 20 | BRIP1
region | 0.762 | 53 | 0.537 | 39 |
| 21 | >1 FA gene
region | 0.6 | 53 | 0.483 | 39 |
To investigate the combined effect of several
variables on patient survival we employed Cox proportional hazards
(PH) regression models in R v. 3.0. (63). We estimated the proportional
hazards for different sets of variables on survival, firstly using
all variables in one Cox PH model. Secondly, we tested all
variables which were significant in the first model. A third test
used backwards elimination, starting with all and deleting the
least significant variable at each step until reaching a stage
where all remaining variables were significant. Since smoking and
alcohol consumption coincided in many patients and both are seen as
mutagenic substances we merged them into one binary variable
(absence/presence) for survival analyses. For analyses of LOH and
survival in Cox PH we evaluated each FA gene region independently.
However, we did not evaluate each microsatellite marker
independently since those showed similar results as the
corresponding FA gene regions in log-rank tests.
Analysis of copy number data
From a published dataset of 106 HNSCC genotyped on
microarrays (18) we extracted
called copy number variants (CNV) that spanned the genes
FANCF, FANCG and BRIP1.
If the reported ploidy deviated from 2 and was
<1.8 or >2.2, we noted a loss or a gain, respectively.
Expression data
We accessed publicly available expression profiles
of three studies on HNSCC tumours and corresponding normal tissues
in NCBI Gene Expression Omnibus. For accessions GDS2520 and GDS3838
we queried the genes of interest directly retrieving lists of
expression values. Fold-change in expression was calculated by
comparing the cancerous and normal tissue per each tested patient.
For GSE55550 we queried for differentially expressed genes by
grouping (pooled) normal tissues and tumours. Then we extracted
array probes corresponding to our genes of interest from the
resulting gene lists and retrieved the fold change in expression
when the raw P-value was <0.05.
Differential gene expression analysis
using TCGA data
We queried the Cancer Genome Atlas data for head and
neck squamous cell carcinoma comprising 521 samples with RNAseq
data (queried in November 2016) using the TCGA Browser v0.9 at
http://tcgabrowser.ethz.ch:3839/TEST/(65). The top 200 differentially expressed
genes for FANCG, FANCF and BRIP1 were
subjected to the gene set enrichment analysis (66). We computed overlaps with hallmarks
gene sets and with gene sets of known molecular function.
In order to identify protein partners that interact
physically with FANCG, FANCF and BRIP1, a
network analysis was performed using GeneMANIA (67).
Results
Detection of LOH
The information content of the microsatellite
markers was high with 68–85% of informative patients per marker, in
53 of 54 patients at least one microsatellite marker was
informative (Table IV). Patient
37 had to be excluded from the analysis of FANCF since none
of the markers was informative here. We detected LOH in 30 of 53
(57%) patients (Table IV), 23
patients had at least one informative marker but did not show
LOH.
LOH was detected most frequently in the gene region
containing FANCG in 40.74% (22/54) of HNSCCs. The gene
region of FANCF showed LOH in 16.98% (9/53) and the
BRIP1 region was affected in 11.11% (6/54) of HNSCC. Patient
116 showed LOH in all three FA gene regions. Patients 26, 458 and
479 showed LOH in FANCG and FANCF regions. Patient
478 showed LOH in FANCG and BRIP1 and patient 14 in
FANCF and BRIP1 regions.
Association analysis
We tested if the frequency of LOH was associated
with age (either below or above 60 years), gender, tumour stages
(T2, T3 and T4), node stage, metastasis stage, histological tumour
grading, smoking or alcohol consumption and recurrent disease. None
of these variables was significantly associated with an increased
or decreased frequency of LOH at the FA gene regions, when
combining several microsatellite markers per region (Fisher's exact
tests and Chi-square tests, data not shown). Individual
microsatellite markers were, moreover, not associated with any
factor except for D17S1607 (in the BRIP1 region) which was
associated with smoking or alcohol consumption (P= 0.029). However,
this association was not significant when corrected for multiple
testing (alpha' of 0.006) and LOH overall was more frequent in
patients without smoking or alcohol consumption.
Survival analysis
We tested for an association of the LOH in regions
of FA genes and overall patient survival using Kaplan-Meier curves
and log-rank tests (Table V).
Higher lymph node stages were most significantly associated with
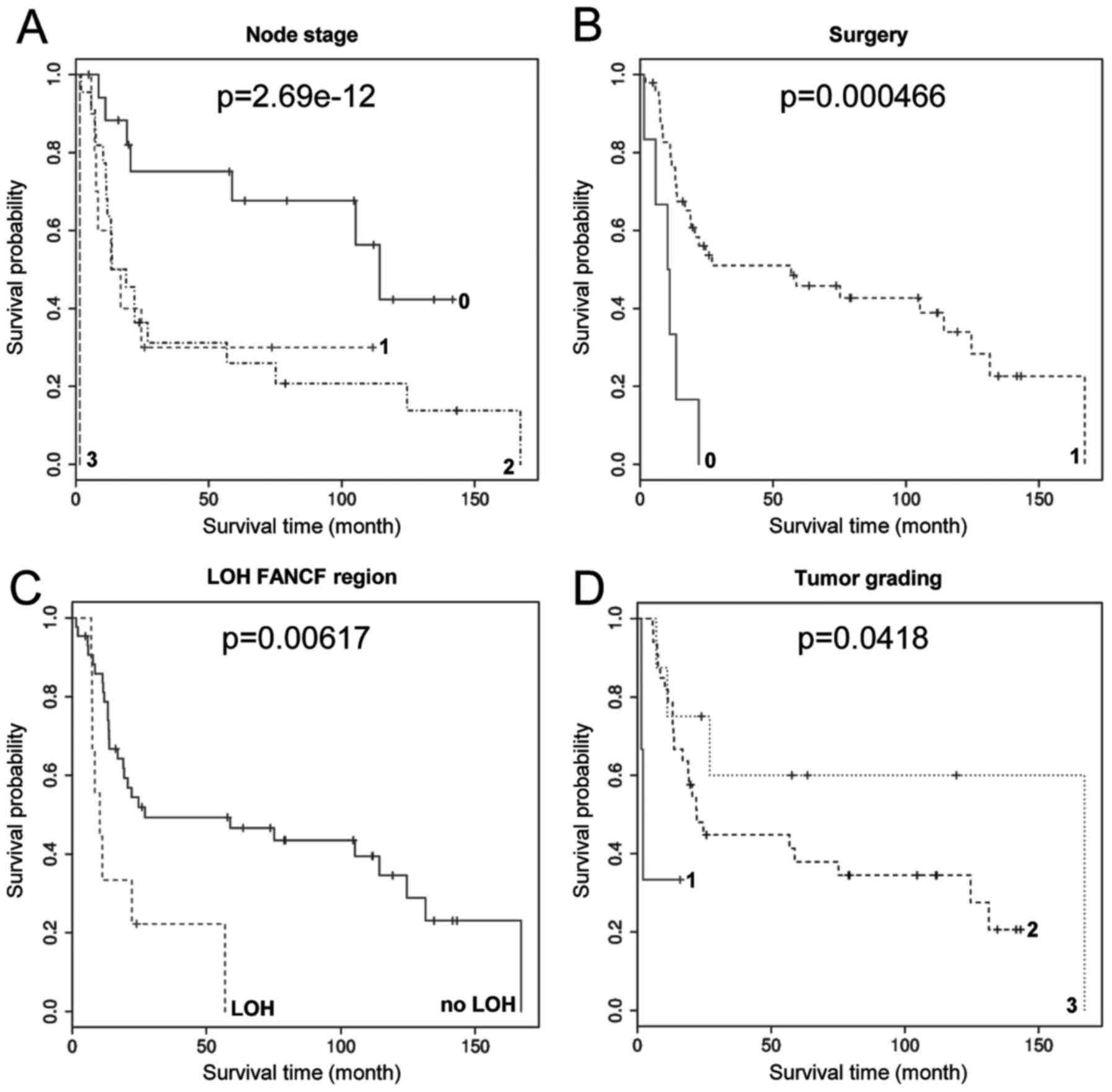
decreased survival (P=2.69e-12) and surgery was most significantly
associated with improved survival (P=0.0004; Fig. 2). Node stage and surgery were
significant also when corrected for multiple testing (21 variables:
alpha'=0.0024 or 13 variables: alpha'=0.0038, see Table V). LOH in the FANCF region
was associated with decreased survival (P=0.006; Fig. 2 and Table I) and higher tumour grading was
associated with increased survival (P=0.0418; Fig. 2). However, these associations were
not significant when corrected for multiple testing. LOH in
FANCG and BRIP1 regions were not significantly
associated with survival, accordingly LOH in more than one FA gene
region was also not significantly associated with survival.
Stratification for patients with tumour stage T4 showed similar
associations. Only node stage remained significantly associated
after correcting for multiple testing (Table V). We did not stratify for T3 and
T2 patients since they were too few (≤10).
To increase our understanding of LOH in FA gene
regions and survival in the context of clinical covariates we
employed Cox PH regression models (Table VI). We evaluated three different
Cox PH models: firstly, a model with all variables, secondly one
with only those variables which were significant in the previous
log-rank tests. We also used backwards elimination of variables to
obtain the third Cox PH model with the most significant likelihood
ratio test, Wald test and score (log-rank) test. Applying a raw
significance level of alpha=0.05, surgery was always significantly
associated with survival (P-values between 0.003 and 0.014) and
tumour grading was significantly associated with survival when
fewer variables were in the model (P-values between 0.047 and
0.019). Node stage was significantly associated only in the Cox PH
model with best likelihood ratio, Wald and score tests (P=0.025).
LOH in FA gene regions were not significantly associated with
survival in the Cox PH models. Moreover, age, gender, tumour stage,
metastatic stage, radiotherapy, chemotherapy and smoking or alcohol
consumption were not significantly associated with survival. When
correcting for multiple testing, applying an alpha' of 0.0167 (due
to the three Cox PH models evaluated), only surgery showed a
significant association with survival, most prominently in the Cox
PH model with the best test statistics.
 | Table VIP-values from Cox proportional
hazards models. |
Table VI
P-values from Cox proportional
hazards models.
| No. | Variables | One Cox PH model
with all variables
| One Cox PH model
with significance in log-rank tests of Kaplan-Meier analysis
| Cox PH model with
best likelihood ratio, Wald and Score tests
|
|---|
| P-values | LCI | UCI | P-values | LCI | UCI | P-values | LCI | UCI |
|---|
| 1 | Gender | 0.88885 | 0.206970 | 39.176 | | | | | | |
| 2 | Tumour stage | 0.57460 | 0.509899 | 33.673 | | | | | | |
| 3 | Node stage | 0.34330 | 0.732887 | 24.415 | 0.1584 | 0.87206 | 23.155 | 0.02539 | 107.130 | 28.550 |
| 4 | Metastasis
stage | 0.21420 | 0.013535 | 26.236 | | | | 0.12550 | 0.01837 | 16.319 |
| 5 | Tumour grading | 0.10808 | 0.084520 | 12.771 | 0.0469 | 0.09622 | 0.9838 | 0.01890 | 0.06185 | 0.7785 |
| 6 | Surgery | 0.00298 | 0.006957 | 0.3615 | 0.0139 | 0.09975 | 0.7709 | 0.00265 | 0.04769 | 0.5267 |
| 7 | Radiotherapy | 0.97083 | 0.155178 | 69.173 | | | | | | |
| 8 | Chemotherapy | 0.53866 | 0.080760 | 37.230 | | | | | | |
| 9 |
Nicotin/Alcohol | 0.07382 | 0.040165 | 11.592 | | | | | | |
| 10 | FANCF region | 0.10518 | 0.631867 | 1262.340 | 0.1723 | 0.74051 | 53.587 | | | |
| 11 | FANCG
region | 0.37524 | 0.393851 | 118.390 | | | | | | |
| 12 | BRIP1
region | 0.86562 | 0.187164 | 73.337 | | | | | | |
| 13 | >1 FA gene
region | 0.09701 | 0.001591 | 17.071 | | | | | | |
| 14 | Age | 0.54531 | 0.919269 | 10.455 | | | | | | |
| Test
statistics: | | | | | | | | | | |
| n | | 40 | | | 42 | | | 41 | |
| Events | | 27 | | | 28 | | | 27 | |
| Rsquare | | 0.509 | | | 0.335 | | | 0.432 | |
| Likelihood ratio
test | 28.42 on 14 df,
P=0.01249 | 17.15 on 4 df,
P=0.001806 | 23.19 on 4 df,
P=0.0001162 |
| Wald test | 24.09 on 14 df,
P=0.04469 | 17.48 on 4 df,
P=0.001561 | 22.93 on 4 df,
P=0.0001308 |
| Score (log-rank)
test | 35.05 on 14 df,
P=0.001444 | 20.63 on 4 df,
P=0.0003744 | 30.97 on 4 df,
P=3.1e-06 |
We did not explore associations of survival and MSI,
because of a very small sample for MSI in only two patients.
Analysis of copy number data
To analyse if LOH in FANCF, FANCG and
BRIP1 may be associated with amplifications or deletions, we
checked if published copy number data of HNSCC (18) showed gains or losses of these genes
(Table VII). Twenty prercent of
HNSCC samples showed copy number variants (CNV) spanning
FANCF; 3.8% (4/105) were losses and 16.2% (17/105) were
gains. FANCG was lost in 28.6% (30/105) and gained in 6.7%
(7/105), in sum it was affected in 35.2% of samples. BRIP1
was lost in 31.6% (31/98) and gained in 4.1% (4/98), in sum
affected in 35.7%. For comparison, TP53 showed losses in 47%
(49/103) of the samples and did not show gains.
 | Table VIILOH, CNV and expression of
FANCF, FANCG and BRIP1 in HNSCC. |
Table VII
LOH, CNV and expression of
FANCF, FANCG and BRIP1 in HNSCC.
| Gene | LOH | CNV (18) | Expression |
|---|
| This study | Sum | Gain | Loss | 22 HNSCC, 22 normal
tissues, dataset GDS2520 (65) | 139 HNSCC, 16
normal tissues, dataset GSE55550 | 17 ESCC, 17 normal
tissues, dataset GDS3838 (66) |
| FANCF | 16.98% | 20% | 16.2% | 3.8% | NA | Underexpressed | Underexpressed
in14/17 |
| FANCG | 40.74% | 35.2% | 6.7% | 28.6% | Overexpressed in
17/22 | Overexpressed | Overexpressed in
14/17 |
| BRIP1 | 11.11% | 35.7% | 4.1% | 31.6% | NA | Overexpressed | Overexpressed in
15/17 |
Our frequencies of LOH correspond to the frequencies
of copy number variants (CNV) recently published in a genome wide
screen of HNSCC for FANCF (17% LOH, 20% CNV) and
FANCG (41% LOH, 35% CNV) (18). The estimates depart for
BRIP1 (11% LOH, 36% CNV) possibly reflecting differences
between the sample sets. However, at least for FANCF and
FANCG we think that our measurement of LOH approximate the
measurement of CNV in array based methods.
Analysis of expression data
One dataset, GDS2520, comprised 22 pairs of HNSCC
corresponding normal tissues (68). Here, FANCG was overexpressed
in 17 HNSCC samples when compared to the corresponding normal
tissue. The highest change in expression was 1.6-fold. Data for
FANCF and BRIP1 were not available in this dataset
(Table VII).
The second dataset (GSE55550, no publication
available) contained gene expression profiles of 139 HNSCC and 16
normal samples. For all of our three genes of interest, there was
an array probe that showed significant differential gene expression
(raw P-value<0.05). Here, FANCF was underexpressed in
HNSCC compared to normal tissues and FANCG and BRIP1
were overexpressed.
A third dataset (GDS3838) compared 17 esophageal
squamous cell carcinomas (ESCC) to corresponding normal tissues
(69). FANCF was
underexpressed in 14 of 17 samples. FANCG and BRIP1
were overexpressed in 14 of 17 and 15 of 17 samples, respectively.
The few samples that showed the opposite pattern (overexpression of
FANCF and underexpression of FANCG or BRIP1),
did not overlap.
Differential gene expression analysis
using TCGA data
In order to identify cellular signaling pathways
that are affected by mutations in FANCG, FANCF and
BRIP1 we performed a differential gene expression analysis
of the TCGA head and neck squamous cell carcinoma samples (n=521)
with subsequent gene set enrichment to identify affected
pathways.
The gene set enrichment analysis for FANCG
revealed a significant overlap with genes that perform transfers of
ubiquitin modifications (FDR q-value 0.00005 genes such as UBE2R2
and UBE2W), a process that has been described before to play a role
in the damage response of FANCG (70).
Differentially expressed genes associated with
FANCG are also of ribonucleotide binding activity (FDR
q-value 0.00006), such as XRCC3 which is together with FANCG
involved in homologous recombination to maintain chromosome
stability and repair DNA damage and also physically interacts with
FANCG (see below).
The gene set enrichment analysis for FANCF
revealed a highly significant overlap with genes that have
transcription factor activity (FDR q-value 4.86E-022), such as a
number of zinc finger proteins. However, these are somewhat
inconclusive as a plethora of transcriptions factors is involved in
the damage response.
The gene set enrichment analysis for BRIP1
revealed highly significant overlaps with genes involved in the
G2/M checkpoint, as in progression through the cell division cycle
(FDR q-value of 2.07e-41), such as e.g. BRCA2 and
BARD1 (BRCA1 associated RING domain 1). Also, genes
differentially expressed in association with BRIP1 are
preferably genes encoding cell cycle related targets of E2F
transcription factors (FDR q-value of 8.14e-30). Some of these,
BRCA2 and BRCA1 also physically interact with
BRIP1 (see below).
In order to complement the gene set enrichment,
protein partners that interact physically with FANCG,
FANCF and BRIP1 were identified in a network analysis
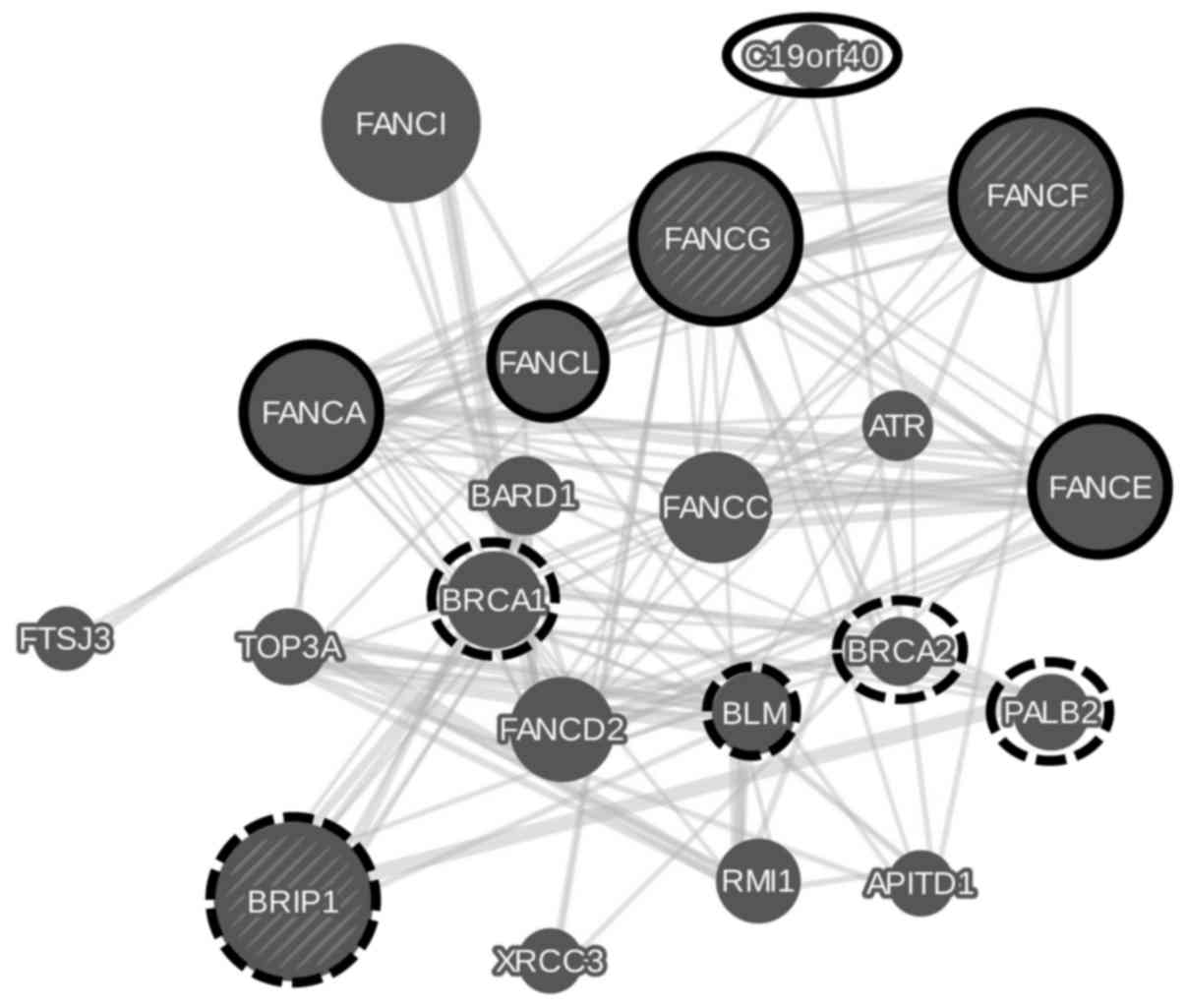
and are shown in Fig. 3. Highly
associated protein sets are members of the Fanconi anemia nuclear
complex (false discovery rate FDR of 9.72e-18) and proteins
involved in double-strand break repair (FDR of 1.79e-3).
Discussion
Frequent LOH in FA gene regions of
HNSCC
The microsatellite markers established here were
informative for almost all patients and allowed us to assess the
frequency of LOH at three FA gene regions in HNSCC tumours. LOH in
at least one of the analysed regions containing the genes
FANCF, FANCG and BRIP1 appears to be a
frequent event in HNSCC development in more than half of the cases
analysed here. Such a high frequency of LOH supports an assumed
role of these genes in HNSCC development, and might also influence
the success of HNSCC therapy.
The frequencies of LOH in the three studied genes is
much higher than that of reported point mutations (0.3–1.1%):
BRIP1 was mutated in 2 of 172 skin cancers, 4 of 173
esophageal cancers and 1 of 113 upper aerodigestive tract cancers,
and the mutational frequencies of FANCG and FANCF
were even lower (71). This
suggests minor role of point mutations, however, non-coding point
mutations have not been studied and may add onto these
frequencies.
Mutations in HNSCC are thought to be induced by
mutagens such as tobacco smoke and alcohol (7–9). In
the present study we observed a mild negative association between
smoking or alcohol consumption and FA mutation for one
microsatellite marker, suggesting that in some cases mutagenesis
induced by smoking or alcohol is not responsible for HNSCC
progression. The frequencies of LOH at FA gene regions reported
here should be seen as a minimum estimate since the distance
between FA genes and the used microsatellite markers was quite
large and result in false negative assignments. The FANCG
region presented with LOH in 41% of the tumours and is thus a
strong candidate for further study. Similarly, the tumour
suppressor CDKN2A on chromosome 9p has been implicated in cancer
development, and the loss of this gene is thought to be a frequent
event in various cancers and in HNSCC with an estimated 25%. A
proposed progression model of HNSCC based on CGH data involved an
early loss of 9p suggested ~80% of the samples (29). The LOH in this region may therefore
be driven by loss of the tumour suppressor CDKN2A rather than that
of FANCG. However, simultaneous loss of DNA repair mediated
by Fancg may promote the accumulation of mutations and a
deregulation of apoptosis at the same time (72, 73). Future studies on cancer genomes
will provide data to test whether heterozygosity is lost in CDKN2A
and FANCG independently, or whether the loss of FANCG
is a result of LOH in CDKN2A.
The markers used previously to study LOH in the
FANCF region on chromosome 11p showed similar frequencies of
LOH, partially using the same microsatellite markers (18–33%)
(74) and also when markers were
2–3 Mb distant from our markers (17%) (32). These studies focused on primary
HNSCCs. A proposed progression model of HNSCC implies that the same
gene region 11p14 is lost in up to 60% of metastatic HNSCC
(29). If the loss of FANCF
predominantly happens during the progression of HNSCC to the
metastatic stage, the gene may already be downregulated via other
mechanisms in primary HNSCC, for instance methylation (40).
Markers previously used on chromosome 17q were more
than 10 Mb distant from the BRIP1 region and showed a higher
LOH frequency (31%) than our estimate (11%), (32). However, another study evaluated
loci around 3 Mb distant from our marker positions and did not
detect LOH (74).
Thus, our data on LOH in the regions of FANCG
and FANCF are in concordance with previous estimates for LOH
in regions containing the studied FA genes. Reports on BRIP1
vary and it is currently difficult to conclude on the role of LOH
adjacent to this gene in HNSCC.
LOH in FA genes and patient survival
Impaired DNA repair in tumour tissue due to mutated
FA genes may increase the sensitivity to DNA damage by radiotherapy
and even more by alkylating agents and may thus prolong patient
survival (72, 73, 75,
76). However, associations of LOH
at FA gene regions and patient survival were not strongly supported
from our data. We observed a mild association of LOH at the
FANCF region and decreased survival in our Kaplan-Meier
curves and log-rank tests. The loss of FANCF as a tumour
suppressor is consistent with decreased survival in the affected
patients and also with decreased expression of the gene in ovary,
breast, lung, cervix and testis cancer (38–42).
However, the weak association for FANCF is not as strong as
that of known predictors for HNSCC survival, such as higher node
stages and surgery (77, 78). The Bonferroni correction may be
overly conservative for the Cox PH models evaluated, as these can
be assumed to be positively correlated and also weaker associations
might be true positive results (79). Thus, in multivariate analyses with
improved statistical power, the weak associations observed here may
be correct.
The observed effect of tumour grading and improved
survival was very weak and may not hold true, as we had a small
sample size for this trait and higher tumour grading has repeatedly
been described as associated with decreased survival (80–82).
Our analysis may also lack statistical power since the analysed
sample of 53 patients had varying clinical data. A rather uniform
collection of tumours from patients with similar clinical data,
e.g. all with surgery and radiotherapy, would result in a strongly
improved study design. A larger sample for only chemotherapy
treated individuals may be necessary to explore a potential link of
chemosensitivity with BRIP1 mutation.
Copy number and expression data
From the queried datasets we found that HNSCC
tumours tend to overexpress FANCG and BRIP1 (Table VII), however, often exhibit
chromosomal aberrations that predominantly involve the loss of
these genes. After the induction of both genes for DNA repair, the
copy number loss in a progressing tumour could remove both genes
and allow the accumulation of mutations. This scenario supports the
assumed role of FANCG and BRIP1 as tumour
suppressors. These data differ from a previous study that found
reduced expression of FANCG and BRIP1 in HNSCC in
tongue carcinoma (26). However, a
loss of FANCG and BRIP1 would diminish expression of
the genes in later stages of HNSCC or in the more aggressive tongue
carcinoma. Also in other cancer types FANCG was lost more
often than gained (breast: 11.5% gains, 24.1% loss; lung: 13.9%
gains, 43.9% losses; and pancreas: 20.3% gains, 39.0% losses)
(71). However, BRIP1 was
gained more often than lost indicating differences of the various
cancer types (breast: 32.8% gains, 12.3% loss; lung: 36.8% gains,
6.9% losses; and pancreas: 27.1% gains, 24.8% losses).
Intergrating copy number data and expression is
somewhat inconclusive as well for FANCF as it shows
underexpression in two sets of HNSCC and is affected by copy number
gains in another set. Copy number gains are not characteristic for
tumour suppressor genes, which FANCF was supposed to be. A
decrease in gene expression, may be explained by point mutations or
methylation of the gene in 15% of HNSCC as previously reported
(40, 83). This could also ameliorate copy
number gains that involve FANCF by chance. In other cancer
types FANCF is affected by CNV in 30–40% of samples,
involving gains and losses (breast: 13.4% gains, 20.4% loss; lung:
13% gains, 27.1% losses; and pancreas: 29.7% gains, 14.8%
losses).
Thus, copy number mutations of FA genes are
frequent in HNSCC and other cancer types. However, an improved
understanding of the mechanisms leading from gene mutations to gene
expression changes and a potential clinical relevance could come
from data on LOH, CNV, point mutations and gene expression obtained
for the same samples.
Differential gene expression analysis
using TCGA data
Our pathway analyses using differential gene
expression analysis with subsequent gene set enrichment and network
analysis showed that physically interacting partners differ for the
studied genes. FANCG and FANCF show some overlap in
interacting with members of the Fanconi anemia nuclear complex
while BRIP1 is rather involved in the double-strand break
repair pathway.
Clinical relevance
As HNSCC is routinely treated with ionizing
radiation and, less frequently, also with chemotherapy, mutations
in DNA repair genes may be relevant for treatment success. Since we
found LOH in FA gene regions in 57% of the patients and other
mutation types may add to this frequency, a substantial proportion
of HNSCC patients may be eligible for poly-adenosine diphosphate
ribose polymerase (PARP) inhibition therapy. PARP inhibition
impairs DNA repair selectively in cancer cells, however, not in
normal somatic cells and may enter the clinics for HNSCC treatment
(33, 84). In theory, cancer cells with an
impaired pathway for homologous recombination (e.g. due to mutated
FA genes) cannot perform double strand break repair. PARP
inhibition then may be used to block the base excision repair
pathway for single strand break repair as well. Unlike normal
cells, cancer cells will then be sensitized to DNA damage induced
by radiation and alkylating chemotherapy (85). In this regard, cells deficient in
FANCA, FANCC or FANCD2 were previously found
to be hypersensitive to PARP inhibition (86). Also human HNSCC cells showed
enhanced cytotoxicity with radiation and PARP inhibition compared
to either agent alone (87). PARP
inhibitors enhanced the effect of radiotherapy in a xenograft model
of human HNSCC leading to reduced tumour volume and enhanced
apoptosis (88). FANCF
knockdown has been shown to induce chemosensitivity in cancer cells
(89, 90). Thus, further studies are needed to
explore the observed association of LOH in FANCF and
decreased HNSCC survival as well as the potential use of FA gene
mutations as an indicator for chemo- and radiosensitivity of head
and neck tumours.
In conclusion, analysing three FA gene regions, we
found LOH in 57% of HNSCC tumours. LOH in FANCF showed a
weak association with survival of radiotherapy and chemotherapy
treated HNSCC patients. Tumours with LOH in FA genes may exhibit an
altered sensitivity to cancer therapy utilizing DNA damaging
agents. Thus, it is worthwhile to perform further studies screening
for other types of mutations in FA genes and involving larger
sample sizes to improve the statistical power of survival
analysis.
Acknowledgments
We thank Diana Jünger for advice on laboratory
work, Susanne Koy for discussion and Jens Plaschke for advice on
statistical data analysis. We thank Janet Kelso and Sebastian
Bittrich for discussion and help in preparing the manuscript.
Abbreviations:
|
SCC
|
squamous cell carcinoma
|
|
LOH
|
loss of heterozygosity
|
|
FA
|
Fanconi anemia
|
|
HPV
|
human papilloma virus
|
|
HNSCC
|
head and neck squamous cell
carcinomas
|
|
PARP
|
polyADP-ribose polymerase
|
|
PCR
|
polymerase chain reaction
|
|
CNV
|
copy number variants
|
|
ESCC
|
esophageal squamous cell
carcinomas
|
|
PH
|
proportional hazards
|
|
MSI
|
microsatellite instability
|
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sadick M, Schoenberg SO, Hoermann K and
Sadick H: Aktuelle Standards und Fortschritte in der onkologischen
Bildgebung von Kopf-Hals-Tumoren. Laryngorhinootologie. 91(Suppl
1): S27–S47. 2012. View Article : Google Scholar
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kademani D: Oral cancer. Mayo Clin Proc.
82:878–887. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bonner JA, Harari PM, Giralt J, Cohen RB,
Jones CU, Sur RK, Raben D, Baselga J, Spencer SA, Zhu J, et al:
Radiotherapy plus cetuximab for locoregionally advanced head and
neck cancer: 5-year survival data from a phase 3 randomised trial,
and relation between cetuximab-induced rash and survival. Lancet
Oncol. 11:21–28. 2010. View Article : Google Scholar
|
|
6
|
Forastiere A, Koch W, Trotti A and
Sidransky D: Head and neck cancer. N Engl J Med. 345:1890–1900.
2001. View Article : Google Scholar
|
|
7
|
Maier H, Dietz A, Gewelke U, Heller WD and
Weidauer H: Tobacco and alcohol and the risk of head and neck
cancer. Clin Investig. 70:320–327. 1992.PubMed/NCBI
|
|
8
|
Lewin F, Norell SE, Johansson H,
Gustavsson P, Wennerberg J, Biörklund A and Rutqvist LE: Smoking
tobacco, oral snuff, and alcohol in the etiology of squamous cell
carcinoma of the head and neck: A population-based case-referent
study in Sweden. Cancer. 82:1367–1375. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Viswanathan H and Wilson JA: Alcohol - the
neglected risk factor in head and neck cancer. Clin Otolaryngol
Allied Sci. 29:295–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fakhry C, Westra WH, Li S, Cmelak A, Ridge
JA, Pinto H, Forastiere A and Gillison ML: Improved survival of
patients with human papillomavirus-positive head and neck squamous
cell carcinoma in a prospective clinical trial. J Natl Cancer Inst.
100:261–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gillison ML, Koch WM, Capone RB, Spafford
M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, et
al: Evidence for a causal association between human papillomavirus
and a subset of head and neck cancers. J Natl Cancer Inst.
92:709–720. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trizna Z and Schantz SP: Hereditary and
environmental factors associated with risk and progression of head
and neck cancer. Otolaryngol Clin North Am. 25:1089–1103.
1992.PubMed/NCBI
|
|
13
|
Hecht F and Hecht BK: Cancer in
ataxia-telangiectasia patients. Cancer Genet Cytogenet. 46:9–19.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Levine AJ, Momand J and Finlay CA: The p53
tumour suppressor gene. Nature. 351:453–456. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blons H and Laurent-Puig P: TP53 and head
and neck neoplasms. Hum Mutat. 21:252–257. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Erber R, Conradt C, Homann N, Enders C,
Finckh M, Dietz A, Weidauer H and Bosch FX: TP53 DNA contact
mutations are selectively associated with allelic loss and have a
strong clinical impact in head and neck cancer. Oncogene.
16:1671–1679. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nylander K, Dabelsteen E and Hall PA: The
p53 molecule and its prognostic role in squamous cell carcinomas of
the head and neck. J Oral Pathol Med. 29:413–425. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Stransky N, Egloff AM, Tward AD, Kostic
AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C,
McKenna A, et al: The mutational landscape of head and neck
squamous cell carcinoma. Science. 333:1157–1160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Agrawal N, Frederick MJ, Pickering CR,
Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et
al: Exome sequencing of head and neck squamous cell carcinoma
reveals inactivating mutations in NOTCH1. Science. 333:1154–1157.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Skinner HD1, Sandulache VC, Ow TJ, Meyn
RE, Yordy JS, Beadle BM, Fitzgerald AL, Giri U, Ang KK and Myers
JN: TP53 disruptive mutations lead to head and neck cancer
treatment failure through inhibition of radiation-induced
senescence. Clin Cancer Res. 18:290–300. 2012. View Article : Google Scholar :
|
|
21
|
Kutler DI, Auerbach AD, Satagopan J,
Giampietro PF, Batish SD, Huvos AG, Goberdhan A, Shah JP and Singh
B: High incidence of head and neck squamous cell carcinoma in
patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg.
129:106–112. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kutler DI, Singh B, Satagopan J, Batish
SD, Berwick M, Giampietro PF, Hanenberg H and Auerbach AD: A
20-year perspective on the International Fanconi Anemia Registry
(IFAR). Blood. 101:1249–1256. 2003. View Article : Google Scholar
|
|
23
|
Rosenberg PS, Socié G, Alter BP and
Gluckman E: Risk of head and neck squamous cell cancer and death in
patients with Fanconi anemia who did and did not receive
transplants. Blood. 105:67–73. 2005. View Article : Google Scholar
|
|
24
|
Walden H and Deans AJ: The Fanconi anemia
DNA repair pathway: Structural and functional insights into a
complex disorder. Annu Rev Biophys. 43:257–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chandrasekharappa SC, Lach FP, Kimble DC,
Kamat A, Teer JK, Donovan FX, Flynn E, Sen SK, Thongthip S, Sanborn
E, et al: NISC Comparative Sequencing Program: Massively parallel
sequencing, aCGH, and RNA-Seq technologies provide a comprehensive
molecular diagnosis of Fanconi anemia. Blood. 121:e138–e148. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wreesmann VB, Estilo C, Eisele DW, Singh B
and Wang SJ: Downregulation of Fanconi anemia genes in sporadic
head and neck squamous cell carcinoma. ORL J Otorhinolaryngol Relat
Spec. 69:218–225. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pfeiffer J, Maier W, Ridder GJ, Zaoui K
and Birkenhäger R: LOH-profiling by SNP-mapping in a case of
multifocal head and neck cancer. World J Clin Oncol. 3:24–28. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sparano A, Quesnelle KM, Kumar MS, Wang Y,
Sylvester AJ, Feldman M, Sewell DA, Weinstein GS and Brose MS:
Genome-wide profiling of oral squamous cell carcinoma by
array-based comparative genomic hybridization. Laryngoscope.
116:735–741. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bockmühl U, Schlüns K, Schmidt S, Matthias
S and Petersen I: Chromosomal alterations during metastasis
formation of head and neck squamous cell carcinoma. Genes
Chromosomes Cancer. 33:29–35. 2002. View Article : Google Scholar
|
|
30
|
Koy S, Hauses M, Appelt H, Friedrich K,
Schackert HK and Eckelt U: Loss of expression of ZAC/LOT1 in
squamous cell carcinomas of head and neck. Head Neck. 26:338–344.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weber F, Xu Y, Zhang L, Patocs A, Shen L,
Platzer P and Eng C: Microenvironmental genomic alterations and
clinicopathological behavior in head and neck squamous cell
carcinoma. JAMA. 297:187–195. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nawroz H, van der Riet P, Hruban RH, Koch
W, Ruppert JM and Sidransky D: Allelotype of head and neck squamous
cell carcinoma. Cancer Res. 54:1152–1155. 1994.PubMed/NCBI
|
|
33
|
Bryant HE, Schultz N, Thomas HD, Parker
KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ and Helleday T:
Specific killing of BRCA2-deficient tumours with inhibitors of
poly(ADP-ribose) polymerase. Nature. 434:913–917. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Duan W, Gao L, Zhao W, Leon M, Sadee W,
Webb A, Resnick K, Wu X, Ramaswamy B, Cohn DE, et al: Assessment of
FANCD2 nuclear foci formation in paraffin-embedded tumors: A
potential patient-enrichment strategy for treatment with DNA
interstrand crosslinking agents. Transl Res. 161:156–164. 2013.
View Article : Google Scholar
|
|
35
|
de Winter JP, Rooimans MA, van Der Weel L,
van Berkel CG, Alon N, Bosnoyan-Collins L, de Groot J, Zhi Y,
Waisfisz Q, Pronk JC, et al: The Fanconi anaemia gene FANCF encodes
a novel protein with homology to ROM. Nat Genet. 24:15–16. 2000.
View Article : Google Scholar
|
|
36
|
Ahmad SI and Kirk SH: Molecular Mechanisms
of Fanconi Anemia. Landes Bioscience/Eurekah.com; 2006
|
|
37
|
de Winter JP, van der Weel L, de Groot J,
Stone S, Waisfisz Q, Arwert F, Scheper RJ, Kruyt FA, Hoatlin ME and
Joenje H: The Fanconi anemia protein FANCF forms a nuclear complex
with FANCA, FANCC and FANCG. Hum Mol Genet. 9:2665–2674. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Taniguchi T, Tischkowitz M, Ameziane N,
Hodgson SV, Mathew CG, Joenje H, Mok SC and D'Andrea AD: Disruption
of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian
tumors. Nat Med. 9:568–574. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Olopade OI and Wei M: FANCF methylation
contributes to chemoselectivity in ovarian cancer. Cancer Cell.
3:417–420. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Marsit CJ, Liu M, Nelson HH, Posner M,
Suzuki M and Kelsey KT: Inactivation of the Fanconi anemia/BRCA
pathway in lung and oral cancers: Implications for treatment and
survival. Oncogene. 23:1000–1004. 2004. View Article : Google Scholar
|
|
41
|
Narayan G, Arias-Pulido H, Nandula SV,
Basso K, Sugirtharaj DD, Vargas H, Mansukhani M, Villella J, Meyer
L, Schneider A, et al: Promoter hypermethylation of FANCF:
Disruption of Fanconi Anemia-BRCA pathway in cervical cancer.
Cancer Res. 64:2994–2997. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koul S, McKiernan JM, Narayan G,
Houldsworth J, Bacik J, Dobrzynski DL, Assaad AM, Mansukhani M,
Reuter VE, Bosl GJ, et al: Role of promoter hypermethylation in
Cisplatin treatment response of male germ cell tumors. Mol Cancer.
3:162004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
de Winter JP, Waisfisz Q, Rooimans MA, van
Berkel CG, Bosnoyan-Collins L, Alon N, Carreau M, Bender O, Demuth
I, Schindler D, et al: The Fanconi anaemia group G gene FANCG is
identical with XRCC9. Nat Genet. 20:281–283. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Demuth I, Wlodarski M, Tipping AJ, Morgan
NV, de Winter JP, Thiel M, Gräsl S, Schindler D, D'Andrea AD, Altay
C, et al: Spectrum of mutations in the Fanconi anaemia group G
gene, FANCG/XRCC9. Eur J Hum Genet. 8:861–868. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gallmeier E, Calhoun ES, Rago C, Brody JR,
Cunningham SC, Hucl T, Gorospe M, Kohli M, Lengauer C and Kern SE:
Targeted disruption of FANCC and FANCG in human cancer provides a
preclinical model for specific therapeutic options.
Gastroenterology. 130:2145–2154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Levitus M, Waisfisz Q, Godthelp BC, de
Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E,
Steltenpool J, Rooimans MA, Pals G, et al: The DNA helicase BRIP1
is defective in Fanconi anemia complementation group J. Nat Genet.
37:934–935. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
47
|
Levran O, Attwooll C, Henry RT, Milton KL,
Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, et al:
The BRCA1-interacting helicase BRIP1 is deficient in Fanconi
anemia. Nat Genet. 37:931–933. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
48
|
Litman R, Peng M, Jin Z, Zhang F, Zhang J,
Powell S, Andreassen PR and Cantor SB: BACH1 is critical for
homologous recombination and appears to be the Fanconi anemia gene
product FANCJ. Cancer Cell. 8:255–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xie J, Peng M, Guillemette S, Quan S,
Maniatis S, Wu Y, Venkatesh A, Shaffer SA, Brosh RM Jr and Cantor
SB: FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage
response. PLoS Genet. 8:e10027862012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cantor SB and Guillemette S: Hereditary
breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future
Oncol. 7:253–261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sobin LH and Fleming ID: TNM
Classification of Malignant Tumors, fifth edition (1997). Union
Internationale Contre le Cancer and the American Joint Committee on
Cancer. Cancer. 80:1803–1804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Idbaih A, Carvalho Silva R, Crinière E,
Marie Y, Carpentier C, Boisselier B, Taillibert S, Rousseau A,
Mokhtari K, Ducray F, et al: Genomic changes in progression of
low-grade gliomas. J Neurooncol. 90:133–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
van Zeeburg HJ, Snijders PJ, Pals G,
Hermsen MA, Rooimans MA, Bagby G, Soulier J, Gluckman E, Wennerberg
J, Leemans CR, et al: Generation and molecular characterization of
head and neck squamous cell lines of fanconi anemia patients.
Cancer Res. 65:1271–1276. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nagao T1, Sugano I, Ishida Y, Tajima Y,
Munakata S, Asoh A, Yamazaki K, Muto H, Konno A, Kondo Y, et al:
Primary large-cell neuroendocrine carcinoma of the parotid gland:
immunohistochemical and molecular analysis of two cases. Mod
Pathol. 13:554–561. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tarmin L, Yin J, Zhou X, Suzuki H, Jiang
HY, Rhyu MG, Abraham JM, Krasna MJ, Cottrell J and Meltzer SJ:
Frequent loss of heterozygosity on chromosome 9 in adenocarcinoma
and squamous cell carcinoma of the esophagus. Cancer Res.
54:6094–6096. 1994.PubMed/NCBI
|
|
56
|
Chen CH, Chang CJ, Yang WS, Chen CL and
Fann CSJ: A genome-wide scan using tree-based association analysis
for candidate loci related to fasting plasma glucose levels. BMC
Genet. 4(Suppl 1): S652003. View Article : Google Scholar
|
|
57
|
Powlesland RM, Charles AK, Malik KTA,
Reynolds PA, Pires S, Boavida M and Brown KW: Loss of
heterozygosity at 7p in Wilms' tumour development. Br J Cancer.
82:323–329. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Orsetti B, Courjal F, Cuny M, Rodriguez C
and Theillet C: 17q21–q25 aberrations in breast cancer: Combined
allelotyping and CGH analysis reveals 5 regions of allelic
imbalance among which two correspond to DNA amplification.
Oncogene. 18:6262–6270. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Indraccolo S, Tisato V, Agata S, Moserle
L, Ferrari S, Callegaro M, Persano L, Palma MD, Scaini MC, Esposito
G, et al: Establishment and characterization of xenografts and
cancer cell cultures derived from BRCA1 −/− epithelial ovarian
cancers. Eur J Cancer. 42:1475–1483. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Canzian F, Salovaara R, Hemminki A, Kristo
P, Chadwick RB, Aaltonen LA and de la Chapelle A: Semiautomated
assessment of loss of heterozygosity and replication error in
tumors. Cancer Res. 56:3331–3337. 1996.PubMed/NCBI
|
|
61
|
Cawkwell L, Bell SM, Lewis FA, Dixon MF,
Taylor GR and Quirke P: Rapid detection of allele loss in
colorectal tumours using microsatellites and fluorescent DNA
technology. Br J Cancer. 67:1262–1267. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hahn M, Wieland I, Koufaki ON, Görgens H,
Sobottka SB, Schackert G and Schackert HK: Genetic alterations of
the tumor suppressor gene PTEN/MMAC1 in human brain metastases.
Clin Cancer Res. 5:2431–2437. 1999.PubMed/NCBI
|
|
63
|
Ihaka R and Gentleman R: R: A language for
data analysis and graphics. J Comput Graph Stat. 5:299–314.
1996.
|
|
64
|
Bland JM and Altman DG: Multiple
significance tests: The Bonferroni method. BMJ. 310:1701995.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Cheng PF, Dummer R and Levesque MP: Data
mining The Cancer Genome Atlas in the era of precision cancer
medicine. Swiss Med Wkly. 145:w141832015.PubMed/NCBI
|
|
66
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic Acids Res. 38(Web Server): W214–W220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kuriakose MA, Chen WT, He ZM, Sikora AG,
Zhang P, Zhang ZY, Qiu WL, Hsu DF, McMunn-Coffran C, Brown SM, et
al: Selection and validation of differentially expressed genes in
head and neck cancer. Cell Mol Life Sci. 61:1372–1383. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hu N, Clifford RJ, Yang HH, Wang C,
Goldstein AM, Ding T, Taylor PR and Lee MP: Genome wide analysis of
DNA copy number neutral loss of heterozygosity (CNNLOH) and its
relation to gene expression in esophageal squamous cell carcinoma.
BMC Genomics. 11:5762010. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhu B, Yan K, Li L, Lin M, Zhang S, He Q,
Zheng D, Yang H and Shao G: K63-linked ubiquitination of FANCG is
required for its association with the Rap80-BRCA1 complex to
modulate homologous recombination repair of DNA interstand
crosslinks. Oncogene. 34:2867–2878. 2015. View Article : Google Scholar
|
|
71
|
Forbes SA, Bindal N, Bamford S, Cole C,
Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al:
COSMIC: Mining complete cancer genomes in the Catalogue of Somatic
Mutations in Cancer. Nucleic Acids Res. 39(Database): D945–D950.
2011. View Article : Google Scholar :
|
|
72
|
Deans AJ and West SC: DNA interstrand
crosslink repair and cancer. Nat Rev Cancer. 11:467–480. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ridet A, Guillouf C, Duchaud E, Cundari E,
Fiore M, Moustacchi E and Rosselli F: Deregulated apoptosis is a
hallmark of the Fanconi anemia syndrome. Cancer Res. 57:1722–1730.
1997.PubMed/NCBI
|
|
74
|
Beder LB, Gunduz M, Ouchida M, Fukushima
K, Gunduz E, Ito S, Sakai A, Nagai N, Nishizaki K and Shimizu K:
Genome-wide analyses on loss of heterozygosity in head and neck
squamous cell carcinomas. Lab Invest. 83:99–105. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
McHugh PJ, Spanswick VJ and Hartley JA:
Repair of DNA inter-strand crosslinks: Molecular mechanisms and
clinical relevance. Lancet Oncol. 2:483–490. 2001. View Article : Google Scholar
|
|
76
|
Bridge WL, Vandenberg CJ, Franklin RJ and
Hiom K: The BRIP1 helicase functions independently of BRCA1 in the
Fanconi anemia pathway for DNA crosslink repair. Nat Genet.
37:953–957. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hoffmann M, Görögh T, Gottschlich S,
Lohrey C, Rittgen W, Ambrosch P, Schwarz E and Kahn T: Human
papillomaviruses in head and neck cancer: 8 year-survival-analysis
of 73 patients. Cancer Lett. 218:199–206. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Sticht C, Hofele C, Flechtenmacher C,
Bosch FX, Freier K, Lichter P and Joos S: Amplification of Cyclin
L1 is associated with lymph node metastases in head and neck
squamous cell carcinoma (HNSCC). Br J Cancer. 92:770–774. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hashemi R and Commenges D: Correction of
the p-value after multiple tests in a Cox proportional hazard
model. Lifetime Data Anal. 8:335–348. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Bryne M, Koppang HS, Lilleng R, Stene T,
Bang G and Dabelsteen E: New malignancy grading is a better
prognostic indicator than Broders' grading in oral squamous cell
carcinomas. J Oral Pathol Med. 18:432–437. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kristensen GB, Abeler VM, Risberg B, Trop
C and Bryne M: Tumor size, depth of invasion, and grading of the
invasive tumor front are the main prognostic factors in early
squamous cell cervical carcinoma. Gynecol Oncol. 74:245–251. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Bryne M, Koppang HS, Lilleng R and
Kjaerheim A: Malignancy grading of the deep invasive margins of
oral squamous cell carcinomas has high prognostic value. J Pathol.
166:375–381. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lim SL, Smith P, Syed N, Coens C, Wong H,
van der Burg M, Szlosarek P, Crook T and Green JA: Promoter
hypermethylation of FANCF and outcome in advanced ovarian cancer.
Br J Cancer. 98:1452–1456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Begg AC: Predicting recurrence after
radiotherapy in head and neck cancer. Semin Radiat Oncol.
22:108–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Curtin NJ and Szabo C: Therapeutic
applications of PARP inhibitors: Anticancer therapy and beyond. Mol
Aspects Med. 34:1217–1256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
McCabe N, Turner NC, Lord CJ, Kluzek K,
Bialkowska A, Swift S, Giavara S, O'Connor MJ, Tutt AN, Zdzienicka
MZ, et al: Deficiency in the repair of DNA damage by homologous
recombination and sensitivity to poly(ADP-ribose) polymerase
inhibition. Cancer Res. 66:8109–8115. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Nowsheen S, Bonner JA and Yang ES: The
poly(ADP-Ribose) polymerase inhibitor ABT-888 reduces
radiation-induced nuclear EGFR and augments head and neck tumor
response to radiotherapy. Radiother Oncol. 99:331–338. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Khan K, Araki K, Wang D, Li G, Li X, Zhang
J, Xu W, Hoover RK, Lauter S, O'Malley B Jr, et al: Head and neck
cancer radiosensitization by the novel poly(ADP-ribose) polymerase
inhibitor GPI-15427. Head Neck. 32:381–391. 2010.
|
|
89
|
Li Y, Zhao L, Sun H, Yu J, Li N, Liang J,
Wang Y, He M, Bai X, Yu Z, et al: Gene silencing of FANCF
potentiates the sensitivity to mitoxantrone through activation of
JNK and p38 signal pathways in breast cancer cells. PLoS One.
7:e442542012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yu J, Zhao L, Li Y, Li N, He M, Bai X, Yu
Z, Zheng Z, Mi X, Wang E, et al: Silencing of fanconi anemia
complementation group f exhibits potent chemosensitization of
mitomycin C activity in breast cancer cells. J Breast Cancer.
16:291–299. 2013. View Article : Google Scholar : PubMed/NCBI
|