Introduction
Esophageal squamous cell carcinoma (ESCC) is one of
the most frequent malignant tumors in China (1). Some advances have been made in the
treatment of ESCC, including surgery, chemotherapy, radiation, or a
combination of these options. However, the associated prognosis of
ESCC patients is still not satisfactory. The five-year overall
survival rate of ESCC patients after surgery is only approximately
14–22% (2–4). Tumor invasion and metastasis are the
major causes of the poor prognosis of ESCC patients (5,6).
Some oncogenic and tumor suppressive factors have been reported to
be associated with ESCC progression. However, only a few of them
are specific and conclusive (7,8).
Therefore, exploring the detailed molecular mechanisms of ESCC cell
progression, invasion, and metastasis will help improve disease
diagnosis and therapy.
MicroRNAs (miRNAs) are endogenously expressed,
small, non-coding RNAs of 19–24 nucleotides that regulate gene
expression by either inhibiting translation or cleaving their
target messenger RNAs (mRNAs) (9,10).
The miRNA target site is considered to be the 3′-untranslated
region (3′-UTR) of mRNA. Mounting evidence shows that miRNAs may
also bind coding or 5′-untranslated regions (5′-UTRs) (11–15).
Evidence also shows that miRNAs are involved in the pathogenesis of
most cancer, such as cell differentiation, proliferation,
oncogenesis, angiogenesis, cell migration, and invasion, where some
can function as tumor suppressors or oncogenes (16–19).
The aberrant regulation of miRNAs has been shown involved in
numerous cancers, including ESCC (4,14).
MicroRNA-34a (miR-34a), which is located in
chromosome 1p36 and belongs to the miR-34 family, is directly
regulated by the p53 transcription factor (20). miR-34a is more commonly
downregulated than the non-malignant tissues in multiple cancers,
such as tongue squamous cell carcinoma (12), non-small cell lung cancer (21), colon cancer (22), pancreatic cancer (23), and others (24–26).
miR-34a in cancer has also been extensively studied in tumor
growth, migration, invasion, and epithelial-to-mesenchymal
transition (EMT). Accordingly, miR-34a has been reported to
suppress cell proliferation, migration, invasion, and EMT in
various cancer cells by targeting oncogenes (22–24,27–31).
miR-34a modulates stem cell self-renewal and difference in multiple
cancers (23,25,32–35).
In addition, miR-34a regulates drug resistance (36–38),
and has been suggested to act as a suppressor that plays a key role
in tumorigenesis. However, the role that miR-34a plays in ESCC is
still in question. One recent study found that miR-34a could
inhibit ESCC cell migration and invasion by targeting Yin Yang-1
(YY-1). YY-1 can suppress the matrix metalloproteinase (MMP)-2 and
-9 expression levels (39).
However, miR-34a is reported to inhibit cell migration and invasion
by directly targeting MMP-9 in tongue squamous cell carcinoma
(12). Therefore, whether miR-34a
can directly target MMP-9 or MMP-2 to regulate ESCC cell migration
and invasion needs to be investigated. The detailed role and
mechanism of miR-34a in ESCC cell regulation needs to be further
elucidated.
This study evaluated the miR-34a and p53 expression
in ESCC tumor tissues and adjacent normal tissues. It shows that
miR-34a and p53 both significantly decreased in the ESCC tissues.
Moreover, the expression of p53 and miR34a has positive correlation
in ESCC tissues. We further verified the functional role for
miR-34a in ESCC cell invasion and migration. The regulative network
of miR-34a in ESCC cancer was also explored. The obtained data
indicate that miR-34a suppressed ESCC cell invasion and migration.
Our study also demonstrates that MMP-2/MMP-9/fibronectin type III
domain containing 3B (FNDC3B) is a direct downstream target of
miR-34a, and miR-34a overexpression in the ESCC cells decreases
both mRNA production and protein expression of MMP-2/MMP-9/FNDC3B.
miR-34a suppressed tumor cell invasion and migration in ESCC by
suppressing MMP-2 and MMP-9/FNDC3B expression levels.
Materials and methods
Ethics statement
The experiments were approved by the Institutional
Ethics Review Board of the First Affiliated Hospital of the Shihezi
University School of Medicine. All samples were obtained from
patients who signed informed consent forms approving the use of
their tissues for research purposes after surgery.
Human tissue samples and cell lines
A total of 15 normal esophageal and 15 human ESCC
tissues were used for real-time polymerase chain reaction (PCR)
analysis. All tissue specimens were obtained from the First
Affiliated Hospital of Shihezi University. All tissues were
formalin-fixed and paraffin-embedded (FFPE) for pathological
diagnosis. The normal esophageal tissues were obtained from a
distance of ≥5 cm from the tumor tissues.
The ESCC cell lines used, including EC9706 and TE-1,
were purchased from the Shanghai Institute of Biochemistry and Cell
Biology (Shanghai, China). HEK293 was a gift from the biochemical
laboratory (American Type Culture Collection) of the Shihezi
University School of Medicine. All cells were cultured in
Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with
10% heat-inactivated fetal bovine serum (FBS; BI), 100 U/ml
penicillin, and 100 µg/ml streptomycin in a humidified
incubator containing 5% CO2 at an atmosphere of
37°C.
RNA isolation and quantitative reverse
transcription PCR (qRT-PCR)
The total RNA, including miRNA, was isolated from
the FFPE and cell samples using the miRNeasy FFPE (no. 73504) and
miRNeasy Mini (no. 217004) kits from Qiagen (Hilden, Germany). The
kits were used according to the manufacturer's instructions. The
concentration and purity of the RNA samples were measured using
NanoDrop2000 (Thermo Scientific). The RNA was reverse transcribed
into cDNA using the miScript II Reverse Transcription (RT) kit (no.
218161) from Qiagen according to the manufacturer's protocol with
1000 ng total RNA. Subsequently, qRT-PCR was conducted using the
SYBR Green PCR kit (miScript SYBR Green PCR kit for miRNA; no.
218073; Qiagen) and the QuantiFast SYBR Green PCR kit for mRNA (no.
204054; Qiagen) containing ROX as the reference dye in the ABI 7500
RT-PCR system. The PCR primers for miR-34a (Hs_miR-34a_1 miScript
primer assay, MS00003318), U6 (Hs_RNU6-2_1 miScript primer assay,
MS00033740), and ACTB (encoding β-actin) (Hs_ ACTB_1_SG,
QT00095431) were purchased from Qiagen. The MMP-2, MMP-9, and
FNDC3B primers were synthesized (Applied Sangon Biotech Co., Ltd.,
Shanghai, China). The sequences were as follows: MMP-2 forward,
5′-TATGGCTTCTGCCCTGAGAC-3′ and MMP-2 reverse
5′-CACACCACATCTTTCCGTCA-3′; MMP-9 forward,
5′-CGAACTTTGACAGCGACAAG-3′ and MMP-9 reverse,
5′-CGGCACTGAGGAATGATCTA-3′; and FNDC3B forward,
5′-TTGGAGAGGGAATGGTGTTT-3′ and FNDC3B reverse
5′-CAGGTCACGCAGCAAGTTAG-3′. Accordingly, U6 and β-actin were used
as internal control for the normalization and quantification of the
miRNA and mRNAs, respectively. Quantification was performed in
triplicate.
miR-34a mimic/inhibitor transfection
The chemically synthesized miR-34a mimic
(Syn-hsa-miR-34a-5p miScript miRNA mimic, MSY0000255), miR-34a
inhibitor (Anti-hsa-miR-34a-5p miScript miRNA inhibitor,
MIN0000255), and negative controls (AllStars Negative Control
siRNA, no. 1027280; miScript inhibitor negative control, no.
1027271) were purchased from Qiagen. The ESCC cells were
transfected with miR-34a mimic (10 nM) for miR-34a overexpression
or with miR-34a inhibitor (50 nM) for miR-34a inhibition by
Hiperfect (no. 301705; Qiagen) according to the manufacturer's
instructions.
Wound healing assay
A total of 30×104 ESCC cells were
cultured in one 6-well culture plate. These cells were transfected
with different concentrations of miR-34a mimic (10 nM) for
overexpression or miR-34a inhibitor (50 nM) for inhibition using
Hiperfect for 24 h. The transfection allowed cells to reach full
confluence. The confluent cells were subsequently wounded by
scraping using a 10 µl pipette tip. The dislodged cells were
removed by washing with a serum-free medium. The cells that
migrated into the wounded area or protruded from the wound border
were captured using an inverted microscope (Olympus BX51; Olympus
Corp., Tokyo, Japan) after 24 h incubation at 37°C with 5%
CO2. The cell migration distances were quantified by
subtracting the distance between the wound edges at 24 h from the
distance measured at 0 h. Each experiment was independently
performed for at least three times.
Cell invasion assays
The 8 µm plain (for migration) or
Matrigel-coated (for invasion) (BD Biosciences, Franklin Lakes, NJ,
USA) pore membrane and Transwell inserts (Corning Costar Corp.,
Corning, NY, USA) were placed in the wells of 24-well culture
plates (Corning Costar Corp.) to assess the migrated or invasive
ability of the cell sublines. Subsequently, EC9706 cells
(9×104 per well) initially transfected with miR-34a
mimic (10 nM) or miR-control (10 nM) were cultured for 24 h. The
ESCC cells were then harvested and re-suspended in 500 µl
serum-free DMEM medium. Accordingly, 150 µl cell suspension
was seeded into the upper chamber. Thereafter, 600 µl DMEM
containing 10% FBS was added to the lower chamber to stimulate cell
penetration. After 24 h incubation at 37°C with 5% CO2,
the DMEM was discarded and washed thrice with 1X PBS. The
non-invasive cells that remained on top of the membrane were then
manually removed with a cotton wool. The invading cells that
adhered to the filter's undersurface were fixed using 4%
paraformaldehyde (Solarbio, Beijing, China) for 30 min at 4°C,
dried under room temperature, and stained with 0.1% crystal violet
for 20 min. The Transwell inserts were thoroughly washed with
water. The invaded cells were counted under a microscope in five
randomly chosen fields. Representative images were then taken. Data
are expressed as the number of invaded cells (means ± standard
deviation) normalized to the number of control cells that migrated.
Each test was performed in triplicate.
Target gene prediction and luciferase
reporter assay
The predicted miR-34a target genes and their target
binding site regions were investigated using a bioinformatics tool
(TargetScan, miRanda, RNA22 software) and a literature review. We
analyzed the biological function and distribution on the tissue of
the target genes. We then chose the migration- and invasion-related
genes and their relationship with the ESCC. We found that MMP-2,
MMP-9, and FNDC3B mRNA contained putative miR-34a target sites. The
miR-34a target sequences in the MMP-9 coding region and the MMP-2
and FNDC3B 3′UTR region were amplified by PCR. These sequences were
then inserted into multiple cloning sites of the pMIR-REPORT that
contained a firefly luciferase reporter gene (Obio Technology,
Shanghai, China). The mutant reporter vectors of the MMP-9 coding
region and the MMP-2 and FNDC3B 3′-UTR regions that lacked the
miR-34a binding sites were obtained through site-directed
mutagenesis. All constructs were verified by DNA sequencing.
For the luciferase reporter assay, the EC9706 and
293T cells (14×104) were seeded into 24-well plates the
day before transfection to ensure 80% confluence at the time of
transfection. Accordingly, 1 µg of firefly luciferase
reporter plasmid, including the wild-type or mutant coding regions
of MMP-9, 3′UTR of MMP-2, and 3′UTR of FNDC3B, and 0.05 µg
of pRL-TK, which is a plasmid expressing the Renilla
luciferase, were transfected into EC9706 and 293T cells cultured in
24-well plates together with 50 nM miR-34a mimic or negative
control and 100 nM of miR-34a inhibitor or negative control using
Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA). The cells were subjected to lysis 48 h post-transfection. The
luciferase activities were determined using the dual luciferase
reporter assay system (Promega) according to the manufacturer's
instructions. The relative firefly luciferase activity (i.e.,
firefly luciferase activity/Renilla luciferase activity) for
each construct was compared to the negative control. The luciferase
activity was averaged in triplicate for each transfection.
Western blot analysis
The ESCC cells were transfected with miR-34a mimic
or miR-34a inhibitor using the previously described method. The
cells were washed three times with 1X PBS 48 h post-transfection
and lysed with RIPA lysis buffer (Solarbio) in ice. The total
proteins were extracted following standard protocol. Equal amounts
of protein from whole cell lysates of each sample were separated on
10% sodium dodecyl sulfate polyacrylamide gel electrophoresis.
These proteins were then transferred to a polyvinylidene difluoride
membrane (Solarbio). The membranes were blocked in 5% non-fat dried
milk at room temperature for 2 h. They were then incubated with
primary antibodies at 4°C overnight and extensively washed. The
membranes were further incubated with a corresponding secondary
antibody at room temperature for 2 h after washing with 1X
Tris-buffered saline buffer containing 0.1% Tween-20 six times (5
min × 6). The primary antibodies against β-actin (diluted 1:1000;
Zhongshan Golden Bridge Biotechnology, Beijing, China), MMP-9
(diluted 1:500; Abcam, Cambridge, MA, USA), MMP-2 (diluted 1:500;
Cell Signaling Technology, Danvers, MA, USA), and FNDC3B (diluted
1:200; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were
employed. The secondary antibodies were used at 1:4000 to 1:5000
concentrations. The relative protein expression levels were
quantified by the Gelpro analyzer software (GelPro32 4.0) using
β-actin as the internal reference.
Statistical analysis
The statistical analyses were performed using SPSS
17.0 (SPSS Inc., Chicago, IL, USA). The experimental data are
presented as mean ± standard deviation based on the results of at
least three repeats. The between-group comparisons were all based
on Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-34a expression is decreased in the
ESCC
The miR-34a expression in the ESCC was confirmed by
determining the miR-34a levels in the ESCC and normal esophageal
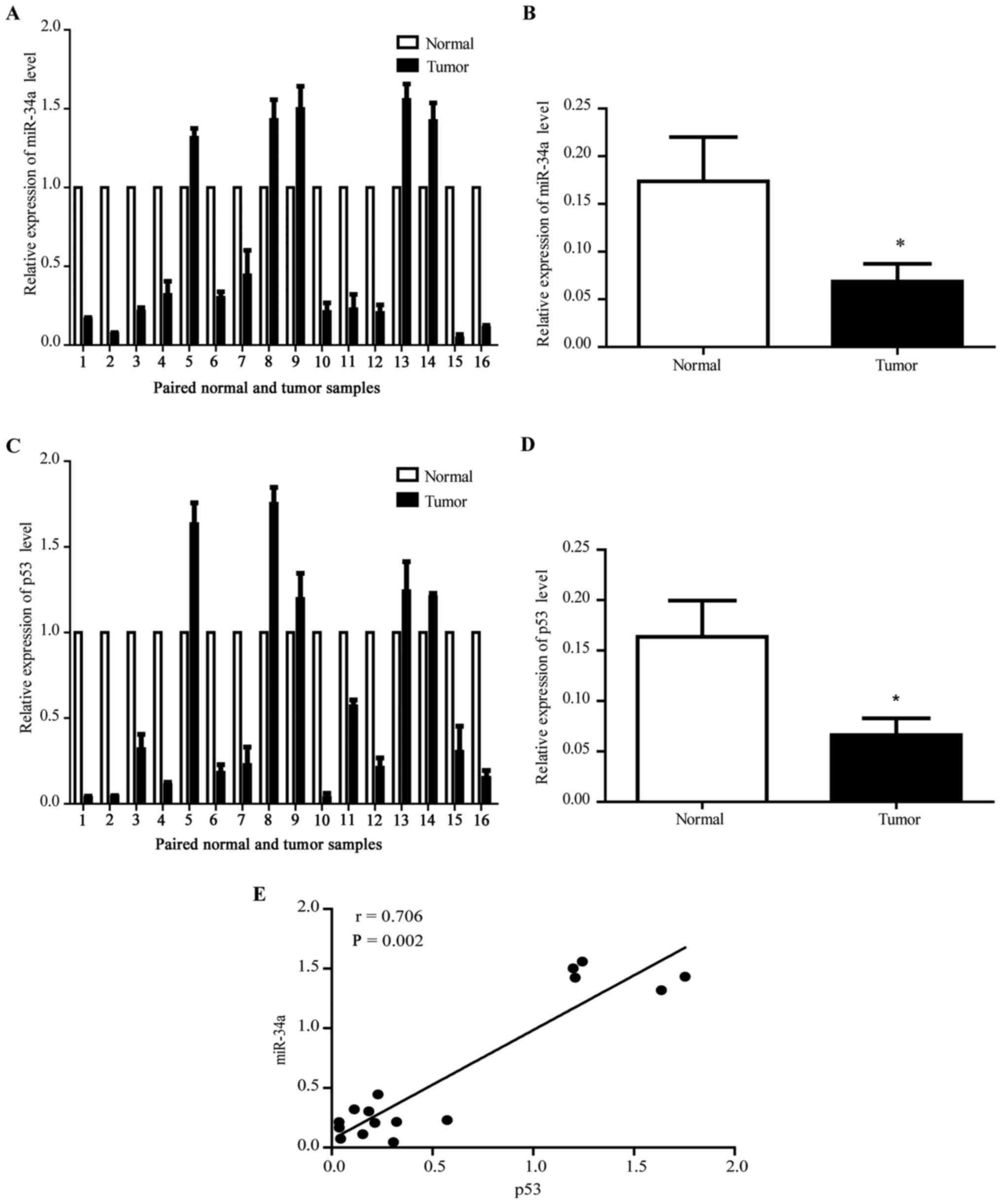
tissues through qRT-PCR using U6 as the internal control. Fig. 1A shows that the miR-34a expression
decreased in 11 of 16 (68.8%) tumor samples. The average miR-34a
expression also decreased in tumor tissues (Fig. 1B). In addition, the expression of
p53 in ESCC tissues declined (Fig. 1C
and D) with the miR34a expression level, the positive
regulation expression between p53 and miR34a (Fig. 1E) indicate that miR34a may be a
downstream target gene of p53 in ESCC similarly to other
cancers.
miR-34a inhibits ESCC cell migration and
invasion
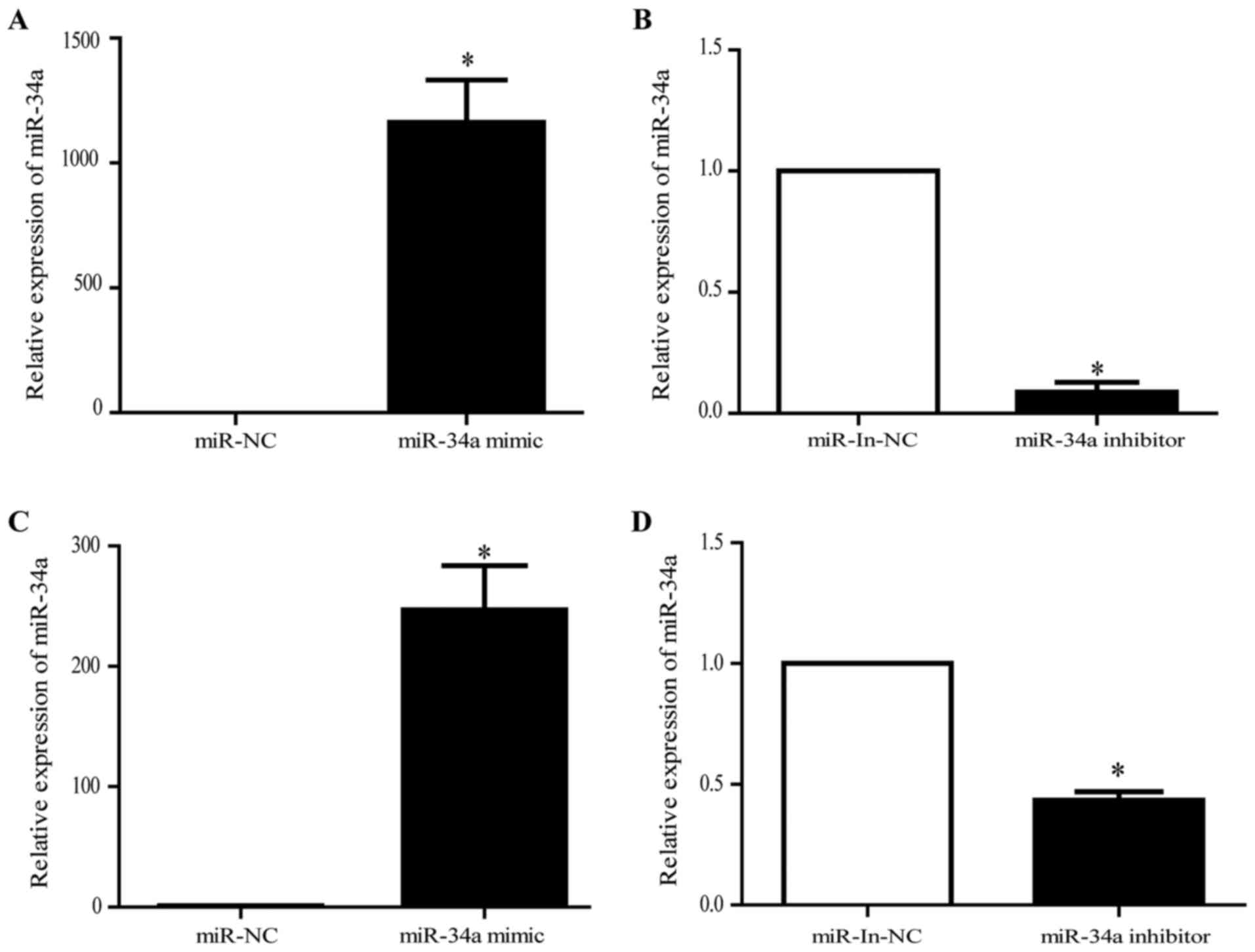
The effects of miR-34a in the ESCC were investigated
by transfecting the miR-34a mimics or the miR-34a inhibitor into
the EC9706 and TE-1. The transfection efficiency was confirmed
using qRT-PCR (Fig. 2). The cell
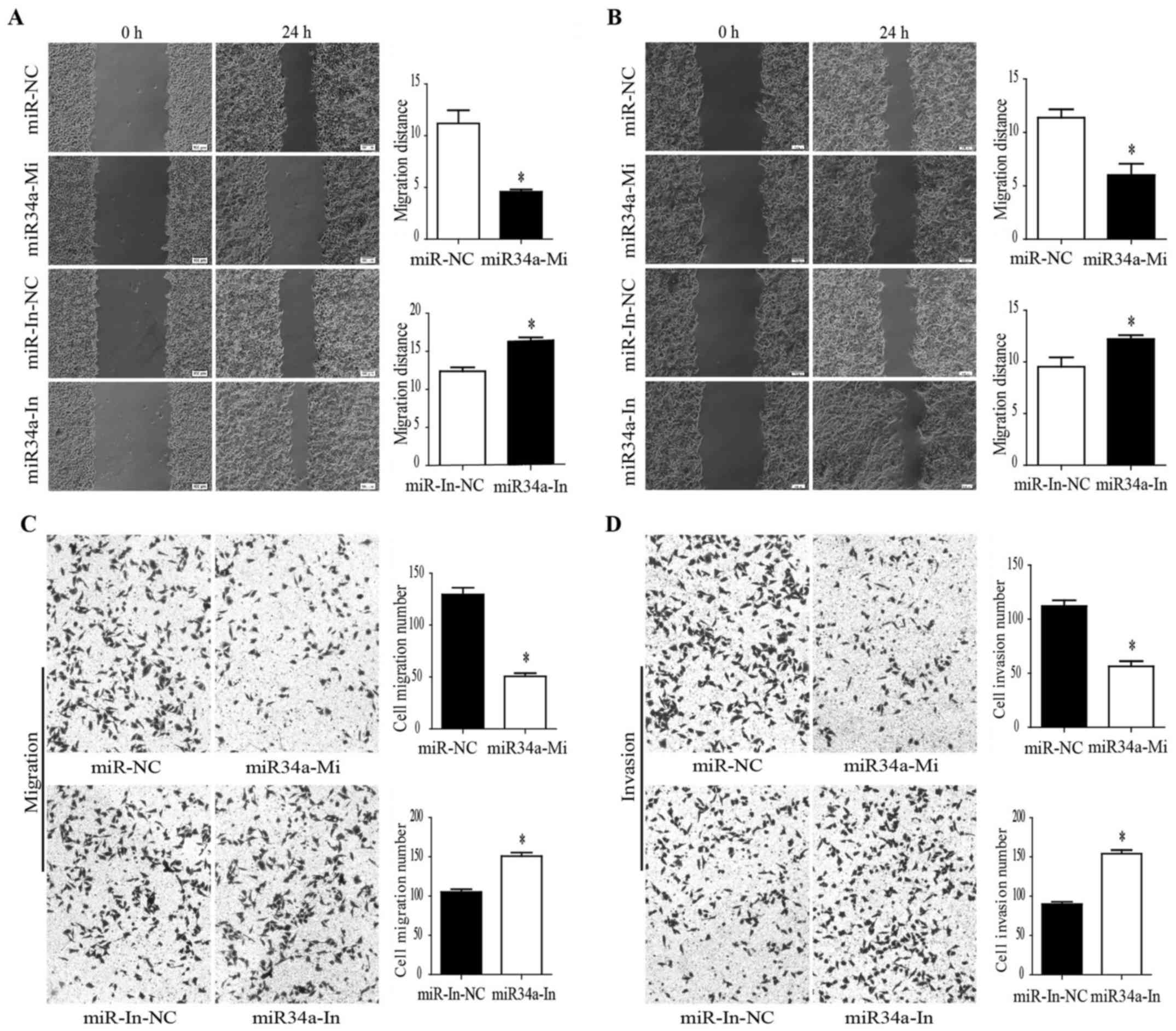
migration assays were then performed within 48 h after miR-34a
mimic transfection. The wound-healing assay showed that cell
migration was significantly inhibited in the miR-34a
mimic-transfected EC9706 cells compared with the negative control.
A comparison to the negative control shows that the inhibited
miR-34a expression significantly promoted EC9706 cell migration
(Fig. 3A). Similarly, cell
migration was significantly inhibited in the miR-34a
mimic-transfected TE-1 cells. miR-34a inhibition also promoted TE-1
cell migration (Fig. 3B). We
conducted Transwell migration and invasion assays. Consequently,
the results demonstrated that EC9706 cell migration and invasion
were inhibited by the miR-34a overexpression. In contrast, the
miR-34a inhibition promoted EC9706 cell migration and invasion
(Fig. 3C and D). These results
suggested that miR-34a could inhibit ESCC cell migration and
invasion, and inhibiting the miR-34a expression can increase ESCC
cell migration and invasion.
miR-34a directly targets and suppresses
MMP-2/MMP-9/FNDC3B in ESCC cells
The prediction results obtained using the
bioinformatics tool and the literature review indicated that the
human miR-34a may target the MMP-9 coding region and the MMP-2 and
FNDC3B 3′-UTR regions. Fig. 4A, C and
E) showed the miR-34a putative binding sites and corresponding
mutant sites of MMP-9, MMP-2, and FNDC3B. We constructed luciferase
reporter plasmids containing putative sequences for MMP-2, MMP-9,
and FNDC3B or their corresponding mutant sequences as controls to
further confirm that miR-34a directly targeted MMP-2, MMP-9, and
FNDC3B. At 48 h post-transfection, the luciferase activity of the
reporter containing the miR-34a-targeted wild-type sequences of
MMP-9 was significantly suppressed in the 293T cells (Fig. 4B) and EC9706 cells (data not shown)
with miR-34a overexpression but not their corresponding mutant
sequences. Similarly, the luciferase activity of the reporter
containing the miR-34a-targeted wild-type sequences of MMP-2 and
FNDC3B was significantly suppressed in the 293T cells (Fig. 4D and F) and EC9706 cells (data not
shown) with miR-34a overexpression but not their corresponding
mutant sequences.
On the contrary, the luciferase activity of the
reporter containing the miR-34a-targeted wild-type sequences of
MMP-2, MMP-9, and FNDC3B increased in the miR-34a
inhibitor-transfected 293T cells (Fig.
4B, D and F) and EC9706 cells (data not shown) but not their
corresponding mutant sequences. The influence of miR-34a on MMP-2,
MMP-9, and FNDC3B expression levels was further confirmed by
measuring the MMP-2, MMP-9, and FNDC3B levels in the EC9706 cells
and TE-1cells with miR-34a overexpression or miR-34a inhibition.
The result showed that MMP-9, MMP-2, and FNDC3B presented an
inverse expression trend to miR-34a in the EC9706 cells (Fig. 5A–C) and TE-1 cells (Fig. 5D–F). On the one hand, western blot
analysis demonstrated that the miR-34a overexpression decreased the
protein levels of MMP-2, MMP-9, and FNDC3B in the EC9706 cells
(Fig. 6A) and TE-1 cells (Fig. 6B). On the other hand, the miR-34a
inhibitor transfection increased the protein levels of MMP-2,
MMP-9, and FNDC3B in the EC9706 cells (Fig. 6A) and TE-1 cells (Fig. 6B). These data demonstrated that
miR-34a directly binds to MMP-2/MMP-9/FNDC3B and represses the
MMP-2/MMP-9/FNDC3B translation in ESCC cells.
 | Figure 5miR-34a overexpression decreases the
MMP-2, MMP-9, and FNDC3B mRNA levels. (A–C) qRT-PCR detection of
MMP-9, MMP-2, and FNDC3B mRNA expression in EC9706 cells
transfected with miR-34a mimic or inhibitor. miR-34a overexpression
decreases the MMP-2, MMP-9, and FNDC3B mRNA levels, whereas miR-34a
inhibition increases them (*P<0.05). (D–F) qRT-PCR
detection of MMP-9, MMP-2, and FNDC3B mRNA expression in TE-1 cells
transfected with miR-34a mimic or inhibitor. miR-34a overexpression
decreases the MMP-2, MMP-9, and FNDC3B mRNA levels, whereas miR-34a
inhibition increases them (*P<0.05). NC, normal
control; In, inhibitor. |
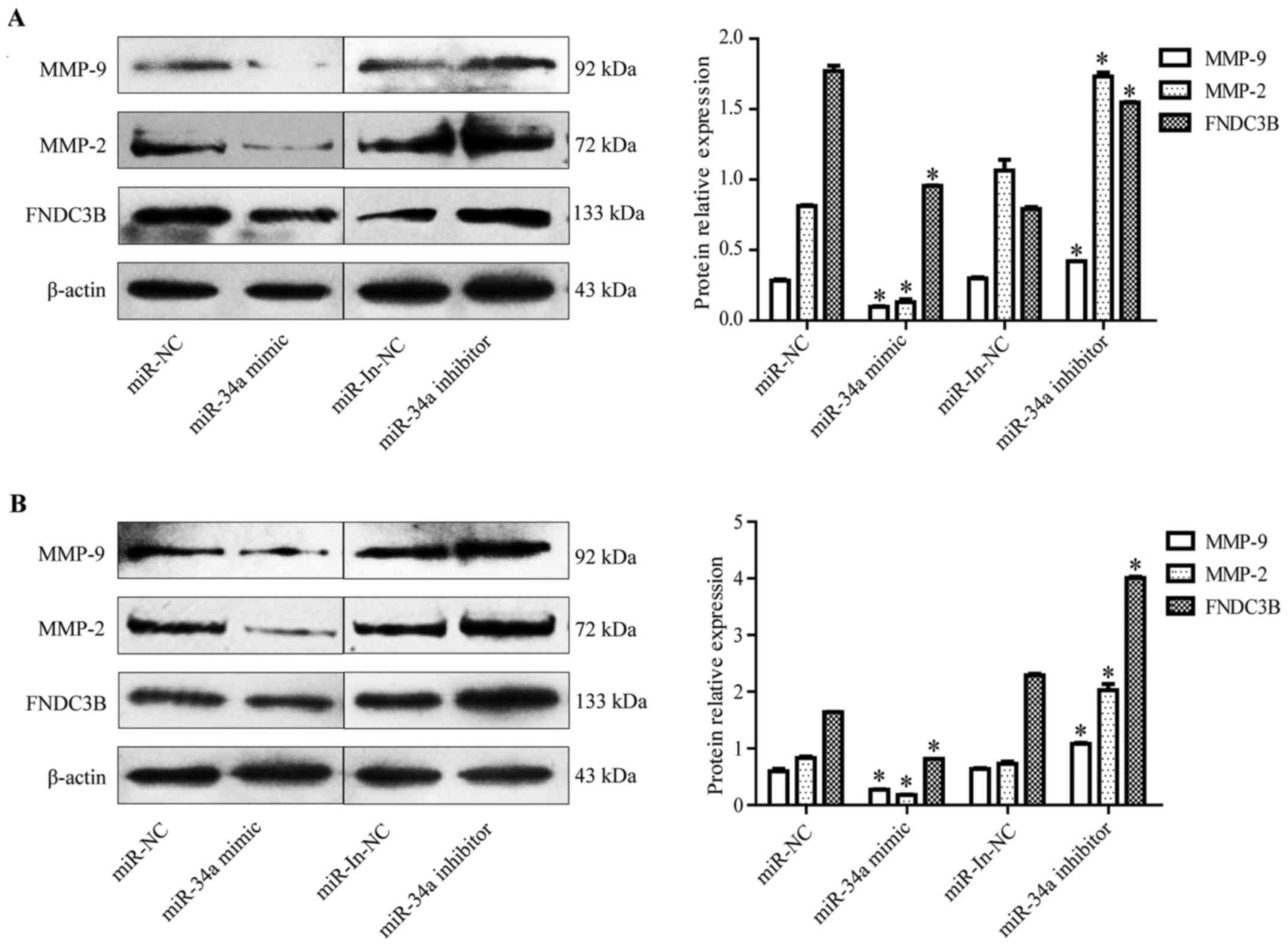 | Figure 6miR-34a overexpression decreases the
MMP-2, MMP-9, and FNDC3B protein levels. (A) The MMP-9, MMP-2, and
FNDC3B protein levels in the EC9706 cells transfected with miR-34a
mimic or miR-34a inhibitor were detected by western blotting.
miR-34a overexpression decreases the MMP-2, MMP-9, and FNDC3B
protein levels, whereas miR-34a inhibition increases them
(*P<0.05). (B) Western blot detection of MMP-9,
MMP-2, and FNDC3B mRNA expression in TE-1 cells transfected with
miR-34a mimic or inhibitor. miR-34a overexpression decreases the
MMP-2, MMP-9, and FNDC3B protein levels, whereas miR-34a inhibition
increases them (*P<0.05). NC, normal control; In,
inhibitor. |
Discussion
ESCC is the leading cause of mortality in digestive
tract malignancies, with a poor five-year overall survival rate
(2–4). Therefore, a better understanding of
the mechanisms involved in ESCC progression is urgent. miRNAs have
demonstrated far-reaching effects on cellular biology and cancer
development (40,41). A number of studies have reported a
relatively low miR-34a expression level in various cancer types and
cancer cell lines, including ESCC (12,21–26,39,42,43).
The present study found that the miR-34a as well as
p53 expression was significantly reduced in human esophageal tumor
tissues compared to adjacent normal tissues. It may be a potential
positive control correlation between p53 and miR-34a in ESCC as
previously reported in other cancers. Epigenetic mechanisms,
including methylation and histone modification, chromosome
deficiency, and transcriptional regulation, can influence the miRNA
expression (44). Furthermore, the
abnormal miR-34a expression is reported in multiple cancers (e.g.,
breast, lung, colon, and bladder cancers, and pancreatic carcinoma)
due to aberrant CpG methylation of its promoter (45,46).
A study found that the miR-34a promoter is more frequently
methylated in the ESCC than in controls. Accordingly, the miR-34a
expression decreases in patients with a high level of methylation
compared to that in normal tissues (42). It indicates that the mechanisms of
miR-34a repression may be aberrant from the CpG methylation of its
promoter in ESCC.
Studies have found that miR-34a overexpression can
suppress cell proliferation, migration, invasion, and EMT (22–24,27–31,43).
Furthermore, miR-34a has significant relationships with node
metastases, clinical stage, and patient mortality in tongue
squamous cell carcinoma (12) and
ESCC (43). This finding indicates
that miR-34a has a crucial role in tumor development, progression,
and prognosis. We observed that miR-34a inhibits ESCC cell
migration and invasion, which is consistent with previous results
(39). Cell migration and invasion
are normal events in cancer processes and are two important
elements that lead to metastases. Metastasis is a major cause of
death in patients with esophageal cancer (6). miR-34a has been reported to suppress
cell migration and invasion by targeting various oncogenes
(22,24,25,30).
Previous studies reveal that miR-34a can inhibit
ESCC cell migration and invasion by targeting YY-1. However,
miR-34a might still directly modulate other genes simultaneously
inhibiting ESCC cell migration and invasion because of the complex
regulative network of miRNAs. We have found that MMP-2 and MMP-9
contain putative miR-34a target sites using a bioinformatics tools
and a literature review. The MMP family, especially MMP-2 and MMP-9
known as gelatinases, is involved in cancer migration and invasion
by degrading type-IV collagen, which is the major component of the
basement membrane. MMP-2 and MMP-9 play vital roles in the early
stages of tumor invasion. They are secreted during tumor growth,
and can affect the surrounding microenvironment, thereby causing
dynamic changes in the tumor bio-behavior (47).
MMP-2 and MMP-9 are reportedly overexpressed in ESCC
tissues compared to that in the paired normal esophageal tissues.
They are also related to tumor invasion and metastasis in the ESCC
(48,49). The luciferase reporter assays in
this study reveal that miR-34a could directly interact with MMP-2
and MMP-9. Both mRNA and protein levels of MMP-2 and MMP-9
significantly decrease when miR-34a is overexpressed in ESCC cells.
This finding is consistent with a report of an indirect negative
correlation of miR-34a with MMP-2 and MMP-9 in ESCC (39). However, our results show that
miR-34a directly targets MMP-2 or MMP-9. This finding seems to be
different from the results of a previous report (39), where miR-34a indirectly
downregulates MMP-2 or MMP-9 suppressing YY-1 in the ESCC. The
current study also confirms that miR-34a directly targets MMP-9 in
tongue squamous cell carcinoma (12) and Fra-1 in colon cancer (22). miR-34a may downregulate mRNA
expression through direct and indirect regulatory mechanisms.
We also found that FNDC3B is the most likely direct
target gene of miR-34a using a bioinformatics software and a
literature review. FNDC3B is located at 3q26.31 and covers a large
area (360 kb). FNDC3B is a member of the fibronectin family
(50) with biological functions
that remain largely unclear. The gene, which was initially
discovered with another name (i.e., factor for adipocyte
differentiation 104), is upregulated in the early stages of
adipocyte differentiation. This upregulation indicates its
potential role as a positive regulator of adipogenesis (51,52).
However, FNDC3B has been recently identified as an important
oncogenic driver gene of the 3q amplicon, thereby adding to the
growing list of oncogenic drivers within this amplified region
(53).
Previous studies have reported that miR-143-targeted
the oncogene FNDC3B, regulating hepatocarcinoma metastasis
(54). Furthermore, FNDC3B
amplification could increase cell proliferation and promote
tumorigenesis of hepatocellular carcinoma (50). The amplification and overexpression
of FNDC3B are found in over 20% of cancers including ESCC (50,53,55).
However, the role of FNDC3B in ESCC is presently still unconfirmed.
Studies have found that the FNDC3B expression is significantly
altered in ESCC and targeted by most miRNAs (56). In addition, FNDC3B overexpression
induces EMT and activates several cancer pathways, including TGFβ1
signaling, which contributes to cancer metastasis (53). miR-34a acts as a suppressor that
regulates TGFβ1 signaling by targeting PDGFRA in glioblastoma
(57) and Smad4 inhibits EMT in
extrahepatic cholangiocarcinoma (58). The TGFβ1 signaling is an activity
in ESCC that could induce EMT and contribute to ESCC metastasis
(59). Therefore, whether miR-34a
has a connection with the TGFβ1 signaling by regulating FNDC3B in
ESCC needs to be determined. We have performed luciferase reporter
assays to confirm that miR-34a could directly interact with FNDC3B.
The results revealed that miR-34a could suppress the luciferase
activity of the reporter containing the miR-34a-targeted wild-type
sequences of FNDC3B. Both mRNA and protein levels of FNDC3B also
significantly decrease when miR-34a is overexpressed in ESCC cells.
In view of these results, FNDC3B is a direct target gene of miR-34a
and may be important in the regulatory network. FNDC3B may also be
involved in the progression of ESCC. We detected that FNDC3B could
promote the ESCC cell invasion and migration (data not shown),
combining this result with previous reports (53,59),
we propose a hypothesis that miR-34a inhibits ESCC cell migration
and invasion by targeting FNDC3B and reduces EMT by inhibiting the
activity of the TGFβ1 signaling pathway. This hypothesis needs
further research.
Previous studies have confirmed that miR-34a is a
downstream target of p53 (60).
However, few studies have reported the downstream targets of
miR-34a in ESCC. Only one study found that miR-34a could directly
target YY-1 in ESCC (39). In the
present study, we found that miR-34a could directly target MMP-2,
MMP-9, and FNDC3B in ESCC. miR-34a can directly and simultaneously
modulate multiple genes in ESCC because of the complex regulative
network of miRNAs.
This study confirmed that miR-34a expression
significantly decreased in ESCC tissues and could inhibit the ESCC
cell line migration and invasion. Accordingly, MMP-2, MMP-9, and
FNDC3B are the genes directly targeted by miR-34a. miR-34a may have
a therapeutic value in ESCC treatment. Therefore, further studies
on the anticancer mechanisms of miR-34a may contribute to the
development of new therapeutic strategies for ESCC.
Abbreviations:
|
miRNA
|
microRNA
|
|
miR-34a
|
microRNA 34a
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
MMP-9
|
matrix metalloproteinase-9
|
|
MMP-2
|
matrix metalloproteinase-2
|
|
FNDC3B
|
fibronectin type III domain containing
3B
|
|
mRNAs
|
messenger RNAs
|
|
3′-UTR
|
3′-untranslated region
|
|
5′-UTRs
|
5′-untranslated regions
|
|
EMT
|
epithelial-mesenchymal transition
|
|
YY-1
|
Yin Yang-1
|
|
PCR
|
polymerase chain reaction
|
|
real-time RT-PCR
|
real-time reverse transcription
PCR
|
|
FFPE
|
formalin-fixed and
paraffin-embedded
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
qRT-PCR
|
quantitative reverse transcription
PCR
|
|
cDNA
|
complementary deoxyribonucleic
acid
|
|
PBS
|
phosphate buffered saline
|
|
TGFβ1
|
transforming growth facor β1
|
|
CDs
|
coding region
|
|
Tris
|
trihydroxymethylaminornethane
|
Acknowledgments
This study was supported in part by the National
Natural Science Foundation of China (grant nos. 81260301, 81560399,
81160301, 81360358, and 81460362). The doctoral grant from the
Xinjiang Production and Construction Corps (grant no. 2014BB019)
and the high-level talent project of Shihezi University (no.
RCZX201533) are also acknowledged. We thank the Biochemical
Laboratory of the Shihezi University School of Medicine for raising
the 293T in this study. The authors would also like to express
their sincere thanks to ShineWrite.com, the professional editing company, for
editing and modifying the English in the manuscript.
References
|
1
|
Chen W, Zheng R, Zeng H and Zhang S: The
updated incidences and mortalities of major cancers in China, 2011.
Chin J Cancer. 34:502–507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gamliel Z and Krasna MJ: Multimodality
treatment of esophageal cancer. Surg Clin North Am. 85:621–630.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee KH, Goan YG, Hsiao M, Lee CH, Jian SH,
Lin JT, Chen YL and Lu PJ: MicroRNA-373 (miR-373)
post-transcriptionally regulates large tumor suppressor, homolog 2
(LATS2) and stimulates proliferation in human esophageal cancer.
Exp Cell Res. 315:2529–2538. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li H, Zheng D, Zhang B, Liu L, Ou J, Chen
W, Xiong S, Gu Y and Yang J: Mir-208 promotes cell proliferation by
repressing SOX6 expression in human esophageal squamous cell
carcinoma. J Transl Med. 12:1962014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang X, Tian X, Liu F, Zhao Y, Sun M, Chen
D, Lu C, Wang Z, Shi X, Zhang Q, et al: Detection of HPV DNA in
esophageal cancer specimens from different regions and ethnic
groups: A descriptive study. BMC Cancer. 10:192010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mizushima T, Nakagawa H, Kamberov YG,
Wilder EL, Klein PS and Rustgi AK: Wnt-1 but not epidermal growth
factor induces beta-catenin/T-cell factor-dependent transcription
in esophageal cancer cells. Cancer Res. 62:277–282. 2002.PubMed/NCBI
|
|
8
|
Yang L, Leung AC, Ko JM, Lo PH, Tang JC,
Srivastava G, Oshimura M, Stanbridge EJ, Daigo Y, Nakamura Y, et
al: Tumor suppressive role of a 2.4 Mb 9q33-q34 critical region and
DEC1 in esophageal squamous cell carcinoma. Oncogene. 24:697–705.
2005. View Article : Google Scholar
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jia LF, Wei SB, Mitchelson K, Gao Y, Zheng
YF, Meng Z, Gan YH and Yu GY: miR-34a inhibits migration and
invasion of tongue squamous cell carcinoma via targeting MMP9 and
MMP14. PLoS One. 9:e1084352014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Phatak P, Byrnes KA, Mansour D, Liu L, Cao
S, Li R, Rao JN, Turner DJ, Wang JY and Donahue JM: Overexpression
of miR-214-3p in esophageal squamous cancer cells enhances
sensitivity to cisplatin by targeting survivin directly and
indirectly through CUG-BP1. Oncogene. 35:2087–2097. 2016.
View Article : Google Scholar :
|
|
15
|
Duursma AM, Kedde M, Schrier M, le Sage C
and Agami R: miR-148 targets human DNMT3b protein coding region.
RNA. 14:872–877. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Blower PE, Chung JH, Verducci JS, Lin S,
Park JK, Dai Z, Liu CG, Schmittgen TD, Reinhold WC, Croce CM, et
al: MicroRNAs modulate the chemosensitivity of tumor cells. Mol
Cancer Ther. 7:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
19
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gallardo E, Navarro A, Viñolas N, Marrades
RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez
J, et al: miR-34a as a prognostic marker of relapse in surgically
resected non-small-cell lung cancer. Carcinogenesis. 30:1903–1909.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu J, Wu G, Lv L, Ren YF, Zhang XJ, Xue
YF, Li G, Lu X, Sun Z and Tang KF: MicroRNA-34a inhibits migration
and invasion of colon cancer cells via targeting to Fra-1.
Carcinogenesis. 33:519–528. 2012. View Article : Google Scholar
|
|
23
|
Nalls D, Tang SN, Rodova M, Srivastava RK
and Shankar S: Targeting epigenetic regulation of miR-34a for
treatment of pancreatic cancer by inhibition of pancreatic cancer
stem cells. PLoS One. 6:e240992011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun H, Tian J, Xian W, Xie T and Yang X:
miR-34a inhibits proliferation and invasion of bladder cancer cells
by targeting orphan nuclear receptor HNF4G. Dis Markers.
2015:8792542015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Corney DC, Hwang CI, Matoso A, Vogt M,
Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH,
Hermeking H, et al: Frequent downregulation of miR-34 family in
human ovarian cancers. Clin Cancer Res. 16:1119–1128. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R,
Sun Z and Zheng X: Downregulation of CCND1 and CDK6 by miR-34a
induces cell cycle arrest. FEBS Lett. 582:1564–1568. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei JS, Song YK, Durinck S, Chen QR, Cheuk
AT, Tsang P, Zhang Q, Thiele CJ, Slack A, Shohet J, et al: The MYCN
oncogene is a direct target of miR-34a. Oncogene. 27:5204–5213.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yan D, Zhou X, Chen X, Hu DN, Dong XD,
Wang J, Lu F, Tu L and Qu J: MicroRNA-34a inhibits uveal melanoma
cell proliferation and migration through downregulation of c-Met.
Invest Ophthalmol Vis Sci. 50:1559–1565. 2009. View Article : Google Scholar
|
|
31
|
Kang J, Kim E, Kim W, Seong KM, Youn H,
Kim JW, Kim J and Youn B: Rhamnetin and cirsiliol induce
radiosensitization and inhibition of epithelial-mesenchymal
transition (EMT) by miR-34a-mediated suppression of Notch-1
expression in non-small cell lung cancer cell lines. J Biol Chem.
288:27343–27357. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guessous F, Zhang Y, Kofman A, Catania A,
Li Y, Schiff D, Purow B and Abounader R: microRNA-34a is tumor
suppressive in brain tumors and glioma stem cells. Cell Cycle.
9:1031–1036. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kang L, Mao J, Tao Y, Song B, Ma W, Lu Y,
Zhao L, Li J, Yang B and Li L: MicroRNA-34a suppresses the breast
cancer stem cell-like characteristics by downregulating Notch1
pathway. Cancer Sci. 106:700–708. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bu P, Chen KY, Chen JH, Wang L, Walters J,
Shin YJ, Goerger JP, Sun J, Witherspoon M, Rakhilin N, et al: A
microRNA miR-34a-regulated bimodal switch targets Notch in colon
cancer stem cells. Cell Stem Cell. 12:602–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park EY, Chang E, Lee EJ, Lee HW, Kang HG,
Chun KH, Woo YM, Kong HK, Ko JY, Suzuki H, et al: Targeting of
miR34a-NOTCH1 axis reduced breast cancer stemness and
chemoresistance. Cancer Res. 74:7573–7582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu MY, Fu J, Xiao X, Wu J and Wu RC:
MiR-34a regulates therapy resistance by targeting HDAC1 and HDAC7
in breast cancer. Cancer Lett. 354:311–319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ,
Zhao JH and Tang JH: MicroRNA-34a modulates chemosensitivity of
breast cancer cells to adriamycin by targeting Notch1. Arch Med
Res. 43:514–521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ghandadi M and Sahebkar A: MicroRNA-34a
and its target genes: Key factors in cancer multidrug resistance.
Curr Pharm Des. 22:933–939. 2016. View Article : Google Scholar
|
|
39
|
Nie J, Ge X, Geng Y, Cao H, Zhu W, Jiao Y,
Wu J, Zhou J and Cao J: miR-34a inhibits the migration and invasion
of esophageal squamous cell carcinoma by targeting Yin Yang-1.
Oncol Rep. 34:311–317. 2015.PubMed/NCBI
|
|
40
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et
al: miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maire G, Martin JW, Yoshimoto M,
Chilton-MacNeill S, Zielenska M and Squire JA: Analysis of
miRNA-gene expression-genomic profiles reveals complex mechanisms
of microRNA deregulation in osteosarcoma. Cancer Genet.
204:138–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cui X, Zhao Z, Liu D, Guo T, Li S, Hu J,
Liu C, Yang L, Cao Y, Jiang J, et al: Inactivation of miR-34a by
aberrant CpG methylation in Kazakh patients with esophageal
carcinoma. J Exp Clin Cancer Res. 33:202014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lin X, Xu XY, Chen QS and Huang C:
Clinical significance of microRNA-34a in esophageal squamous cell
carcinoma. Genet Mol Res. 14:17684–17691. 2015. View Article : Google Scholar
|
|
44
|
Choi JD and Lee JS: Interplay between
epigenetics and genetics in cancer. Genomics Inform. 11:164–173.
2013. View Article : Google Scholar
|
|
45
|
Lodygin D, Tarasov V, Epanchintsev A,
Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J and Hermeking
H: Inactivation of miR-34a by aberrant CpG methylation in multiple
types of cancer. Cell Cycle. 7:2591–2600. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chim CS, Wong KY, Qi Y, Loong F, Lam WL,
Wong LG, Jin DY, Costello JF and Liang R: Epigenetic inactivation
of the miR-34a in hematological malignancies. Carcinogenesis.
31:745–750. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Duffy MJ, Maguire TM, Hill A, McDermott E
and O'Higgins N: Metalloproteinases: Role in breast carcinogenesis,
invasion and metastasis. Breast Cancer Res. 2:252–257. 2000.
View Article : Google Scholar
|
|
48
|
Samantaray S, Sharma R, Chattopadhyaya TK,
Gupta SD and Ralhan R: Increased expression of MMP-2 and MMP-9 in
esophageal squamous cell carcinoma. J Cancer Res Clin Oncol.
130:37–44. 2004. View Article : Google Scholar
|
|
49
|
Li Y, Ma J, Guo Q, Duan F, Tang F, Zheng
P, Zhao Z and Lu G: Overexpression of MMP-2 and MMP-9 in esophageal
squamous cell carcinoma. Dis Esophagus. 22:664–667. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen CF, Hsu EC, Lin KT, Tu PH, Chang HW,
Lin CH, Chen YJ, Gu DL, Lin CH, Wu JY, et al: Overlapping
high-resolution copy number alterations in cancer genomes
identified putative cancer genes in hepatocellular carcinoma.
Hepatology. 52:1690–1701. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tominaga K, Kondo C, Johmura Y, Nishizuka
M and Imagawa M: The novel gene fad104, containing a fibronectin
type III domain, has a significant role in adipogenesis. FEBS Lett.
577:49–54. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nishizuka S, Ramalingam S, Spurrier B,
Washburn FL, Krishna R, Honkanen P, Young L, Tsutomu S, Steeg PS
and Austin J: Quantitative protein network monitoring in response
to DNA damage. J Proteome Res. 7:803–808. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cai C, Rajaram M, Zhou X, Liu Q, Marchica
J, Li J and Powers RS: Activation of multiple cancer pathways and
tumor maintenance function of the 3q amplified oncogene FNDC3B.
Cell Cycle. 11:1773–1781. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fan X, Chen X, Deng W, Zhong G, Cai Q and
Lin T: Up-regulated microRNA-143 in cancer stem cells
differentiation promotes prostate cancer cells metastasis by
modulating FNDC3B expression. BMC Cancer. 13:612013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lu Y, Yi Y, Liu P, Wen W, James M, Wang D
and You M: Common human cancer genes discovered by integrated
gene-expression analysis. PLoS One. 2:e11492007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yang Y, Li D, Yang Y and Jiang G: An
integrated analysis of the effects of microRNA and mRNA on
esophageal squamous cell carcinoma. Mol Med Rep. 12:945–952.
2015.PubMed/NCBI
|
|
57
|
Genovese G, Ergun A, Shukla SA, Campos B,
Hanna J, Ghosh P, Quayle SN, Rai K, Colla S, Ying H, et al:
microRNA regulatory network inference identifies miR-34a as a novel
regulator of TGF-β signaling in glioblastoma. Cancer Discov.
2:736–749. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Qiao P, Li G, Bi W, Yang L, Yao L and Wu
D: microRNA-34a inhibits epithelial mesenchymal transition in human
cholangio-carcinoma by targeting Smad4 through transforming growth
factor-beta/Smad pathway. BMC Cancer. 15:4692015. View Article : Google Scholar
|
|
59
|
Zhou Q, Dong Wang L, Du F, Zhou Y, Rui
Zhang Y, Liu B, Wei Feng C, Gao SS, Fan ZM, Yang CS, et al: Changes
of TGFbeta1 and TGFbetaRII expression in esophageal precancerous
and cancerous lesions: A study of a high-risk population in Henan,
northern China. Dis Esophagus. 15:74–79. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chang TC, Wentzel EA, Kent OA,
Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M,
Ferlito M, Lowenstein CJ, et al: Transactivation of miR-34a by p53
broadly influences gene expression and promotes apoptosis. Mol
Cell. 26:745–752. 2007. View Article : Google Scholar : PubMed/NCBI
|