Introduction
Renal cell carcinoma (RCC) represents the leading
cause of death due to urological malignancies (1) and clear cell renal cell carcinoma
(ccRCC) is the most common histological subtype of RCC. Despite
improvements in available treatments, the prognosis remains poor
for locally advanced and metastatic ccRCC (2). Therefore, understanding the molecular
basis of ccRCC is crucial to design a novel therapeutic drug that
will improve survival rate.
MicroRNAs (miRNAs) are non-coding RNAs that regulate
gene expression, mainly at the translational level (3). Anomalous changes in miRNA expression
have been shown to be associated with several human cancers
including ccRCC (4–6). Various studies have reported the
miRNA profile in ccRCC specimens using microarray and RNA
sequencing technologies (7–11).
Although altered miRNA expression has been reported in ccRCC, the
mechanisms through which miRNAs contribute to ccRCC pathogenesis
remain poorly understood.
MicroRNA-122 (miR-122), a known tumour suppressor in
hepatocellular carcinoma (HCC), functions by targeting oncogenes
such as cyclin G1 (12), CDK4
(13), and AKT3 (14). However, several studies have also
shown that miR-122 is highly expressed in ccRCC (10,15,16),
suggesting that miR-122 functions as an onco-microRNA (oncomiR) in
renal cancer (17,18). Whilst the physiological role of
miR-122 in cancer appears dependent on cancer types, the precise
mechanism of miR-122 on ccRCC progression is largely unknown.
Tight junctions govern the permeability of
epithelial and endothelial cells and are the most topical structure
of these cell types (19–21). It is thought that the interaction
with and penetration of the endothelium by cancer cells is a key
step in the formation of metastasis (22–24).
Occludin, a component of tight junctions, possesses four
transmembrane domains, two extracellular loops and two
intracellular domains (25).
Numerous studies have emphasized the important role of occludin in
the assembly and maintenance of tight junctions (26–28).
Moreover, several studies suggest that occludin also plays a tumour
suppressor role. Raf-1 disrupts tight junctions through the
downregulation of occludin, and over-expression of occludin
suppresses Raf-1-induced tumour growth in rat salivary gland
epithelial cells (29). Occludin
has also been shown to regulate oncogenic properties in several
cancers (30–34). In ccRCC, loss of function of von
Hippel-Lindau (VHL), a tumour suppressor gene, is known to
downregulate the expression of occludin and lead to disruption of
tight junction assembly (35).
Herein, we showed that highly expressed miR-122
correlated with low progression-free survival compared to low
miR-122 expression in ccRCC clinical specimens. We further
demonstrated that miR-122 promoted malignant phenotypes partially
via targeting occludin in ccRCC cells. Finally we showed that
occludin was significantly downregulated in ccRCC specimens
compared to normal kidney tissues, and this correlated with the
expression levels of miR-122. Our findings identify that miR-122 is
as a potent regulator of malignant phenotypes and functions as an
oncomiR in ccRCC, leading to a novel therapeutic strategy for
treatment of ccRCC.
Materials and methods
Chemicals and antibodies
miRIDIAN miRNA hairpin inhibitor negative control,
miRIDIAN miRNA mimic negative control, miRIDIAN hairpin inhibitor
or miRIDIAN miRNA mimic for human hsa-miR-122 (MIMAT0000421) were
purchased from Thermo Scientific Dharmacon (Waltham, MA, USA). The
miRIDIAN hairpin inhibitors and mimics were used at a concentration
of 50 nM. Polyclonal anti-occludin and monoclonal anti-β-tubulin
antibodies were purchased from Sigma (St. Louis, MO, USA).
Polyclonal Anti-ACTIVE p38 (Thr180/Tyr182)
and Anti-ACTIVE JNK1/2 (Thr183/Tyr185)
antibodies were purchased from Promega (Madison, WI, USA).
Monoclonal anti-phospho-Erk1/2
(Thr202/Tyr204), anti-Erk1/2, and anti-p38
antibodies were purchased from Cell Signaling Technology (Beverly,
MA, USA). Monoclonal anti-JNK1/2 antibody was purchased from BD
Transduction Laboratories (San Jose, CA, USA).
Clinical specimens
ccRCC specimens were obtained from patients while
they underwent primary curative resection at the Osaka University
Medical Hospital, Japan. Tumour-associated normal renal tissue was
also obtained from a subset of these patients when possible. Prior
written and informed consent was obtained from each patient, and
the study was approved by the ethics review board of the Osaka
University Medical Hospital and the methods were carried out in
accordance with the approved guidelines.
Quantitative real-time PCR
Following excision, tissue samples were immediately
immersed in RNAlater (Qiagen, Valencia, CA, USA) and stored at
−20°C until RNA extraction. miRNAs were purified using the miRNeasy
mini kit (Qiagen). Real-time PCR analysis was conducted to validate
miR-122 expression in ccRCC using 80 tumour samples and 10 adjacent
normal renal samples (for Fig.
1B–D) and another distinct 122 tumour tissue samples (for
Fig. 1E and F) using the Mir-X
miRNA first-strand synthesis kit (Takara, Shiga, Japan). Thermal
cycling conditions included an initial step at 98°C for 30 sec, and
40 cycles at 95°C for 2 sec and 66°C for 5 sec using an miR-122
specific primer (5′-tggagtgtgacaatggtgtttg-3′), and 98°C for 30
sec, and 40 cycles at 95°C for 2 sec and at 63°C for 5 sec by using
an U6 snRNA primer (Takara). Clinical and pathological data related
to the clinical samples are presented in Table I.
 | Table IClinical and pathological data
related to the clinical samples. |
Table I
Clinical and pathological data
related to the clinical samples.
|
Characteristics | Validation cohort
for miR-122 expression | For the PFS
analysis |
|---|
| Age (year) | | |
| Mean | 62.6 | 63.8 |
| Range | 35–86 | 34–82 |
| Gender | | |
| Male | 58 | 38 |
| Female | 31 | 83 |
| Unknown | 1 | 1 |
| Pathologic
stage | | |
| Normal | 10 | 0 |
| pT1a | 11 | 56 |
| pT1b | 69 | 65 |
| Unknown | 0 | 1 |
| Pathologic
grade | | |
| G1 | 23 | 47 |
| G2 | 52 | 72 |
| G3 | 5 | 2 |
| Unknown | 0 | 1 |
| Lymphatic
invasion | | |
| − | 74 | 127 |
| + | 6 | 10 |
Cell culture
Three human ccRCC cell lines (Caki-1, Caki-2 and,
ACHN), obtained from the American Type Culture Collection (ATCC),
were cultured in RPMI-1640 medium (Wako, Osaka, Japan) supplemented
with 10% fetal bovine serum, 100 U/ml penicillin G, and 0.1
µg/ml streptomycin.
Western blotting
Protein samples were separated by sodium dodecyl
sulphate (SDS)-polyacrylamide gel electrophoresis (PAGE) on a
7.5–15% SDS-polyacrylamide gel and then transferred to a
polyvinylidene difluoride membrane using the Bio-Rad semi-dry
transfer system (1 h, 12 V). Immunoreactive proteins reacting with
the antibodies described above were visualized by treatment with a
detection reagent (ECL Prime western blotting detection reagent; GE
Healthcare, Lafayette, CO, USA). Densitometric analysis was
performed using NIH ImageJ software.
Luciferase reporter assay
A pmirGLO dual-luciferase miRNA target expression
vector was used for the 3′-UTR luciferase reporter assay (Promega).
The following oligonucleotides were used for the evaluation of
efficacy of the miR-122 hairpin inhibitor on occludin 3′-UTR:
5′-CTA GCG GCC GCT AGT TCA ACT GGG CTG AAC ACT CCA G-3′ and 5′-TCG
ACT GGA GTG TTC AGC CCA GTT GAA CTA GCG GCC GCT AGA GCT-3′.
Luciferase activity was determined using a luminometer (Turner
Biosystems 20/20 n luminometer, Promega).
Wound healing assay
Cell migration was examined by wound healing assay.
In brief, Caki-2 cells transfected with the miRIDIAN hairpin
inhibitor or the negative control inhibitor, and ACHN cells
transfected with the miRIDIAN microRNA mimic or the negative
control mimic were seeded in a 24-well plate (Caki-2 cells:
2.0×104 cells/well, ACHN cells: 4.0×104
cells/well) and incubated for 72 h. A wound was created in a
monolayer of approximately 90% confluent Caki-2 or ACHN cells using
a sterile 1 ml pipette tip. Cell migration pictures were recorded
at 0 and 12 h after wound creation using an Olympus IX71
fluorescence microscope.
Water-soluble tetrazolium salt-1 (WST-1)
cell proliferation assay
Cell proliferation was examined by WST-1 assay.
Caki-2 cells transfected with the miRIDIAN hairpin inhibitor or the
negative control inhibitor, and ACHN cells transfected with the
miRIDIAN miRNA mimic or the negative control mimic were seeded in a
96-well plate (0.3×104 cells/well) and incubated for 72
h. After incubation for 2 h with the WST-1 reagent (Dojindo, Osaka,
Japan) at 37°C and 5% CO2, the optical density was read
at a wavelength of 450/630 nm (Ex/Em).
Soft agar colony formation assay
Caki-2 or ACHN cells were seeded at
1.0×104 cells/well in 6-well plates in RPMI-1640 medium
(supplemented with 10% FBS and 0.3% agarose with 0.4% agarose
underlay). Dishes were incubated at 37°C and 5% CO2.
After 14 days, colonies were stained with crystal violet (Wako) and
counted.
Cell invasion assay
The BioCoat tumour invasion system (Corning Inc.,
Corning, NY, USA) was used to perform the cell invasion assay.
Caki-2 cells transfected with the miRIDIAN hairpin inhibitor or the
negative control inhibitor were seeded in a 96-well plate
(3×104 cells/well). Following incubation for 12 h, the
cells were labelled with calcein AM (4 µg/ml, Takara), and
the fluorescence of the invaded cells was read at a wavelength of
494/517 nm (Ex/Em).
Fluorescence microscopy
Cells plated on coverslips were washed with
phosphate-buffered saline (PBS). The cells were fixed and
permeabilized in 4% formaldehyde for 15 min at room temperature and
then washed twice with PBS. After blocking with 2% bovine serum
albumin in PBS for 1 h, the coverslips were incubated with the
anti-occludin antibody (1:500) at 4°C. The coverslips were washed
twice with PBS and incubated with the fluorochrome-conjugated
secondary antibody for 1 h at room temperature. The cells were then
examined under the fluorescence microscope, Bio-Zero (Keyence,
Osaka, Japan).
Immunohistochemistry
The expression of occludin was determined by
immunohistochemical staining of paraffin-embedded tissues of ccRCC
samples with the highest and lowest levels of miR-122 expression
samples (10 samples of each). Formalin-fixed paraffin-embedded
sections (5 µm in thickness) were deparaffinized and
rehydrated. After the slides were steamed for 20 min in 10 mM
citrate buffer (pH 6.0) for antigen retrieval, endogenous
peroxidase was blocked using 3% H2O2.
Immunohistochemical staining for occludin was performed using
anti-occludin (1:500) and the EnVision+ Detection System
(Dako, Santa Clara, CA, USA) was used according to the
manufacturer's instructions. Primary antibodies were incubated
overnight at 4°C and counterstained with hematoxylin. The levels of
occludin staining were classified into four groups according to the
staining intensity of the positive cells (staining score, 0–3;
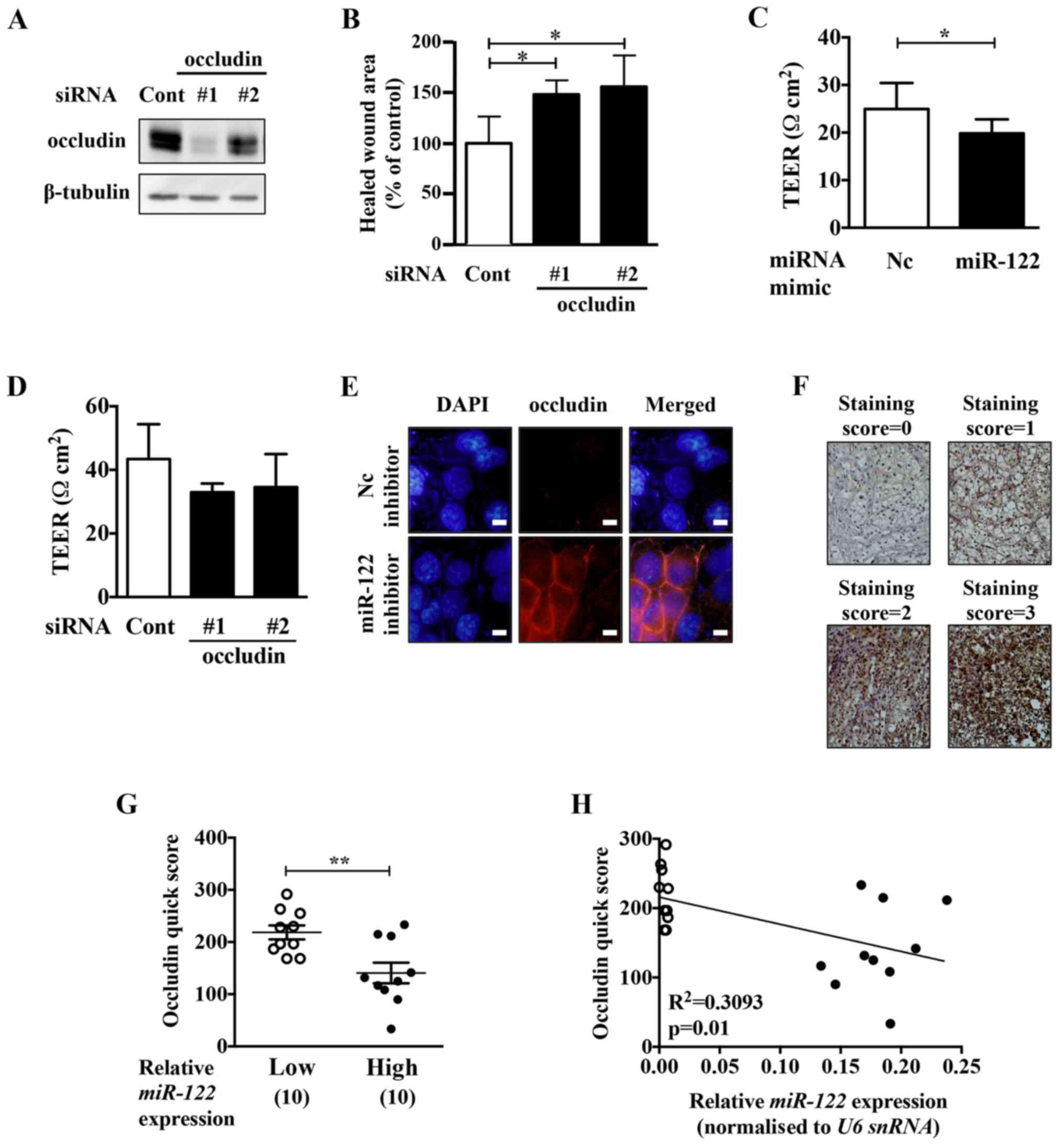
Fig. 5F) and Quick score was
calculated as follows; Quick score = (percentage of occludin
positive cells) × (staining score). For each ccRCC sample, three
points were calculated.
Transendothelial electrical resistance
(TEER) measurement
ACHN cells transfected with occludin siRNAs or
miR-122 mimic for 48 h were reseeded (1.0×105
cells/well) on a 0.4 µm pore size 24-well cell culture
insert (Thermo Scientific Dharmacon) and incubated for 1 day. TEER
was measured using a Millicell® ERS-2 voltohmmeter
(Millipore, Billerica, MA, USA). TEER was calculated as follows:
(sample well Ω − empty well Ω) × (cultured area cm2) =
TEER (Ω cm2).
Statistics
The results are expressed as the mean ± standard
deviation (SD). Differences between the values were statistically
analysed using the Student's t-test or one-way analysis of variance
(ANOVA) with Bonferroni post-hoc tests. Correlations between the
values were analysed using Pearson correlation analysis (GraphPad
Prism 6.0, GraphPad software, San Diego, CA, USA). A P-value of
<0.05 was considered statistically significant.
Results
High miR-122 expression correlates with
poor progression-free survival in ccRCC
In our previous microarray analysis identifying
unique miRNA expression signatures (GEO accession number: GSE55138)
(8), aberrantly high expression of
miR-122 was detected in ccRCC tissues compared with adjacent normal
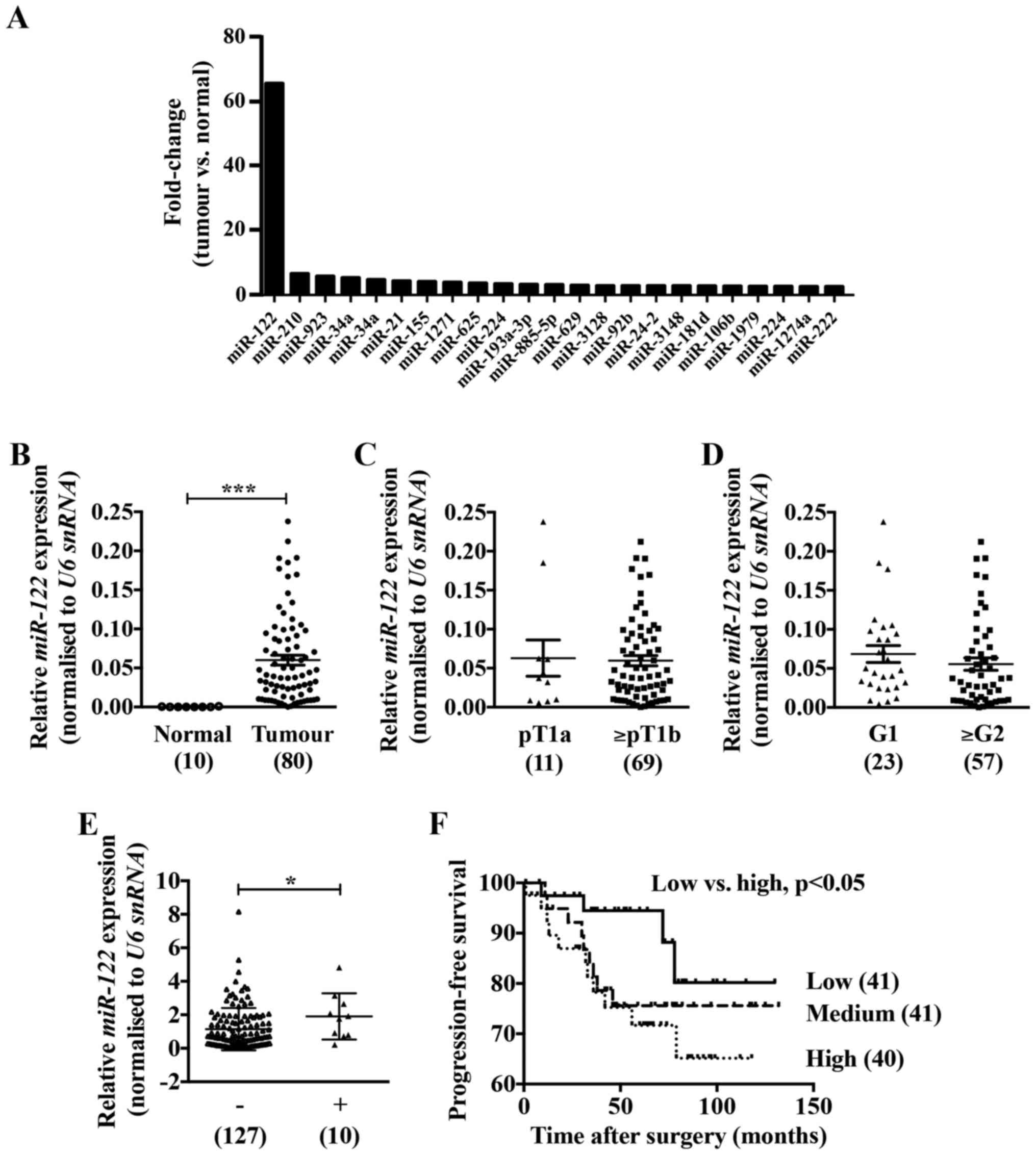
renal tissues (Fig. 1A). To
confirm the expression of miR-122 in ccRCC specimens we performed
real-time PCR analysis. Although there was no correlation with
pathological stages or grades (Fig. 1C
and D), the expression of miR-122 was approximately 300-fold
higher in the ccRCC tissues compared to the adjacent normal renal
tissues (Fig. 1B) and was
significantly high in ccRCC tissues accompanied by lymphatic
invasion compared with those without lymphatic invasion (Fig. 1E). Noteworthy, ccRCC patients with
high miR-122 expression had significantly poorer progression-free
survival compared with those with low miR-122 expression (Fig. 1F).
miR-122 inhibitor reduced migration and
invasion activities of Caki-2 cells
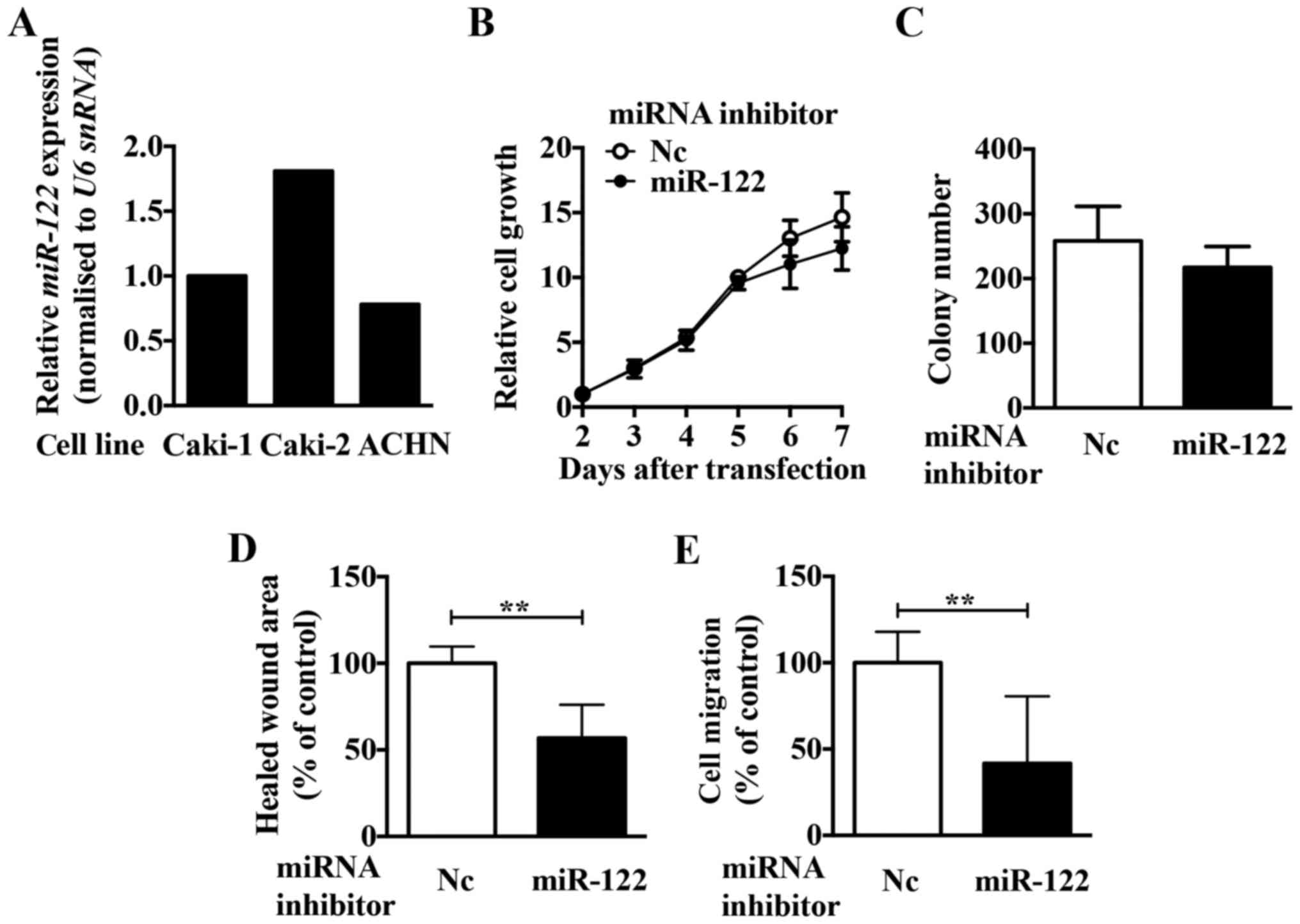
To investigate the role of miR-122 in ccRCC, we
first compared the expression levels of miR-122 in three ccRCC cell
lines (Caki-1, Caki-2, and ACHN, Fig.
2A). Since the expression of miR-122 was highest in Caki-2
cells, we first analysed the function of miR-122 using Caki-2
cells. To investigate the function of miR-122, we examined the
effect of a miR-122 inhibitor on growth, migration, and invasion in
Caki-2 cells. Although the miR-122 inhibitor had no significant
effect on the cell growth (Fig. 2B and
C), migration and invasion activities were significantly
reduced in Caki-2 cells (Fig. 2D and
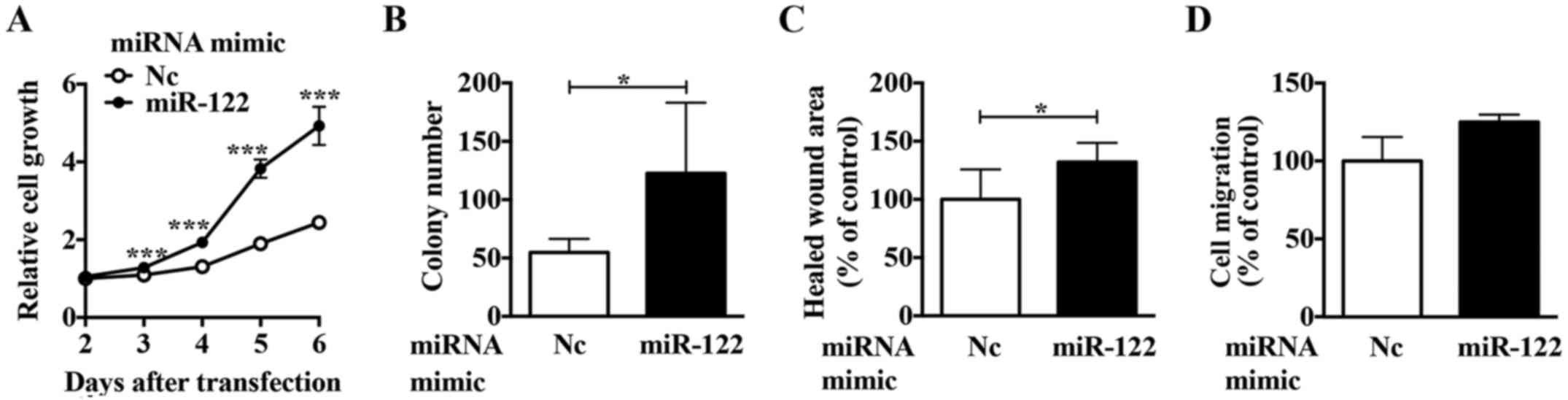
E). Of the cell lines examined, ACHN cells had the lowest
miR-122 expression. Using a miR-122 mimic in ACHN cells, we
observed upregulated proliferation (Fig. 3A and B), migration (Fig. 3C), and invasion activities
(Fig. 3D). These results suggest
that miR-122 is involved in regulating the expression of crucial
molecule/s in ccRCC cells.
miR-122 regulates occludin expression in
ccRCC cells
We used target prediction programs (miRbase,
TargetScan, and miRanda) to identify the potential miR-122 target
in ccRCC cells. We then focused on occludin as a potential target,
because it is a component of tight junctions and functions as a
tumour suppressor in several cancers (30,31).
TargetScan predicts that the 3′-untranslated region (UTR) of
occludin mRNA contains a complementary site for the seed
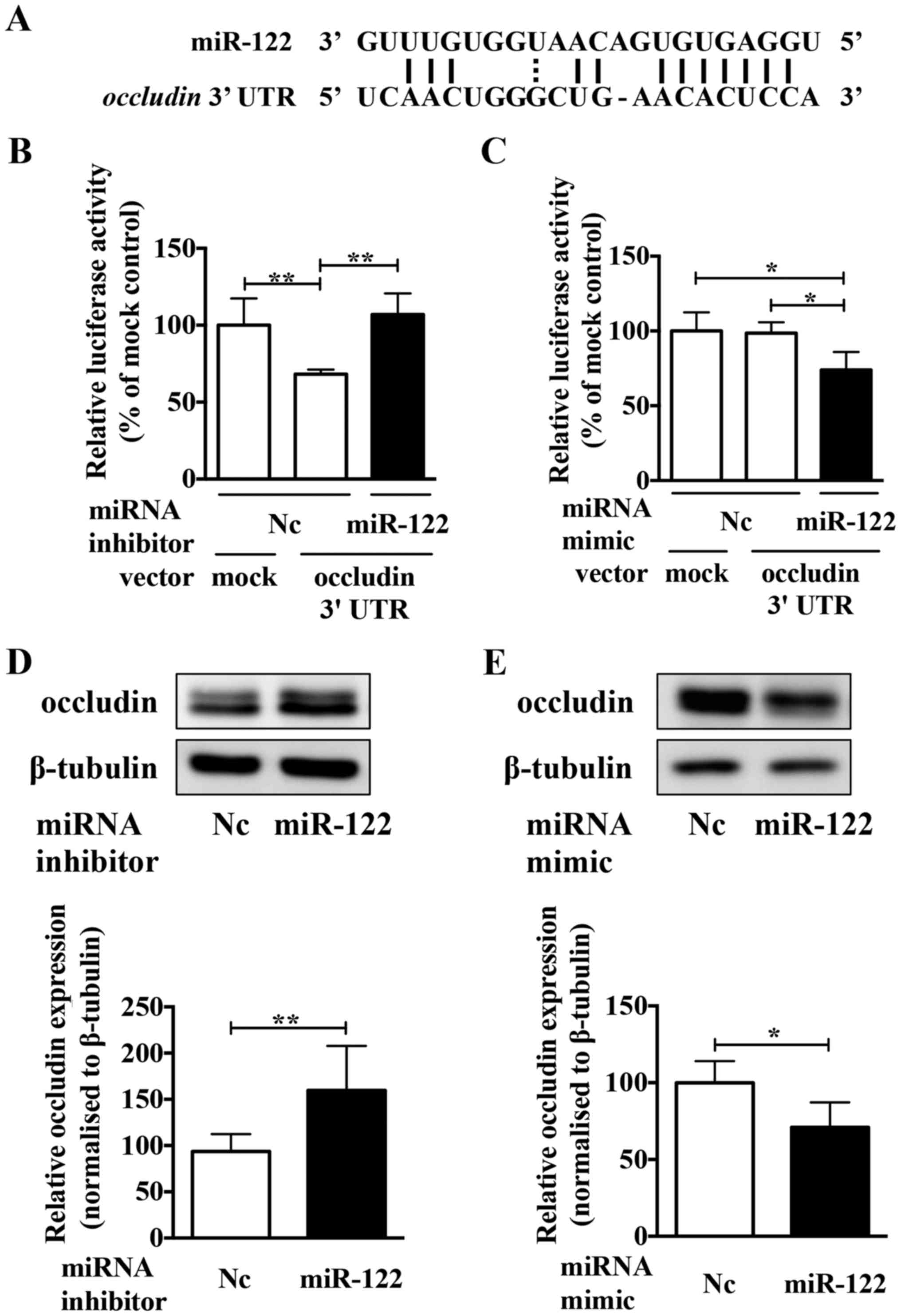
region of miR-122 (Fig. 4A). To
confirm whether or not occludin was a direct target of miR-122, the
miR-122 inhibitor and a luciferase reporter plasmid containing the
predicted miR-122 binding site (within the 3′-UTR of the human
occludin gene) were co-transfection into Caki-2 cells.
Luciferase activity confirmed that the reporter construct was
working appropriately (Fig. 4B).
The luciferase activity in cells co-transfected with the miR-122
inhibitor increased significantly compared to those co-transfected
with the negative control inhibitor. Conversely, compared to the
negative control mimic, the miR-122 mimic significantly decreased
the luciferase activity (Fig. 4C).
Moreover, the miR-122 inhibitor upregulated the expression of
occludin protein in Caki-2 cells (Fig.
4D), and the miR-122 mimic reduced occludin protein expression
in ACHN cells (Fig. 4E). These
results indicated that occludin is a direct target of miR-122 in
ccRCC cells.
Occludin knockdown upregulates cell
migration activity in ACHN cells
To examine whether the malignant phenotypes
upregulated by the miR-122 mimic in ccRCC cells were due to the
decrease in occludin expression, we performed occludin knockdown
experiments in ACHN cells (Fig. 5A and
B). Occludin knockdown upregulated the migration activity of
ACHN cells. The motility of ACHN cells was upregulated by occludin
knockdown. Transepithelial electrical resistance (TEER) analysis
showed that miR-122 mimic and occludin knockdown downregulated the
TEER in the cells (Fig. 5C and D).
TEER is a quantitative method to measure the integrity of tight
junctions in cultured cells. These results suggest that miR-122
might upregulate the permeability of tight junctions via occludin
reduction, leading to an increase in the motility of ACHN
cells.
miR-122 expression and occludin protein
expression have an inverse correlation in ccRCC specimens
The miR-122 inhibitor upregulated the expression of
occludin at the cell-cell adhesive surface in Caki-2 cells
(Fig. 5E). To examine whether
miR-122 expression and occludin protein expression are negatively
correlated, immunohistochemistry analysis was performed on the
ccRCC samples with the lowest and highest miR-122-expression (10
samples for each). ccRCC tissues with high levels of miR-122
expression had a significantly weak occludin quick score compared
to those with low levels of miR-122 expression (Fig. 5F and G). Moreover, miR-122 and
occludin protein expression showed a negative correlation in the
ccRCC tissues (Fig. 5H).
miR-122 promotes MAPK signaling in ccRCC
cell lines
Although the miR-122 mimic upregulated the cell
proliferation, motility, and invasion activities, occludin
knockdown only upregulated cell motility. Target prediction
programs predict that putative miR-122 binding sequences exist in
other genes involved in cell growth [e.g., p27 (36), PHD3 (37,38),
and DUSP2 (39)] and
invasion [e.g., CPEB1 (40), FOXP2 (41), and SOX17 (42)]. Occludin is known to mediate the
MAPK signalling pathway via oncogenic proteins such as Raf
(29). Therefore, we examined the
effect of miR-122 on the MAPK pathway by miR-122 overexpression and
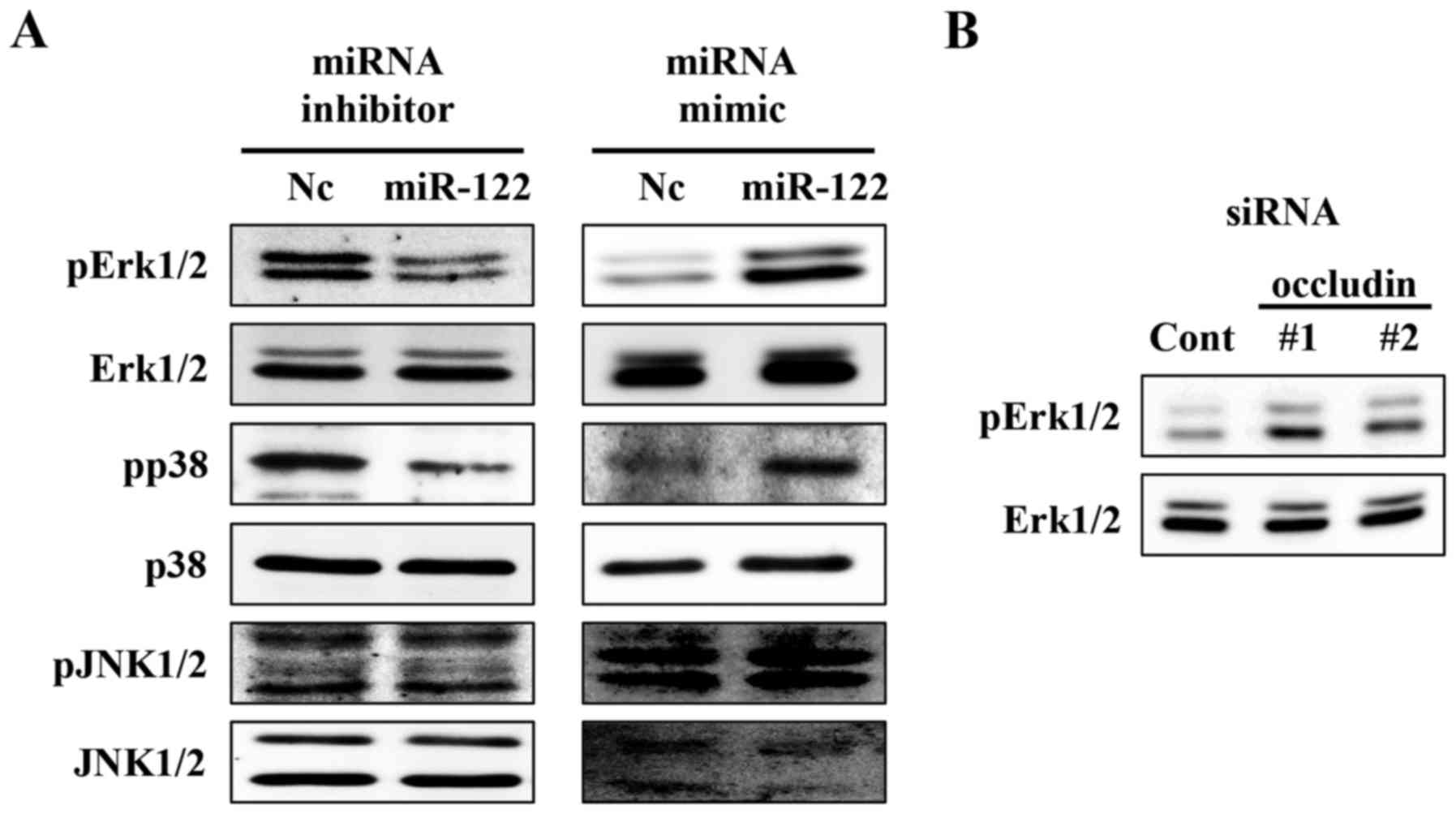
loss of function experiments. We found that miR-122 affected the
phosphorylation status of Erk1/2 and p38 (Fig. 6A). Likewise, occludin knockdown
upregulated the phosphorylation status of Erk1/2 (Fig. 6B). Therefore, miR-122 might also
mediate ccRCC cell growth and invasion via occludin and other
target molecules including MAPK signalling components.
Discussion
In the present study, we found that high expression
of miR-122 was significantly correlated with poor progression-free
survival in ccRCC. Overexpression and loss of function experiments
revealed that miR-122 functions as an oncomiR by upregulating
malignant phenotypes in ccRCC cells. We identified that miR-122
directly targets occludin in ccRCC cells. Moreover, we showed that
the miR-122 expression levels and occludin protein levels are
negatively correlated in ccRCC clinical specimens.
Although miR-122 mimic significantly downregulated
the TEER in ACHN cells, occludin knockdown only slightly suppressed
the TEER (Fig. 5D). Claudin1 and
PVRL1, which play roles in the organization of adherence junctions
and tight junctions (43,44), respectively, also have potential
miR-122 binding sites within their 3′UTRs. Therefore, miR-122 might
upregulate cell motility of these proteins as well as occludin in
ccRCC cells.
Although miR-122 expression was not correlated with
pathological stages and grades, miR-122 expression was
significantly high in ccRCC tissues accompanied by lymphatic
invasion compared with those without lymphatic invasion (Fig. 1E). Lymphovascular invasion
correlates with adverse outcome in several malignancies (45–47).
In ccRCC, lymphovascular invasion is associated with
metastasis-free survival, disease-specific survival, and overall
survival (48,49). While the precise mechanism needs to
be investigated further, high miR-122 expression might lead to
increased lymphovascular invasion via upregulated cell motility and
invasion activities, leading to a poorer prognosis for ccRCC
patients.
Originally miR-122 was reported as a tumour
suppressor gene in hepatocellular carcinoma (HCC) (12–14).
However, in breast cancer, miR-122 has been reported as a tumour
suppressor targeting insulin-like growth factor 1 receptor (IGF1R)
(50), and as an oncomiR by
reprogramming the glucose metabolism in the tumour microenvironment
via the exosome (18). The present
study demonstrates for the first time that, unlike that in HCC or
breast cancer, miR-122 functions as an oncomiR in ccRCC in both
cell lines and clinical specimens. Lian et al showed that
miR-122 expression led to upregulated cancer cell malignancy
phenotypes in RCC (17),
consistent with our present study in ccRCC. The phase II clinical
study of miR-122-targeted locked nucleic acid (LNA), miravirsen,
inhibits viral proliferation, resulting in a long-lasting antiviral
effect against HCV without evidence of viral resistance (51). Therefore, a miR-122-targeted
bridged nucleic acid might be a promising molecular-targeted drug
for the treatment of ccRCC.
Abbreviations:
|
miR-122
|
microRNA-122
|
|
ccRCC
|
clear cell renal cell carcinoma
|
|
oncomiR
|
onco-microRNA
|
Acknowledgments
This study was supported by a Grant-in-Aid for
Scientific Research (25670025) from the Ministry of Education,
Culture, Sports, Science and Technology of Japan and by Project
MEET, Osaka University Graduate School of Medicine. The TEER
measurement with the Millicell® ERS-2 voltohmmeter was
kindly supported by Dr Takefumi Doi and Dr Yoshiaki Okada (Graduate
School of Pharmaceutical Sciences, Osaka University). N. Nonomura
received commercial research grants from Takeda Pharmaceutical,
Novartis Pharma, and Astra Zeneca.
References
|
1
|
Jonasch E, Futreal PA, Davis IJ, Bailey
ST, Kim WY, Brugarolas J, Giaccia AJ, Kurban G, Pause A, Frydman J,
et al: State of the science: An update on renal cell carcinoma. Mol
Cancer Res. 10:859–880. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dutcher JP: Recent developments in the
treatment of renal cell carcinoma. Ther Adv Urol. 5:338–353. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li M, Wang Y, Song Y, Bu R, Yin B, Fei X,
Guo Q and Wu B: MicroRNAs in renal cell carcinoma: A systematic
review of clinical implications (Review). Oncol Rep. 33:1571–1578.
2015.PubMed/NCBI
|
|
7
|
Weng L, Wu X, Gao H, Mu B, Li X, Wang JH,
Guo C, Jin JM, Chen Z, Covarrubias M, et al: MicroRNA profiling of
clear cell renal cell carcinoma by whole-genome small RNA deep
sequencing of paired frozen and formalin-fixed, paraffin-embedded
tissue specimens. J Pathol. 222:41–51. 2010.PubMed/NCBI
|
|
8
|
Jingushi K, Ueda Y, Kitae K, Hase H, Egawa
H, Ohshio I, Kawakami R, Kashiwagi Y, Tsukada Y, Kobayashi T, et
al: miR-629 targets TRIM33 to promote TGFβ/Smad signaling and
metastatic phenotypes in ccRCC. Mol Cancer Res. 13:565–574. 2015.
View Article : Google Scholar
|
|
9
|
Nakata W, Uemura M, Sato M, Fujita K,
Jingushi K, Ueda Y, Kitae K, Tsujikawa K and Nonomura N: Expression
of miR-27a-3p is an independent predictive factor for recurrence in
clear cell renal cell carcinoma. Oncotarget. 6:21645–21654. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Osanto S, Qin Y, Buermans HP, Berkers J,
Lerut E, Goeman JJ and van Poppel H: Genome-wide microRNA
expression analysis of clear cell renal cell carcinoma by next
generation deep sequencing. PLoS One. 7:e382982012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang K and Xu H: Prognostic value of
meta-signature miRNAs in renal cell carcinoma: An integrated miRNA
expression profiling analysis. Sci Rep. 5:102722015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang F, Zhang L, Wang F, Wang Y, Huo XS,
Yin YX, Wang YQ, Zhang L and Sun SH: Modulation of the unfolded
protein response is the core of microRNA-122-involved sensitivity
to chemotherapy in hepatocellular carcinoma. Neoplasia. 13:590–600.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nassirpour R, Mehta PP and Yin MJ: miR-122
regulates tumorigenesis in hepatocellular carcinoma by targeting
AKT3. PLoS One. 8:e796552013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chow TF, Youssef YM, Lianidou E, Romaschin
AD, Honey RJ, Stewart R, Pace KT and Yousef GM: Differential
expression profiling of microRNAs and their potential involvement
in renal cell carcinoma pathogenesis. Clin Biochem. 43:150–158.
2010. View Article : Google Scholar
|
|
16
|
Chen J, Zhang D, Zhang W, Tang Y, Yan W,
Guo L and Shen B: Clear cell renal cell carcinoma associated
microRNA expression signatures identified by an integrated
bioinformatics analysis. J Transl Med. 11:1692013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lian JH, Wang WH, Wang JQ, Zhang YH and Li
Y: MicroRNA-122 promotes proliferation, invasion and migration of
renal cell carcinoma cells through the PI3K/Akt signaling pathway.
Asian Pac J Cancer Prev. 14:5017–5021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fong MY, Zhou W, Liu L, Alontaga AY,
Chandra M, Ashby J, Chow A, O'Connor ST, Li S, Chin AR, et al:
Breast-cancer-secreted miR-122 reprograms glucose metabolism in
premetastatic niche to promote metastasis. Nat Cell Biol.
17:183–194. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsukita S and Furuse M: Occludin and
claudins in tight-junction strands: Leading or supporting players?
Trends Cell Biol. 9:268–273. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang WG, Martin TA, Matsumoto K, Nakamura
T and Mansel RE: Hepatocyte growth factor/scatter factor decreases
the expression of occludin and transendothelial resistance (TER)
and increases paracellular permeability in human vascular
endothelial cells. J Cell Physiol. 181:319–329. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang WG, Bryce RP, Horrobin DF and Mansel
RE: Regulation of tight junction permeability and occludin
expression by polyunsaturated fatty acids. Biochem Biophys Res
Commun. 244:414–420. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gopalakrishnan S, Raman N, Atkinson SJ and
Marrs JA: Rho GTPase signaling regulates tight junction assembly
and protects tight junctions during ATP depletion. Am J Physiol.
275:C798–C809. 1998.PubMed/NCBI
|
|
23
|
van ZF: Krupitza G and Mikulits W: Initial
steps of metastasis: Cell invasion and endothelial transmigration.
Mutat Res. 728:23–34. 2011. View Article : Google Scholar
|
|
24
|
Stoletov K, Kato H, Zardouzian E, Kelber
J, Yang J, Shattil S and Klemke R: Visualizing extravasation
dynamics of metastatic tumor cells. J Cell Sci. 123:2332–2341.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shin K, Fogg VC and Margolis B: Tight
junctions and cell polarity. Annu Rev Cell Dev Biol. 22:207–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rao R: Occludin phosphorylation in
regulation of epithelial tight junctions. Ann NY Acad Sci.
1165:62–68. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Van Itallie CM, Fanning AS, Holmes J and
Anderson JM: Occludin is required for cytokine-induced regulation
of tight junction barriers. J Cell Sci. 123:2844–2852. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Raleigh DR, Boe DM, Yu D, Weber CR,
Marchiando AM, Bradford EM, Wang Y, Wu L, Schneeberger EE, Shen L,
et al: Occludin S408 phosphorylation regulates tight junction
protein interactions and barrier function. J Cell Biol.
193:565–582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li D and Mrsny RJ: Oncogenic Raf-1
disrupts epithelial tight junctions via downregulation of occludin.
J Cell Biol. 148:791–800. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Runkle EA, Rice SJ, Qi J, Masser D,
Antonetti DA, Winslow MM and Mu D: Occludin is a direct target of
thyroid transcription factor-1 (TTF-1/NKX2-1). J Biol Chem.
287:28790–28801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Osanai M, Murata M, Nishikiori N, Chiba H,
Kojima T and Sawada N: Epigenetic silencing of occludin promotes
tumorigenic and metastatic properties of cancer cells via
modulations of unique sets of apoptosis-associated genes. Cancer
Res. 66:9125–9133. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Osanai M, Murata M, Nishikiori N, Chiba H,
Kojima T and Sawada N: Occludin-mediated premature senescence is a
fail-safe mechanism against tumorigenesis in breast carcinoma
cells. Cancer Sci. 98:1027–1034. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Long H, Crean CD, Lee WH, Cummings OW and
Gabig TG: Expression of Clostridium perfringens enterotoxin
receptors claudin-3 and claudin-4 in prostate cancer epithelium.
Cancer Res. 61:7878–7881. 2001.PubMed/NCBI
|
|
34
|
Tzelepi VN, Tsamandas AC, Vlotinou HD,
Vagianos CE and Scopa CD: Tight junctions in thyroid
carcinogenesis: Diverse expression of claudin-1, claudin-4,
claudin-7 and occludin in thyroid neoplasms. Mod Pathol. 21:22–30.
2008. View Article : Google Scholar
|
|
35
|
Harten SK, Shukla D, Barod R, Hergovich A,
Balda MS, Matter K, Esteban MA and Maxwell PH: Regulation of renal
epithelial tight junctions by the von Hippel-Lindau tumor
suppressor gene involves occludin and claudin 1 and is independent
of E-cadherin. Mol Biol Cell. 20:1089–1101. 2009. View Article : Google Scholar :
|
|
36
|
Blain SW, Scher HI, Cordon-Cardo C and
Koff A: p27 as a target for cancer therapeutics. Cancer Cell.
3:111–115. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Henze AT, Garvalov BK, Seidel S, Cuesta
AM, Ritter M, Filatova A, Foss F, Dopeso H, Essmann CL, Maxwell PH,
et al: Loss of PHD3 allows tumours to overcome hypoxic growth
inhibition and sustain proliferation through EGFR. Nat Commun.
5:55822014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Garvalov BK, Foss F, Henze AT, Bethani I,
Gräf-Höchst S, Singh D, Filatova A, Dopeso H, Seidel S, Damm M, et
al: PHD3 regulates EGFR internalization and signalling in tumours.
Nat Commun. 5:55772014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lin SC, Chien CW, Lee JC, Yeh YC, Hsu KF,
Lai YY, Lin SC and Tsai SJ: Suppression of dual-specificity
phosphatase-2 by hypoxia increases chemoresistance and malignancy
in human cancer cells. J Clin Invest. 121:1905–1916. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nagaoka K, Fujii K, Zhang H, Usuda K,
Watanabe G, Ivshina M and Richter JD: CPEB1 mediates
epithelial-to-mesenchyme transition and breast cancer metastasis.
Oncogene. 35:2893–2901. 2016. View Article : Google Scholar :
|
|
41
|
Yan X, Zhou H and Zhang T, Xu P, Zhang S,
Huang W, Yang L, Gu X, Ni R and Zhang T: Downregulation of FOXP2
promoter human hepatocellular carcinoma cell invasion. Tumour Biol.
36:9611–9619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kuo IY, Wu CC, Chang JM, Huang YL, Lin CH,
Yan JJ, Sheu BS, Lu PJ, Chang WL, Lai WW, et al: Low SOX17
expression is a prognostic factor and drives transcriptional
dysregulation and esophageal cancer progression. Int J Cancer.
135:563–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Günzel D and Yu AS: Claudins and the
modulation of tight junction permeability. Physiol Rev. 93:525–569.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Takai Y and Nakanishi H: Nectin and
afadin: Novel organizers of intercellular junctions. J Cell Sci.
116:17–27. 2003. View Article : Google Scholar
|
|
45
|
Cheng L, Montironi R, Davidson DD and
Lopez-Beltran A: Staging and reporting of urothelial carcinoma of
the urinary bladder. Mod Pathol. 22(Suppl 2): S70–S95. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lim SB, Yu CS, Jang SJ, Kim TW, Kim JH and
Kim JC: Prognostic significance of lymphovascular invasion in
sporadic colorectal cancer. Dis Colon Rectum. 53:377–384. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Cheng L, Jones TD, Lin H, Eble JN, Zeng G,
Carr MD and Koch MO: Lymphovascular invasion is an independent
prognostic factor in prostatic adenocarcinoma. J Urol.
174:2181–2185. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Katz MD, Serrano MF, Humphrey PA, Grubb RL
III, Skolarus TA, Gao F and Kibel AS: The role of lymphovascular
space invasion in renal cell carcinoma as a prognostic marker of
survival after curative resection. Urol Oncol. 29:738–744. 2011.
View Article : Google Scholar
|
|
49
|
Belsante M, Darwish O, Youssef R, Bagrodia
A, Kapur P, Sagalowsky AI, Lotan Y and Margulis V: Lymphovascular
invasion in clear cell renal cell carcinoma - association with
disease-free and cancer-specific survival. Urol Oncol.
32:30.e23–30.e28. 2014. View Article : Google Scholar
|
|
50
|
Wang B, Wang H and Yang Z: MiR-122
inhibits cell proliferation and tumorigenesis of breast cancer by
targeting IGF1R. PLoS One. 7:e470532012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Janssen HL, Reesink HW, Lawitz EJ, Zeuzem
S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A,
Zhou Y, et al: Treatment of HCV infection by targeting microRNA. N
Engl J Med. 368:1685–1694. 2013. View Article : Google Scholar : PubMed/NCBI
|