Introduction
A central nervous system tumor is one of the most
complicated human high-risk malignant tumors. Most primary brain
tumors derived from glial cells or precursor cells are named glioma
which can be subdivided into astrocytoma, oligodendroglioma,
ependymoma and glioblastoma in light of the pathological type
(1). According to the clinical
malignant grade of tumors from the World Health Organization (WHO)
(2), grades I and II are the least
malignant phenotypes, such as oligodendroglioma and astrocytoma,
grade III comprises the moderate malignant gliomas, such as
anaplastic astrocytoma and anaplastic oligoastrocytoma, whereas
grade IV (glioblastoma multiforme, GBM) is the most malignant type
of gliomas (3). GBM is the most
common and devastating primary brain tumor. Despite treatment with
surgical section, chemotherapy with temozolomide (TMZ) and
radiation, the median survival of patients is between 12 to 15
months (4) because of the high
infiltrating capacity and near-universal tumor recurrence (5). Even with recent advances in cancer
diagnostic methodology and treatment, the prognosis of GBM has not
improved. Glioma stem cells (GSC) have been verified to be one of
the major reasons for poor prognosis in GBM and other high grade
primary brain tumors, making it nearly impossible to remove or kill
all of these tumor cells by conventional therapies (6,7).
GSCs express normal neural stem cell (NSC) markers which are
characteristic of self-renewal and multi-differentiation potential.
GSCs can promote tumor proliferation, blood vessel growth and
radiation and chemotherapy resistance, which all means GSCs play
important roles in tumor recurrence and stemness maintenance of GBM
(8,9). Despite the progress in understanding
of the molecular mechanisms involved in the genesis and progression
of glioma, the prognosis and treatment of this tumor is still
unsatisfactory (10).
CUEDC2 is a CUE domain containing protein, a small
ubiquitin-binding motif with approximately 40 amino acid residues
in many eukaryotic proteins. It has a dual role in monoubiquitin
and polyubiquitin recognition, as well as in facilitating
intramolecular monoubiquitination (11,12).
Recent studies have suggested that CUEDC2 played critical roles in
many biological processes, such as cell cycle, inflammation and
tumorigenesis (12).
As a multifunctional protein, the function of CUEDC2
in cancer is debated. Previous studies have indicated that CUEDC2
possesses both oncogenic and tumor-suppressive properties. It was
reported that the CUEDC2 caused earlier activation of APC/cdc20 to
promote the metaphase-anaphase transition that led to chromosome
missegregation and aneuploidy which might contribute to tumor
development (13). Zhang et
al (14) indicated that CUEDC2
inhibited APC/cdh1 to promote the G1-S transition which might
stimulate the proceeding of cell cycle in skin cancer where the
expression of CUEDC2 was significantly elevated. Also, in breast
cancer, CUEDC2 expression is elevated which promotes the
degradation of estrogen receptor-α (ERα) and progesterone receptor
(PR) and further leads to the resistance to endocrine therapy by
tamoxifen and early relapse (15–17).
In ovarian serous carcinoma, CUEDC2 may be a promising biomarker
and CUEDC2-positive expression was found to be associated with a
shorter disease-free survival time (18).
However, other studies shown that the existence of
CUEDC2 was beneficial to many kinds of normal tissues. High
expression of CUEDC2 reduced colonic inflammatory reactions,
decreased the expression of pro-inflammatory cytokines,
significantly inhibited the activation of signaling pathways such
as NF-κB and JAK1-STAT3 and prevented excessive proliferation of
the inflammatory mucous epithelial cells which directly accelerated
the occurrence of colorectal cancer (18). In lung adenocarcinoma cells, the
promotion of proliferation by decreased CUEDC2 was associated with
activation of the PI3K/Akt pathway, which leads to a poor clinical
outcome and a shorter survival time in patients (19). Moreover, CUEDC2 could inhibit the
NF-κB signaling pathway to increase imatinib sensitivity in chronic
myeloid leukemia (CML) cells (20). CUEDC2 could act as an adaptor
protein to target IκB kinase (IKK) for dephosphorylation and
inactivation by recruiting protein phosphatase (PP1), and thus,
repressed the activation of NF-κB, signal pathway which played
pivotal roles in inflammatory responses and tumorigenesis (15). Furthermore, CUEDC2 was found to
regulate JAK1/STAT3 signaling pathway and to inhibit this pathway
by increasing the stability of SOCS3 (suppressors of cytokine
signaling 3) (21).
Although the roles of CUEDC2 in the development of
many different cancers have been studied, its role in gliomas,
especially in GBM is still unknown. However, this study showed that
CUEDC2 was markedly downregulated in surgical specimens of GBM and
glioma cell lines, especially in GSCs isolated from glioma cell
line, the expression of CUEDC2 is extremely low. Overexpression of
CUEDC2 inhibits proliferation, migration and invasion as well as
arrests cell cycle at G1 phase of U251 glioma cells. In contrast,
knockdown of CUEDC2 promoted cell proliferation, migration and
invasion, as well as accumulation of cell cycle at G1 phase.
Further studies showed that overexpression of CUEDC2 suppressed the
activation and translocation of STAT3 and NF-κB from the cell
cytoplasm to the nucleus. Thus, our results suggested that the
CUEDC2 played a tumor-suppressive role in glioma development.
Materials and methods
Preparation of glioma tissue samples
Glioma tissue samples were obtained during surgical
removal of tumors from patients histopathologically diagnosed with
different clinical pathology classification, and normal tissues
were obtained from brain surgery from the Affiliated Hospital of
Xuzhou Medical University. The histological characterization and
clinicopathological staging of the samples were performed according
to the World Health Organization (WHO) criteria which is the most
widely accepted classification scheme for the diffuse gliomas.
Lentivirus package and constructions of
stable cell lines
The 293T packaging cells were transfected with
GV-CUEDC2/GV-vector, Helper1.0, Helper2.0 using liposome-based
transfection method and the packaged virus-containing supernatant
was collected and used to infect U251 cells. Stable overexpression
of CUEDC2 and its control cell lines were obtained by flow
cytometry sorting. Additionally, downregulation of CUEDC2 and its
control cell lines were infected by lentivirus of GV-CUEDC2-RNAi
and GV-RNAi-vector packaged virus. Then through puromycin lasting
one week, stable cell lines were obtained. The cells stably
overexpressing CUEDC2 or carrying the vector alone were named as
'U251-CUEDC2' cells, while those stably expressing the
CUEDC2-specific RNAi were designated as 'RNAi-CUEDC2' or
'RNAi-vector' cells.
Identification of glioma stem cells
isolated from U251
U251 cells were cultured normally, and then adding
EGF (20 ng/ml), bFGF (20 ng/ml), LIF (10 ng/ml), B27 factors (×50)
in serum-free conditions to culture U251 stem cells. GSCs were
identified by labeling with rabbit anti-CD133 (polyclonal, 1:100;
Proteintech Group, Inc., Rosemont, IL, USA) and mouse anti-Nestin
(monoclonal, 1:100). GSCs were seeded in 48-well plates covered
with polylysine and cultured for 48 h. Then the old medium was
discarded, washed 3 times with phosphate-buffered saline (PBS) and
then cells were fixed for 30 min at room temperature in 4%
formaldehyde. Fixed cells were permeabilized and blocked for 30 min
at room temperature using 0.3% Triton X-100 and 5% goat serum/PBS,
and then wash 3 times with PBS and immunostained using CD133 and
Nestin antibody overnight at 4°C. The next day, washed 3 times with
PBS, the secondary antibody (1:100; Vicmed Biotech Co., Ltd.,
Xuzhou, China), rhodamine (TRITC)-conjugated goat anti-mouse and
Alexa Fluor488-conjugated AffiniPure goat anti-rabbit, were
incubated for 2 h at room temperature in the dark. Nuclei were
stained with 4′6-diamidino-2-phenylindole (DAPI). Fluorescence
images were captured with fluorescent inverted microscope.
For glioma sphere formation assay, the U251 cells
(5×104/well) were inoculated on 24-well plate with
serum-free DMEM F12 containing EGF, bFGF, LIF and B27 and cultured
for 6 days, then images were taken.
Flow cytometric analysis of cell
cycle
Cell cycle distribution was examined by flow
cytometry according to the standard method. Cells were harvested by
trypsinization and washed with cold PBS 3 times and then fixed with
70% cold ethanol at 4°C overnight. Before staining, the cells were
centrifuged in a cooled centrifuge and suspended in 100 µl
of RNase A (Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China).
After 30 min of 37°C incubation, incubation at 4°C followed for 30
min after adding 400 µl propidium iodide (PI; Nanjing KeyGen
Biotech). Samples were checked through FACScan cytometry
(Becton-Dickinson, San Jose, CA, USA) and data were analyzed by
FlowJo software (Tree Star, Inc., Ashland, OR, USA). Three
independent experiments were carried out for each cell line.
Cell proliferation assay
Cell proliferation was detected via the Cell
Counting Kit-8 assay (CCK-8; Dojindo Laboratories, Kumamoto,
Japan). Cells were inoculated on a 96-well flat-bottomed
microplate, with a density of 4,000/100 µl. After incubating
12 h until the cells grew to 50% confluence, 10 µl CCK-8 dye
was added to each well. Then incubated for 30, 60 and 90 min at
37°C and the absorbance at the wavelength of 450 nm was measured by
a UVmax kinetic microplate reader (Molecular Devices, Wokingham,
UK). The above results were regarded as the starting value (0 day),
and then we checked the CCK-8 results at the indicated time-points
(0, 1, 2, 3 and 4 days) and recorded the 30, 60 and 90 min OD450
value.
In vitro migration and invasion
assay
Cell migration was evaluated by scratch wound assay.
Briefly, cells (106/well) were plated in a 6-well plate
and cultured for 48 h to yield a confluent monolayer. The culture
solution was changed to serum-free Dulbecco's modified Eagle's
medium (DMEM)/F12 at 6 h before wounding. Then wounded with a
10-µl pipette tip. The remaining cells were washed twice
with PBS and cultured with serum-free DMEM/F12. Photographs were
taken at 0, 24 and 48 h. At the indicated times, migrating cells at
the front of the wound were photographed and the percentage of the
cleaned area at each time-point was compared with the area at time
0. Each area was measured using Image-Pro Plus version 6.0
software.
Cell invasion was evaluated using a
Transwell Matrigel invasion assay
Before the assay, 80 µl of the diluted
Matrigel was put into the upper chamber of 24-well Transwell
chamber (Corning, Inc., Corning, NY, USA) and incubated at 37°C for
gelling. The U251 cells were harvested and the cells suspended with
serum-free DMEM/F12. Then inoculate cells into the upper chamber
with a density of 2×104, 4×104 and
8×104, respectively. Lower champer of the Transwell was
filled with 600 µl of 10% fetal bovine serum (FBS) culture
medium, then incubated at 37°C for 20 h and removed the Transwell
from 24-well plates. Cells were fixed by 4% formaldehyde and
stained by crystal violet staining solution.
Verification of quantitative real-time
PCR
Total RNA of different cells groups was isolated
using TRIzol reagent (Life technogies) according to the instruction
manual. An ultraviolet spectrophotometer was used to measure the
concentration and purity of total RNA. The first-strand cDNA was
synthesized by Vic qRT Super kit (Vicmed Biotech) and qPCR using
SYBR® qPCR Mix (Roche, Basel, Switzerland) and
implemented on LightCycler® 480 real-time fluorescence
quantitative real-time PCR (Roche). qRT-PCR conditions were as
follows: 30 sec at 95°C, 30 sec at 59°C and 1 min at 72°C for 40
cycles; 95°C for 2 min, followed by 40 cycles of 95°C for 1.5 sec
and 60°C for 60 sec. GAPDH was used as reference gene and qRT-PCR
was performed with three technical replicates for each sample on
the same plate. Dissolution curve was used to analyze the
characteristics of the PCR products. Relative expression levels of
CUEDC2 gene were calculated by the 2−ΔΔCt formula.
Analysis of p-STAT3 and NF-κB expression
and translocation
Coverglass was put into 24-well plates then
different groups of U251 cells were seeded into wells and cultured
for 2 days to make sure of good growth. Then immunofluorescence
staining experiment was done as described above. The cells were
stained by labeling with rabbit anti-p-STAT3 (1:500; Bioworld
Technology, Inc., St. Louis Park, MN, USA) or anti-1κB. The
secondary antibody (1:100; Vicmed Biotech), Alexa Flour
594-conjugated goat anti-rabbit was incubated for 2 h at room
temperature in the dark. After staining with DAPI, glycerol was
used to seal and fluorescence images were captured with Olympus
Bx43 microscope. The U251 cells overexpressed or knocked down of
CUEDC2 were seeded in the 6-well plates and then the protein was
extracted for the western blot analysis. The antibodies used were:
p-STAT3, STAT3, NF-κB and GAPDH (Cell Signaling Technology,
Danvers, MA, USA).
Statistical analysis
All data were processed with the SPSS 16.0 (SPSS,
Inc., Chicago, IL, USA). The measurement data are expressed as mean
± standard deviation (SD). Independent sample t-test was used to
determine significant differences in the mean values between the
two groups. One-way analysis of variance (ANOVA) was used to
compare the mean values of multiple samples. P<0.05 was
considered statistically significant for all tests.
Results
The expression of CUEDC2 in grade IV
human glioma samples and cell lines is low compared with normal
human brain tissue and asctrocyte cells, especially in GSC isolated
from glioma cell line
Several independent microarray databases from
Oncomine database was used to investigate the expression of CUEDC2
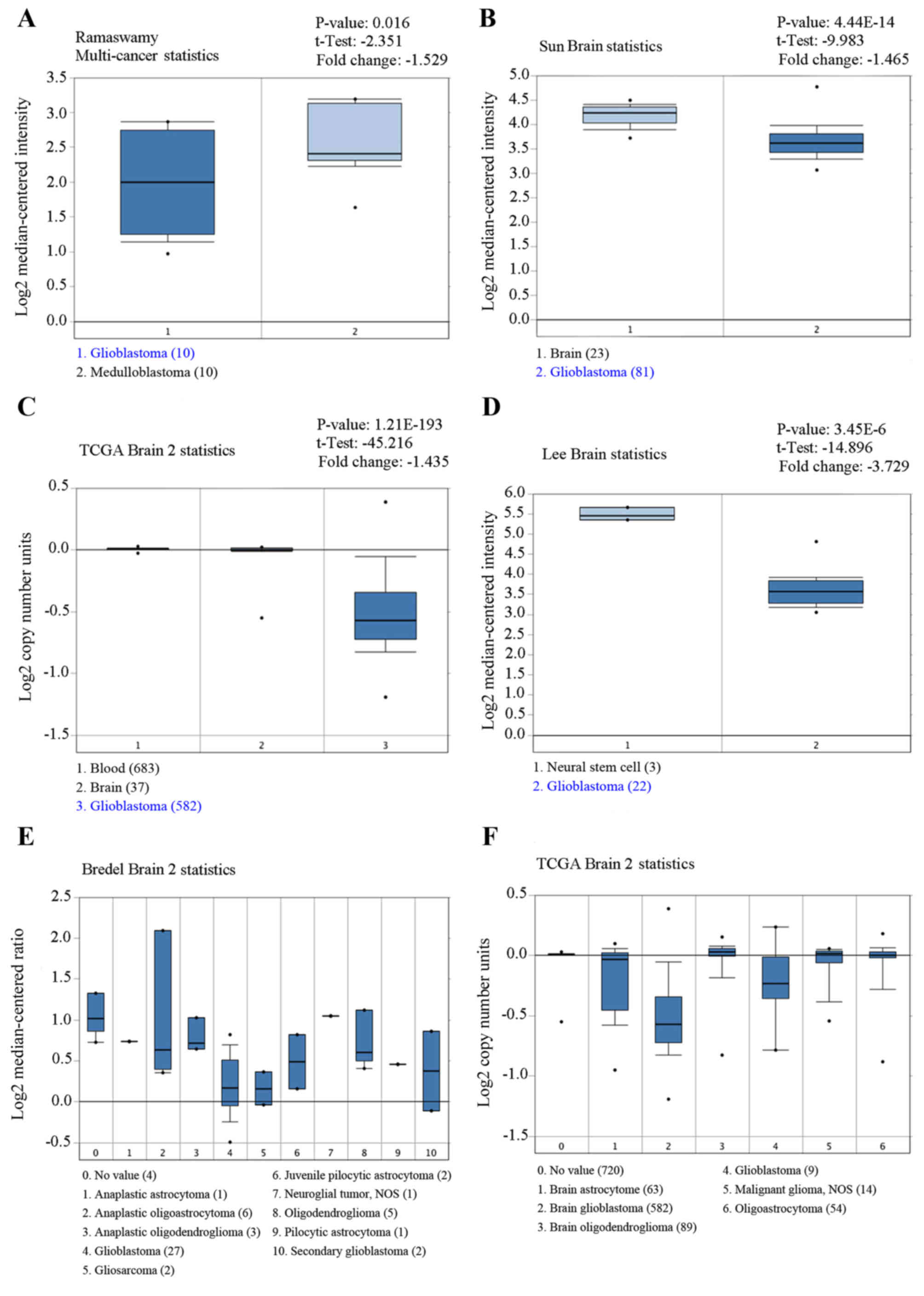
in gliomas. As shown in Fig. 1,
the expression of CUEDC2 is low in glioblastoma compared to normal
brain (P<0.05). The expression of CUEDC2 in GBM tumors was the
lowest among various types of glioma.
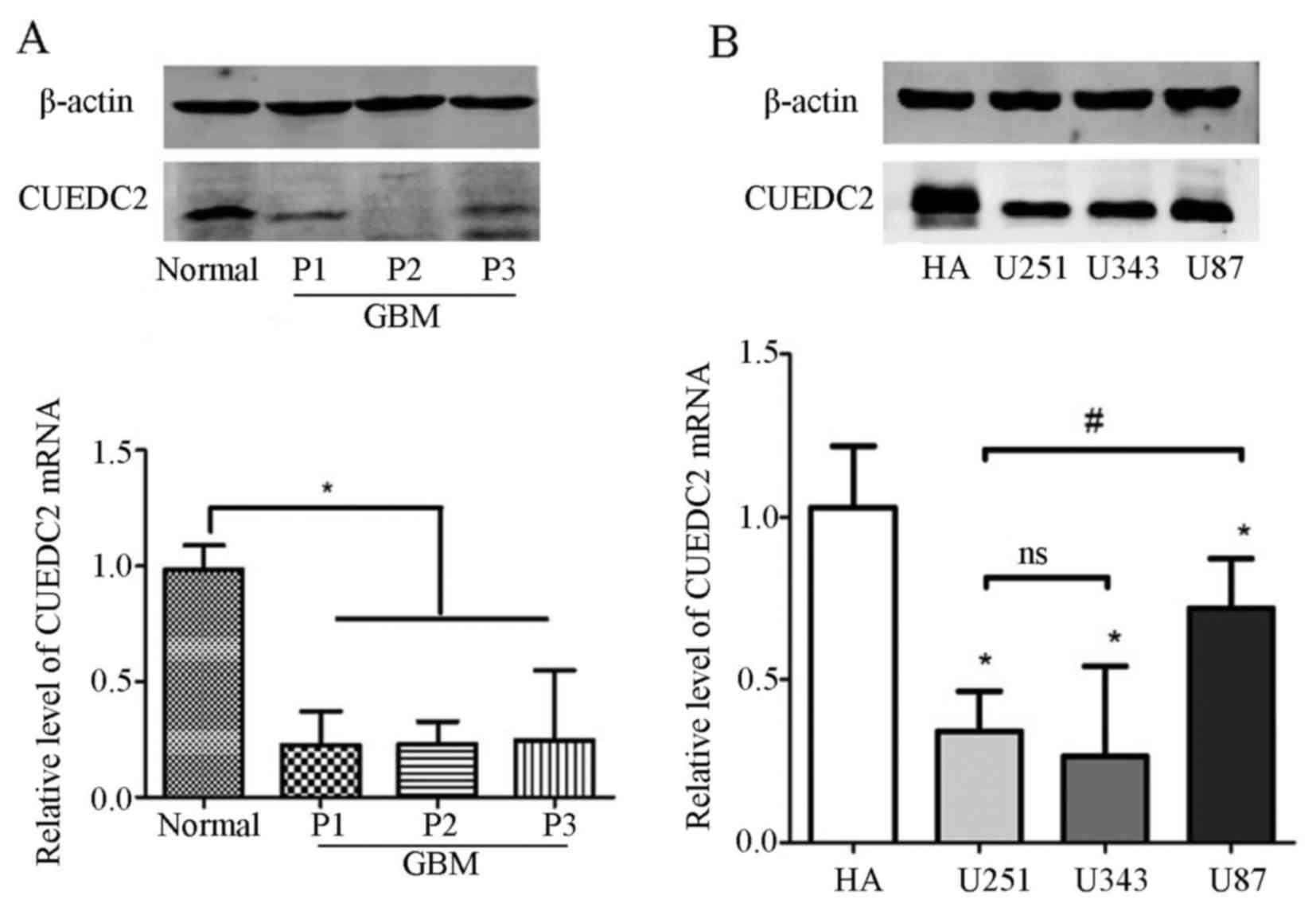
Furthermore, the expression of CUEDC2 was detected
in the clinical samples. Our results indicated that the mRNA and
protein expression levels of CUEDC2 (Fig. 2A) were much lower in high grade
glioma tissues than that of normal brain tissues. According the
follow-up records from hospitals, most of the GBM cases (grade IV)
which had low expression of CUEDC2 lead to shorter lifetime after
surgical removal. Moreover, the expression of CUEDC2 in glioma cell
lines was also analyzed and the results showed that there was lower
expression of CUEDC2 in glioma cell lines than that of normal human
asctrocyte (NHA) cells (Fig.
2B).
The expression of CUEDC2 in GSC isolated from U251
cells by the serum-free medium induced method was also determined.
The isolated GSCs were identified using stem cells markers, CD133
and Nestin via immunofluorescence (Fig. 3A). Then, western blot analysis
showed that the expression of CUEDC2 is extremely low (Fig. 3B) compared to U251 cells. The
results demonstrated that low expression of CUEDC2 might be related
to the poor prognosis in high grade glioma, especially in GBMs and
involved in the development of the GBM and the stemness of GSC.
Overexpression of CUEDC2 inhibits the
proliferation of U251 cells
In order to evaluate the roles of CUEDC2 in high
grade glioma, the stable CUEDC2 overexpression (CUEDC2-OE) or
CUEDC2 knockdown U251 cell line (CUEDC2-KD) was constructed by
lentivirus, and screened by the puromycin and flow cytometry
sorting. The western blot analysis and RT-PCR experiments verified
the stable CUEDC2 overexpression or knockdown U251 cell line was
obtained. The expression of CUEDC2 in protein and mRNA levels in
CUEDC2-OE group was significantly higher than U251-vector control
groups (Fig. 4A). Furthermore, the
expression of CUEDC2 in CUEDC2-KD was decreased obviously (Fig. 4B). The RNAi sequence which has the
best knockdown efficiency was chosen for the following
experiments.
CCK-8 assay was applied to investigate the effects
of CUEDC2 on proliferation. Compared with the control group, the
growth rate of CUEDC2-OE U251 cells was evidently decreased on the
fourth day (Fig. 4C). However,
knockdown of the expression of CUEDC2 promoted proliferation of
U251 cells (Fig. 4D). These
results suggested that overexpression of CUEDC2 suppressed the
proliferation of U251 cells. Downregulation of CUEDC2 promotes U251
cell G1-S and S-G2 phase transition, while overexpression of CUEDC2
arrests G1 phase. Cell proliferation is closely related to the cell
cycle progression (22). To
evaluate the effects of CUEDC2 on cell cycle, the cell cycle
distribution of the stable cell lines overexpressed or knocked down
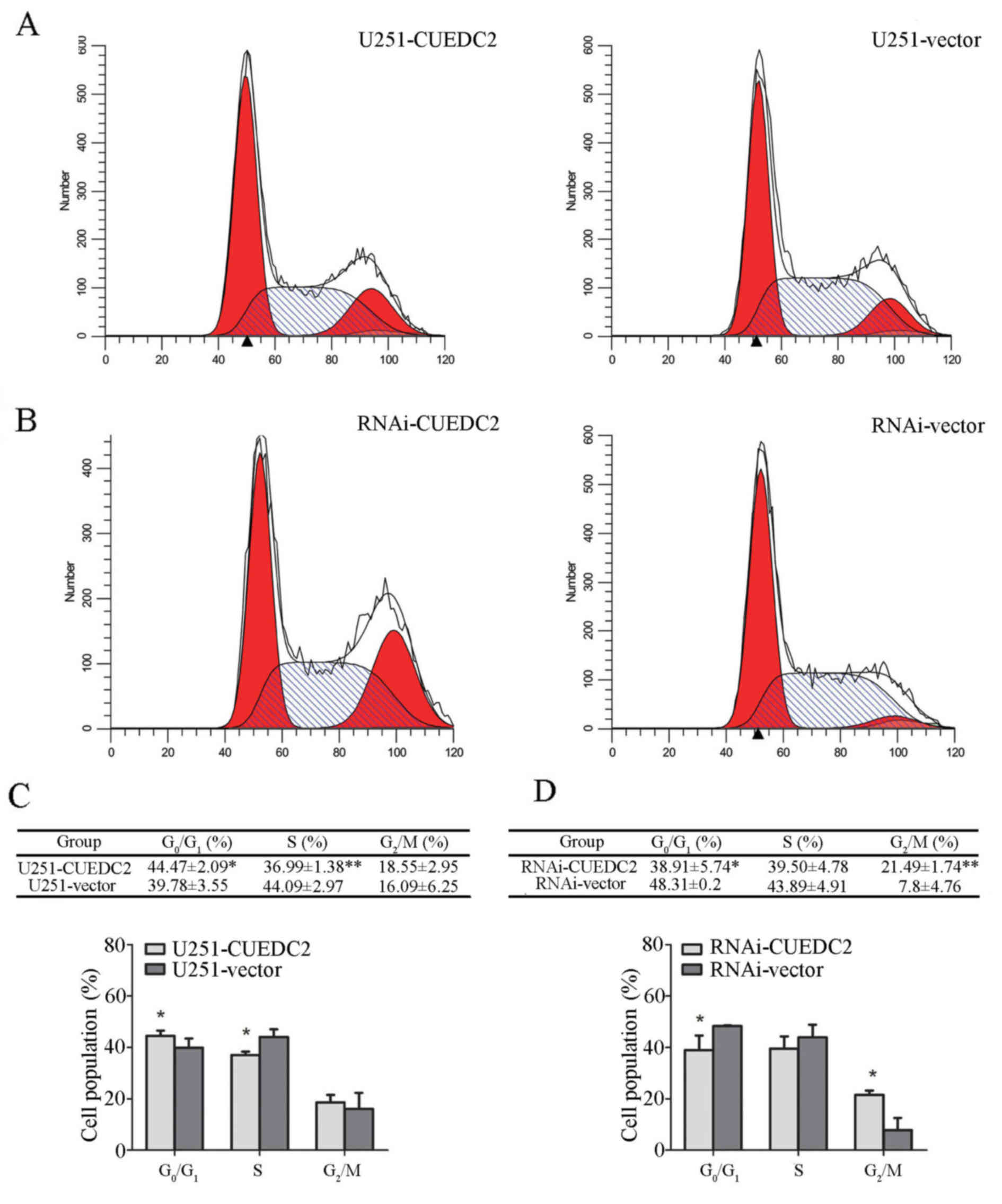
of CUEDC2 was measured using flow cytometry. Our results showed
that overexpression of CUEDC2 caused cell cycle arrest at G1 phase
compared to U251-vector cells (Fig. 5A
and B), the population of U251-CUEDC2 cells in G1 phase was
44.47±2.09 compared to 39.78±3.55% of U251-vector group cells
(P<0.05). Knockdown of the expression of CUEDC2 promoted the
G1-S and S-G2 phase transition. In particular, the percentage of G2
phase cells with U251-CUEDC2-KD group (21.5±1.7%) was much higher
than that of control group (7.8±4.7%) (Fig. 5C and D). Taken together, these
results suggested that inhibiting the expression of CUEDC2 promoted
DNA synthesis and the G1 to S phase cell cycle transition and lead
to more cells entering into G2/M phase.
Knockdown of the expression of CUEDC2
promotes U251 cell migration and invasion
The effect of CUEDC2 on glioma cell migration and
invasion was detected. The scratch wound assay revealed that
overexpression of CUEDC2 decreased the migration of U251-CUEDC2
cells compared with the U251 control group (Fig. 6A). Moreover, knockdown of the
expression of CUEDC2 increased the U251 cell migration (Fig. 6B).
Subsequently, in vitro invasion experiment
was explored by Transwell assay to analyze the effects of CUEDC2 on
glioma cells. Our results demonstrated that overexpression of
CUEDC2 decreased the invasion of U251 cells. Different glioma cell
density gradient: 2, 4 and 8 million cells separately showed the
same trends (Fig. 7). All these
results verified that CUEDC2 played an important role by
suppressing migration and invasion.
Overexpression of CUEDC2 inhibits GSC
neurosphere formatiom and knockdown of the expression of CUEDC2
promotes GSC sphere formation
The recurrence and incurability of glioma is related
to the maintenance of GSC. In order to investigate the effects of
CUEDC2 on the behavior of the GSC, the CUEDC2 stable overexpression
or CUEDC2 knockdown GSC from CUEDC2-OE and CUEDC2-KD cells was
obtained by serum-free medium culture. The glioma stem cells sphere
formation experiment was carried out for about one week. The size
of GSC from CUEDC2-OE is smaller, but larger GSC was obtained from
CUEDC2-KD than that of control GSC from untransfected U251 cells
(Fig. 8). These results indicated
that CUEDC2 might inhibit the stemness of GSC and stem cell sphere
formation.
Overexpression of CUEDC2 inhibits the
activation of JAK1-STAT3 and NF-κB pathways
The activation of JAK1-STAT3 pathway has been
verified to play an important role in the progression of human
gliomas. Activation of this pathway predicts poor prognosis of
patients with gliomas (23).
Therefore, whether CUEDC2 expression affects the JAK1-STAT3 pathway
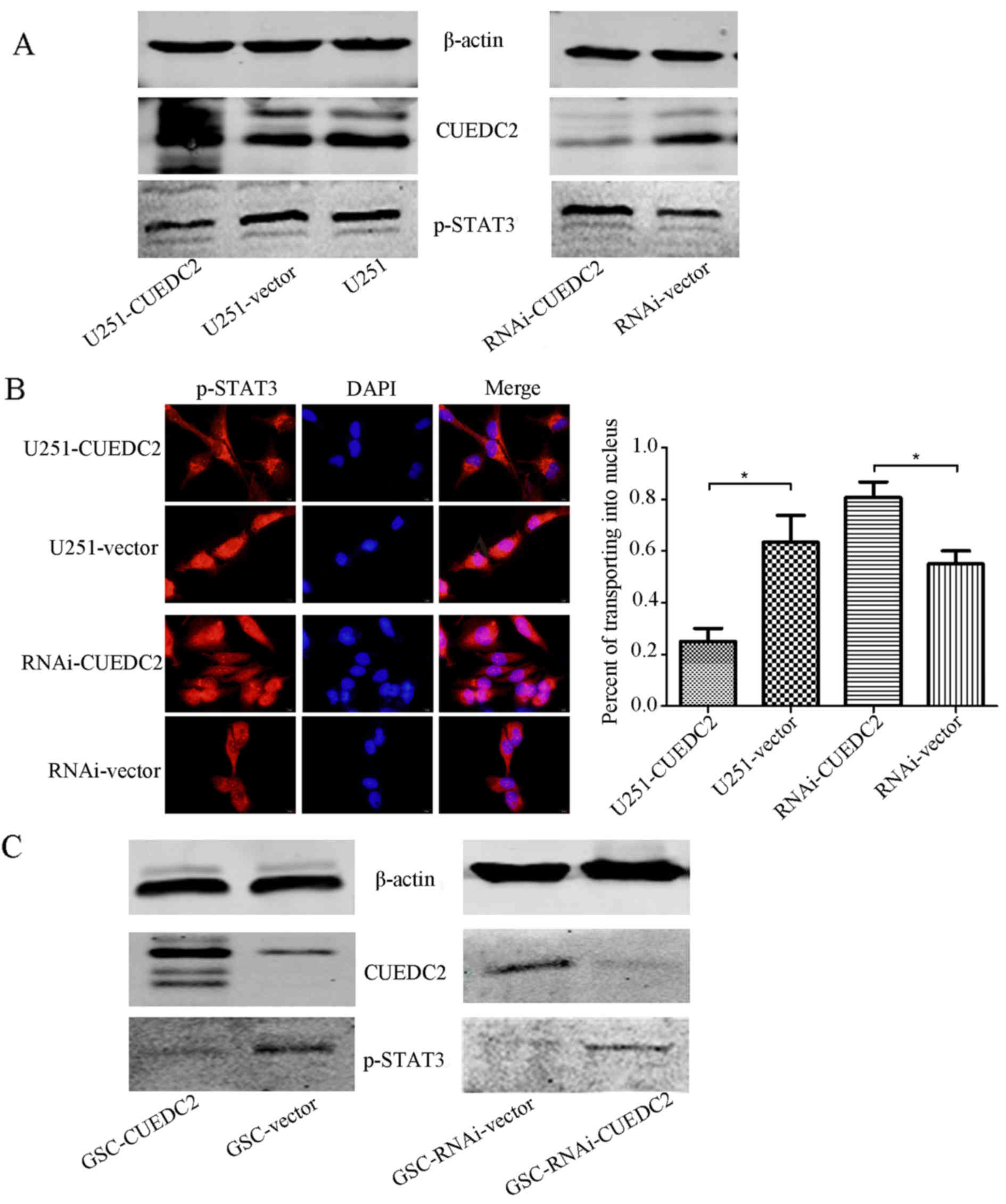
in transgenic U251 cells was investigated. As shown in Fig. 9A, overexpression of CUEDC2
inhibited the activation of JAK1-STAT3 signal pathway in U251
cells. The phosphorylation of STAT3 was increased in CUEDC2-RNAI
cells compared to control U251 cells. The immunofluorescence
experiments indicated that in the U251-CUEDC2 cells, STAT3 protein
is mainly located in cytoplasm, while in the U251-CUEDC2-KD cells,
an increase of STAT3 translocated to the nucleus was observed
(Fig. 9B).
Furthermore, as is known, the existence of GSC is
one of the origins of GBM tumorigenesis. The JAK1-STAT3 signaling
pathway participated in the maintenance of self-renewal of GSC.
Thus, the effect of CUEDC2 on this pathway in GSC was also
detected. Overexpression of CUEDC2 in the GSC also decreased the
activation of p-STAT3 and JAK1-STAT3 signal pathway (Fig. 9C).
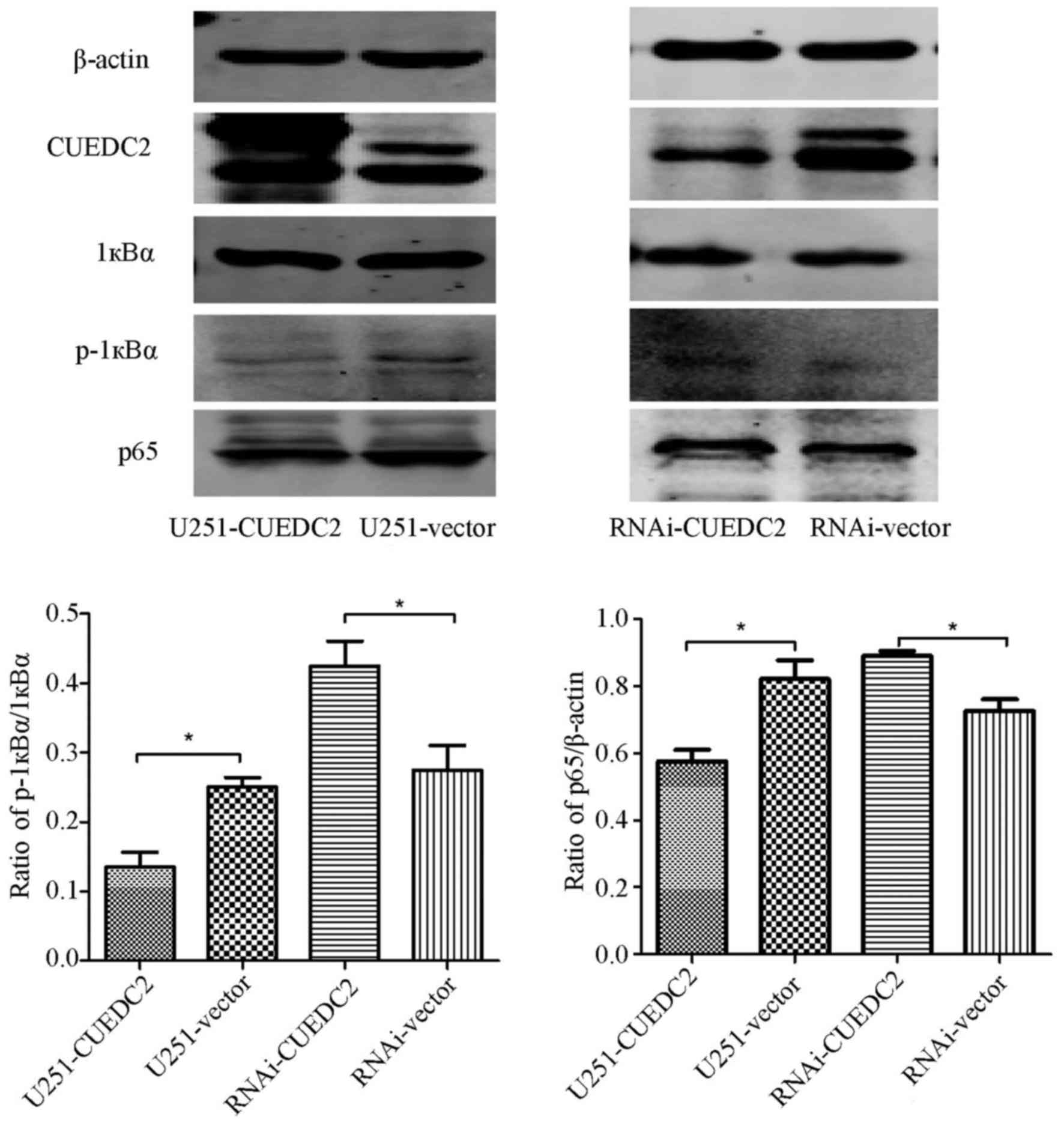
NF-κB is an important transcription
factor participating in inflammation and tumorigenesis
The activation of NF-κB was also analyzed. We found
that the phosphorylation level of 1κB is elevated after knockdown
of CUEDC2 (Fig. 10) by western
blot analysis. Moreover, the immunofluorescence assay indicated
that overexpression of CUEDC2 inhibits the nuclear translocation of
NF-κB and more NF-κB was mainly concentrated in the cytosol. More
NF-κB flocked into the nucleus in the knockdown of CUEDC2 (Fig. 11). These results suggested that
the downregulation of CUEDC2 in glioma may be related to the
activation of JAK1-STAT3 and NF-κB pathway which plays an important
role in tumorigenesis and malignancy.
Discussion
GSC has drawn research attention due to its roles in
the high infiltrating capacity and near-universal malignant glioma
recurrence (5). The major
limitations of glioma treatment are the prevalence of recurrence
after surgery, infiltration into surrounding tissues and intrinsic
or acquired resistance to chemo- and radiotherapy. Intrinsic
resistance to chemo- and radiotherapy is highly reminiscent of the
recently emerging cancer stem-like cell theory, which holds that
the cells with potent tumorigenic capacity are the same as those
that contribute to tumor maintenance and recurrence. Cancer
stem-like cells (CSCs), also called tumor-initiating cells, are a
small population of tumor cells that self-renew like normal stem
cells and are capable of inducing tumorigenesis. CSCs were first
recognized in leukemia. The existence of CSCs was then identified
in solid tumors such as brain, breast, colon, lung and other
tissues. Notably, several independent reports have demonstrated the
existence of brain tumor stem-like cells in different patient
tissues. These cells are thought to be responsible for resistance
to chemo- and radiotherapy, induction of tumor angiogenesis and
tumorigenic capacity under hypoxic conditions. For this reason,
CSCs are regarded as good potential therapeutic targets; however,
critical molecular mechanisms that maintain the 'stemness' of CSCs
are largely unknown. The molecular studies on GSCs such as
gene-expression and differential protein analyzing bring out a nice
prospect in classification of malignant gliomas and their molecular
therapy (24,25).
The Notch, Wnt and Sonic hedgehog, signaling
pathways for development, are known to have important roles in
maintaining stemness in the stem cell niche. These signaling
pathway activation or inactivation may promote the development of
glioma or maintain the stem cell stemness. Several markers, most of
them previously described for neuroprogenitor cells, have been
reported to identify GSC. Also, with the technical development,
many genes, such as NANOG, OCT4, SOX2, EGFR, P53, SOCS1, STAT3 and
JAK1 were proven to be critically involved in the regulation of
self-renewal and expansion of many different types of normal stem
cells, including neural stem cells. Recently, CUEDC2 has drawn
attention for its roles in different cancers such as breast, skin
cancer, ovarian serous carcinoma and chronic myeloid leukemia.
Furthermore, this gene was also proven to play important roles in
the mataining of stemness of the breast cancer stem cells. Although
the roles of this gene have been studied in the different cancers
and stem cells, whether it played important roles in the
development of the glioma and maintain the stemness in glioma stem
cells is still unclear. In order to investigate the roles of CUEDC2
in the development of the glioma and the maintenance of the
stemness in the glioma stem cells, the expression of CUEDC2 in the
patients and the cell line was determined. The results from the
Oncomine database showed that the expression of CUEDC2 in the GBMs
from the patients was much lower than that of normal brains.
Furthermore, this database also suggested that the expression of
CUEDC2 in the patients had an inverse relationship with the glioma
grade, and thus the expression of the CUEDC2 in the patients might
be used as a prognostic indicator of gliomas or as a biomarker for
the pathological level of glioma.
CUEDC2 is a CUE domain containing protein, located
on chromosome 10, roles of which in the development of cancer are
still debated. It has a role in monoubiquitin and polyubiquitin
recognition, as well as in facilitating intramolecular
monoubiquitination (11,26). The roles of the CUEDC2 have been
studied in many different cancers, the functions of CUEDC2 seemed
to have a dual function, either in tumor promotion or tumor
suppression. Some research groups inferred the CUEDC2 as a tumor
promoter; Gao et al (13)
reported that CUEDC2 caused earlier activation of APC/Cdc20 to
promote the metaphase-anaphase transition that led to chromosome
missegregation and aneuploidy which might contribute to tumor
development. Pan et al (16) further showed that CUEDC2 led to the
resistance to endocrine therapy in breast cancer by promoting the
degradation of estrogen receptor-α (ERα) and progesterone receptor
(PR). Furthermore, Zhang et al (14) showed that CUEDC2 degradation was
elevated for UV light-induced G1 arrest in skin cancer. Wang et
al (18) also showed that
CUEDC2 may be a promising biomarker and CUEDC2-positive expression
was associated with a shorter disease-free survival time in ovarian
serous carcinoma. Other research groups proved the CUEDC2 as a
tumor suppressor, Sun et al (19) reported that the decreased CUEDC2 in
lung adenocarcinoma cells led to a poor clinical outcome and a
shorter survival time in patients. In order to determine whether
the CUEDC2 played important roles in the development of glioma as
the expression of CUEDC2 in the glioma grade indicated, the stable
overexpression and downregulation of CUEDC2 cell lines,
respectively, was constructed. Our results indicated that
overexpression of CUEDC2 inhibits cell proliferation, migration and
invasion. Then, we further analyzed the effect of CUEDC2 on cell
cycle and GSCs sphere formation, which also suggested
downregulation of CUEDC2 promoted G1-S and S-G2 phase transition
and GSCs sphere formation. All in all, our results indicated that
the CUEDC2 might be a tumor suppressor in the development of glioma
which was consistence with Sun et al (19).
Although this study indicated that CUEDC2 played
important roles in inhibiting the development of the glioma, the
mechanism of how CUEDC2 affects development of the glioma was still
unclear. As is known, hyper-activation of JAK1-STAT3 signaling
pathway has close relationship to the development of certain types
of human tumors. Previous studies reported that gliomas patients
who possessed high-activation of JAK1 and STAT3 had lower overall
survival rates than those with low JAK1 and STAT3 activation
(23,27). When the JAK1-STAT3 pathway is
activated, STAT3 will be phosphorylated and phosphorylated-STAT3
(p-STAT3) forms dimers and translocates into the nucleus to
regulate expression of genes by binding to elements of their
promoters. It regulates many pathways which play important roles in
tumorigenesis, including cell cycle progression, apoptosis, and
tumor angiogenesis, metastasis, as well as tumor cell invasion of
the immune system (28). The study
by Wang et al (18) showed
that CUEDC2 reduced colonic inflammatory reaction and inhibited the
excessive proliferation of the inflammatory mucous epithelial cell
through inhibiting the activation of NF-κB and STAT3. CUEDC2 has
been reported to act as an adaptor protein to target IκB kinase
(IKK) for dephosphorylation and inactivation by recruiting protein
phosphatase (PP1), and have an inhibitory role in the activation of
transcription factor NF-κB signaling pathway, which play pivotal
roles in inflammatory responses and tumorigenesis (9). In the present study, the expression
of IKBα was detected when CUEDC2 was overexpressed, our results
implied that level of IKBα phosphorylation was decreased, when
CUEDC2 is overexpressed in U251 cells. IKBα is considered as an
inhibitor of NF-κB signaling pathway, when phosphorylated, leading
to the elimination or decreased NF-κB signaling. The expression of
p65 reduced and the amount of p65 transporting into the nucleus.
All these results suggested that CUEDC2 inhibited the development
of the glioma partially by affecting the NF-κB signaling pathway.
Accumulated evidence has shown that JAK1-STAT3 pathway activation
plays an important role in tumorigenesis and progression (29,30).
Inhibiting the excessive activation of this pathway may become a
therapeutic focus for treating GBM. Studies of Zhang et al
(21) indicated that CUEDC2 and
SOCS3 cooperate to negatively regulate JAK1-STAT3 pathway. In this
study, our results indicated that overexpression of CUEDC2
inhibited the activation of the JAK1-STAT3 pathway and the STAT3
nucleus translocation. Thus, CUEDC2 may also inhibit the
development of the glioma partially by affecting the JAK1-STAT3
signaling pathway. In total, our results implied that CUEDC2
affected the development of the glioma by influencing the
activation of NF-κB and JAK1-STAT3 signaling pathways.
In the present study, the roles of CUEDC2 in the
development of glioma was investigated, indicating overexpression
of CUEDC2 inhibits cell proliferation, migration and invasion and
GSC sphere formation. Our results also suggested that CUEDC2
affected the development of the glioma by influencing the
activation of NF-κB and JAK1-STAT3 signaling pathway. Combined with
the expression of CUEDC2 in the patient and the cell lines, all
these result implied that CUEDC2 might be a tumor inhibitor in
malignant glioblastoma. Thus, CUEDC2 may be considered as a
potential diagnostic indicator and therapeutic targets for GBM
treatment.
Acknowledgments
The present study was supported by grants from the
National Natural Science Foundation of China (no. 81402073 and no.
81570136), the Natural Science Foundation of Jiangsu Province
(BK20130218), the Program of the China Postdoctoral Science
Foundation (2014M551663), the Foundation of Jiangsu Province Six
Talents Peak (JY-061), the Key Project Supported by Jiangsu
Province Universities (15KJA320005) and the Project Supported by
Jiangsu Province Universities (15KJB310023).
Glossary
Abbreviations
Abbreviations:
|
GBM
|
glioblastoma multiforme
|
|
p-STAT3
|
phosphorylated-STAT3
|
|
TMZ
|
temozolomide
|
|
GSC
|
glioma stem cells
|
|
NSC
|
neural stem cell
|
|
ERα
|
estrogen receptor-α
|
|
PR
|
progesterone receptor
|
|
IKK
|
IκB kinase
|
|
PP1
|
protein phosphatase
|
|
SOCS3
|
suppressors of cytokine signaling
3
|
|
WHO
|
World Health Organization
|
|
DAPI
|
4′6-diamidino-2-phenylindole
|
|
PI
|
propidium iodide
|
|
CSCs
|
cancer stem-like cells
|
References
|
1
|
Fuller GN and Scheithauer BW: The 2007
Revised World Health Organization (WHO) Classification of Tumours
of the Central Nervous System: Newly codified entities. Brain
Pathol. 17:304–307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feng H, Liu KW, Guo P, Zhang P, Cheng T,
McNiven MA, Johnson GR, Hu B and Cheng SY: Dynamin 2 mediates
PDGFRα-SHP-2-promoted glioblastoma growth and invasion. Oncogene.
31:2691–2702. 2012. View Article : Google Scholar
|
|
4
|
Helseth R, Helseth E, Johannesen TB,
Langberg CW, Lote K, Rønning P, Scheie D, Vik A and Meling TR:
Overall survival, prognostic factors, and repeated surgery in a
consecutive series of 516 patients with glioblastoma multiforme.
Acta Neurol Scand. 122:159–167. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guryanova OA, Wu Q, Cheng L, Lathia JD,
Huang Z, Yang J, MacSwords J, Eyler CE, McLendon RE, Heddleston JM,
et al: Nonreceptor tyrosine kinase BMX maintains self-renewal and
tumorigenic potential of glioblastoma stem cells by activating
STAT3. Cancer Cell. 19:498–511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liebelt BD, Shingu T, Zhou X, Ren J, Shin
SA and Hu J: Glioma stem cells: Signaling, microenvironment, and
therapy. Stem Cells Int. 2016:78498902016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bao S, Wu Q, McLendon RE, Hao Y, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bao S, Wu Q, Sathornsumetee S, Hao Y, Li
Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD and Rich JN: Stem
cell-like glioma cells promote tumor angiogenesis through vascular
endothelial growth factor. Cancer Res. 66:7843–7848. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Z, Bao S, Wu Q, Wang H, Eyler C,
Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, et al:
Hypoxia-inducible factors regulate tumorigenic capacity of glioma
stem cells. Cancer Cell. 15:501–513. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peñuelas S, Anido J, Prieto-Sánchez RM,
Folch G, Barba I, Cuartas I, García-Dorado D, Poca MA, Sahuquillo
J, Baselga J, et al: TGF-beta increases glioma-initiating cell
self-renewal through the induction of LIF in human glioblastoma.
Cancer Cell. 15:315–327. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Donaldson KM, Yin H, Gekakis N, Supek F
and Joazeiro CA: Ubiquitin signals protein trafficking via
interaction with a novel ubiquitin binding domain in the membrane
fusion regulator, Vps9p. Curr Biol. 13:258–262. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Man J and Zhang X: CUEDC2: An emerging key
player in inflammation and tumorigenesis. Protein Cell. 2:699–703.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao YF, Li T, Chang Y, Wang YB, Zhang WN,
Li WH, He K, Mu R, Zhen C, Man JH, et al: Cdk1-phosphorylated
CUEDC2 promotes spindle checkpoint inactivation and chromosomal
instability. Nat Cell Biol. 13:924–933. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang WN, Zhou J, Zhou T, Li AL, Wang N,
Xu JJ, Chang Y, Man JH, Pan X, Li T, et al:
Phosphorylation-triggered CUEDC2 degradation promotes UV-induced G1
arrest through APC/CCdh1 regulation. Proc Natl Acad Sci
USA. 110:11017–11022. 2013. View Article : Google Scholar
|
|
15
|
Li HY, Liu H, Wang CH, Zhang JY, Man JH,
Gao YF, Zhang PJ, Li WH, Zhao J, Pan X, et al: Deactivation of the
kinase IKK by CUEDC2 through recruitment of the phosphatase PP1.
Nat Immunol. 9:533–541. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pan X, Zhou T, Tai YH, Wang C, Zhao J, Cao
Y, Chen Y, Zhang PJ, Yu M, Zhen C, et al: Elevated expression of
CUEDC2 protein confers endocrine resistance in breast cancer. Nat
Med. 17:708–714. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang PJ, Zhao J, Li HY, Man JH, He K,
Zhou T, Pan X, Li AL, Gong WL, Jin BF, et al: CUE domain containing
2 regulates degradation of progesterone receptor by
ubiquitin-proteasome. EMBO J. 26:1831–1842. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang S, Pu J, Li N, Li C, Li C, Yu L, Wang
X, Fu S and Cui L: CUEDC2 protects against experimental colitis and
suppresses excessive proliferation of intestinal mucosa. Dig Dis
Sci. 60:3603–3609. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun L, Bai L, Lin G, Wang R, Liu Y, Cai J,
Guo Y, Zhu Z and Xie C: CUEDC2 down-regulation is associated with
tumor growth and poor prognosis in lung adenocarcinoma. Oncotarget.
6:20685–20696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang H, Chang G, Wang J, Lin Y, Ma L and
Pang T: CUEDC2 sensitizes chronic myeloid leukemic cells to
imatinib treatment. Leuk Res. 37:1583–1591. 2013h. View Article : Google Scholar
|
|
21
|
Zhang WN, Wang L, Wang Q, Luo X, Fang DF,
Chen Y, Pan X, Man JH, Xia Q, Jin BF, et al: CUEDC2 (CUE
domain-containing 2) and SOCS3 (suppressors of cytokine signaling
3) cooperate to negatively regulate Janus kinase 1/signal
transducers and activators of transcription 3 signaling. J Biol
Chem. 287:382–392. 2012. View Article : Google Scholar :
|
|
22
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tu Y, Zhong Y, Fu J, Cao Y, Fu G, Tian X
and Wang B: Activation of JAK/STAT signal pathway predicts poor
prognosis of patients with gliomas. Med Oncol. 28:15–23. 2011.
View Article : Google Scholar
|
|
24
|
Phillips HS, Kharbanda S, Chen R, Forrest
WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et
al: Molecular subclasses of high-grade glioma predict prognosis,
delineate a pattern of disease progression, and resemble stages in
neurogenesis. Cancer Cell. 9:157–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Collins VP: Mechanisms of disease: Genetic
predictors of response to treatment in brain tumors. Nat Clin Pract
Oncol. 4:362–374. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shih SC, Prag G, Francis SA, Sutanto MA,
Hurley JH and Hicke L: A ubiquitin-binding motif required for
intramolecular monoubiquitylation, the CUE domain. EMBO J.
22:1273–1281. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Carro MS, Lim WK, Alvarez MJ, Bollo RJ,
Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al:
The transcriptional network for mesenchymal transformation of brain
tumours. Nature. 463:318–325. 2010. View Article : Google Scholar
|
|
28
|
Osugi T, Oshima Y, Fujio Y, Funamoto M,
Yamashita A, Negoro S, Kunisada K, Izumi M, Nakaoka Y, Hirota H, et
al: Cardiac-specific activation of signal transducer and activator
of transcription 3 promotes vascular formation in the heart. J Biol
Chem. 277:6676–6681. 2002. View Article : Google Scholar
|
|
29
|
Yu H and Jove R: The STATs of cancer - new
molecular targets come of age. Nat Rev Cancer. 4:97–105. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee H, Deng J, Kujawski M, Yang C, Liu Y,
Herrmann A, Kortylewski M, Horne D, Somlo G, Forman S, et al:
STAT3-induced S1PR1 expression is crucial for persistent STAT3
activation in tumors. Nat Med. 16:1421–1428. 2010. View Article : Google Scholar : PubMed/NCBI
|