1. Introduction
Colorectal cancer (CRC) is the 3rd most common
cancer in men and women in the United States. There are
approximately 135,000 new cases of colorectal cancer diagnosed per
year (1). Approximately one out of
three cases of colorectal cancer is located in the rectum and
categorized as rectal adenocarcinoma. Common treatments for
localized (non-metastatic) rectal cancer include surgery,
chemotherapy and radiation therapy. Neoadjuvant (pre-operative)
chemoradiation has been shown in multiple randomized clinical
trials to improve clinical outcomes and toxicity profiles compared
to post-operative chemoradiation for locally-advanced rectal
cancer. This approach has become the current standard of care, and
typically consists of 5-fluorouracil based chemoradiation over 5.5
weeks, followed by total mesorectal excision 6–10 weeks later
(2,3). In ~10–20% of cases, pathological
complete response (pCR) is observed in the surgical resection
specimen after neoadjuvant chemoradiation (4–8). In
addition, up to 25–50% of patients develop a clinical complete
response (cCR) after neoadjuvant chemoradiation, which is
determined by endoscopic or imaging-based assessments (9). The discrepancy between rates of pCR
and cCR results from the presence of microscopic tumor deposits
which are undetectable using clinical techniques, but are
discovered with meticulous pathologic assessment following
resection. Notaby, a study published in 2004 showed that for
patients who experienced a pCR or cCR after chemoradiation,
subsequent surgery had no effect on disease-free survival or cancer
control (10). The potential for
avoiding surgery, and thereby improving quality of life for these
patients, has become an active area of research, particularly in
patients who otherwise would require an abdominoperineal resection
(APR) and permanent colostomy. Indeed, there have been multiple
publications over the last several years highlighting this 'watch
and wait' approach, commonly called non-operative management (NOM)
or selective surgery (9,11-13).
Using this approach, rates of avoiding pelvic surgery in patients
treated with definitive chemoradiation have been reported to be as
high as ~70% in some studies, while still maintaining equivalent
cancer control (10).
Additionally, local control rates remain as high as 95% with the
use of salvage surgery when a local recurrence is detected. While
these results are promising, the challenge still remains to
prospectively identify which patients are best suited for a
non-operative approach.
Because less than half of patients experience a pCR
or cCR after chemoradiation, the present study is focused on
pre-therapeutic biomarkers that may predict which patients are more
likely to achieve a complete response. Many different types of
biomarkers have been studied in the hope of identifying patients
who would be best treated with a non-operative approach. Many of
these molecular profiling studies have focused on DNA, looking for
genetic mutations in specific tumor suppressor genes and/or
oncogenes, such as APC or TP53, to predict response to therapy. One
of the most studied cancer genes is KRAS, a GTPase which is
implicated in mediating resistance to the anti-EGFR agent cetuximab
(14). KRAS has also been
theorized to confer radioresistance, but the results from studies
have been mixed. Some studies (15,16)
have linked KRAS mutations to lower rates of pCR in patients
receiving chemoradiation therapy, while other studies have found
that KRAS mutations have no consistent utility in predicting the
probability of pCR (17,18). Some articles have postulated that
this inconsistency may be due to the fact that mutations
specifically in codon 13 of the KRAS gene are also more likely to
have concurrent TP53 mutations, potentially explaining why KRAS
gene mutations may be associated with radioresistance (since TP53
mutations have also been linked to radioresistance) (19). Other studies have identified other
genes such as the DNA repair gene SMC1 (20), the apoptotic gene LUM (21) and the DNA repair gene XRCC3
(22) that predict response to
chemoradiation. However, many of these genes are rarely replicated
across studies, resulting in the identification of many
non-overlapping genes that may predict radiation sensitivity or
resistance that is beyond the scope of the present review (23).
Due to the difficulty with identifying genetic
aberrations consistently conferring radiation resistance, other
genetic biomarkers such as methylation status and non-coding RNA
are now being investigated. For example, a 2013 study found that
methylation of the TIMP3 gene correlated with chemo-radiation
resistance (24). In addition, a
2014 study found the expression of long non-coding RNA (lncRNA)
lincRNA-p21 to be correlated with improved response to
chemoradiation (25). Additional
analyses of methylation status and lncRNA biomarkers are
ongoing.
In recent years, another type of non-coding RNA,
micro-RNA (miRNA), has been increasingly studied in cancer, along
with their possible radio-sensitizing and/or radio-resistant
properties. miRNA begins as a DNA transcript called pri-miRNA in
the nucleus, where DGCR8 and Drosha then cut it into pre-miRNA,
which subsequently leaves the nucleus (26). In the cytoplasm, an enzyme called
DICER cuts the pre-miRNA hairpin into the mature miRNA, which is
then loaded onto the RNA-induced silencing complex (RISC). RISC
delivers miRNA to particular messenger RNA (mRNA) in order to
silence those transcripts. The miRNA binds to the untranslated
region of mRNA to prevent it from being able to enter the ribosome
and be translated (26). miRNA
dysregulation is a well-documented contributing factor to
carcinogenesis with loss of normal function resulting in altered
expression of important oncogenes and/or tumor suppressor genes
(27). Finally, since miRNA can be
secreted into bodily fluids with minimal degradation (unlike mRNA),
miRNAs have the potential to serve as stable, and relatively
non-invasive biomarkers for prognosis and prediction of therapeutic
response (28,29).
2. Patient studies evaluating miRNAs and
response in rectal cancer
In recent years, the role of miRNA dysregulation in
cancer has been better elucidated as more studies are identifying
particular miRNAs that predict response to treatments such as
radiation and chemoradiation. As non-operative management for
rectal cancer continues to gain momentum amongst patients and
practitioners, it will be especially important to integrate
reliable methods of predicting disease response to ensure proper
patient selection for this approach. We performed a literature
review, and to date, twelve studies have analyzed rectal cancer
tumor tissue to evaluate the role of various miRNAs (miRs) in
predicting therapeutic response (Table
I). The results of these studies are listed in Table I. Each of these studies, except for
one, included pathological staging, as it has been shown to
correlate with prognosis better than clinical downstaging after
pre-operative chemoradiation (30). In addition, the majority of these
studies performed unbiased screening of hundreds of miRNAs using
various platforms (e.g. TaqMan microRNA, miScript assay, Agilent
SurePrint Technology Rel 12.0), rather than studying a few miRNAs
in a hypothesis-driven (i.e. a priori) manner. As such, these
studies may be confounded by type I error resulting from multiple
comparison testing methodology. The only studies that were driven
by a priori evaluation of certain miRNAs were the studies by
Drebber et al (31) Carames
et al (36) and Svoboda
et al (45). Many of the
miRNAs identified in these studies have been shown to impact DNA
damage response, cell cycle and apoptotic signaling pathways. The
associations of some of these miRNAs with various protein mediators
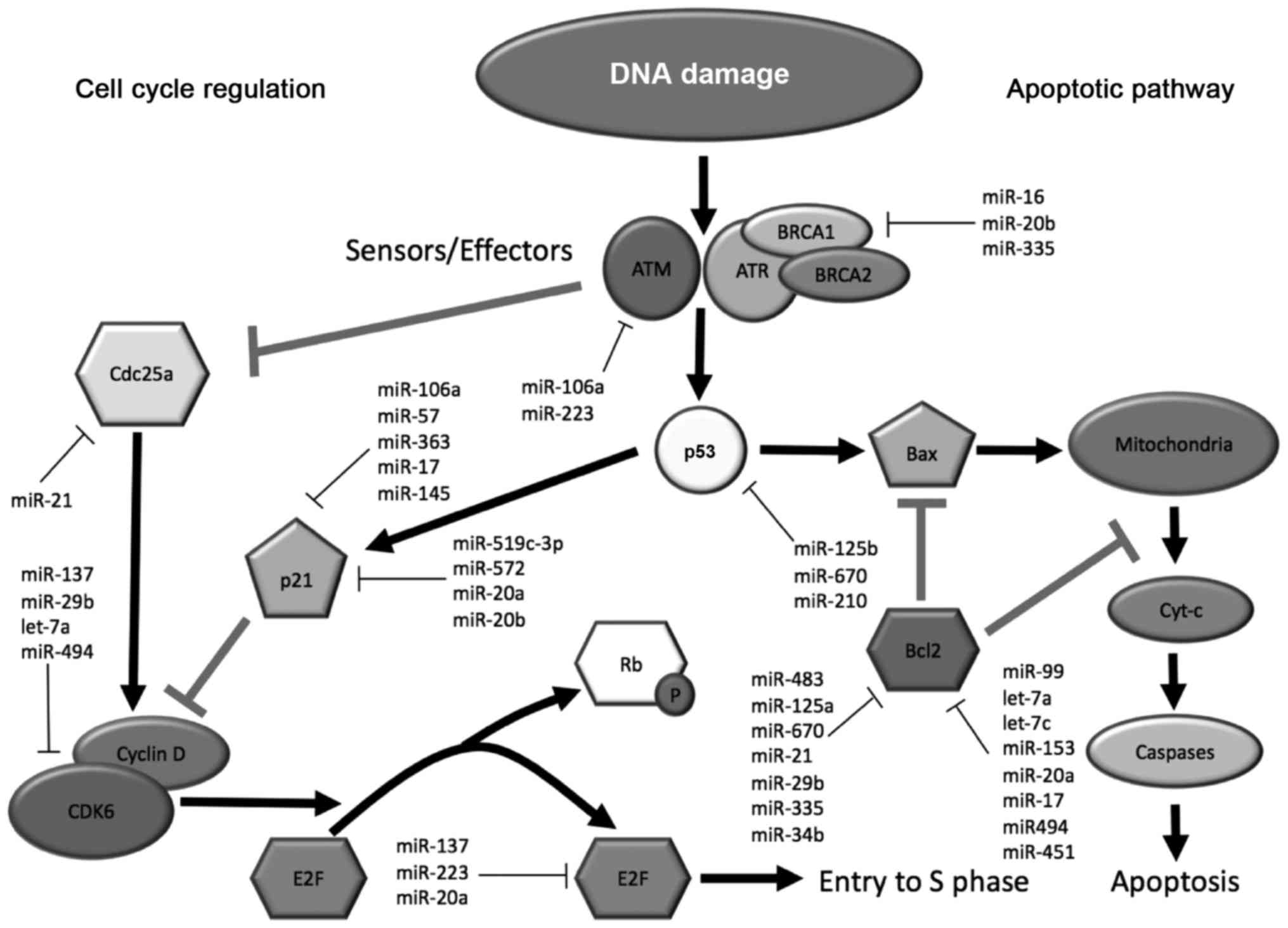
of these pathways are depicted in Fig.
1.
 | Table IRectal cancer patient studies
investigating the relationship between specific miRs and response
to therapy. |
Table I
Rectal cancer patient studies
investigating the relationship between specific miRs and response
to therapy.
| Study | No. of
patients | Radiation dose and
chemotherapy | Response
assessment | No. of miRNAs
examined | miRNA plaform | Identihed
miRNAs |
|---|
| Svoboda et
al (45) | 35 | 50.4 Gy
capecitabine | Dworak regression
grade | 9 | TaqMan MicroRNA
assay | Upregulated
(intratherapy)
Poor response: miR-125b, miR-137 |
|
| Drebber et
al (31) | 40 | 50.4 Gy 5-FU | WHO
classification | 3 | miScript assay | Downregulated
Poor response: miR-145 |
|
| Della Vittoria
Scarpati et al (42) | 35 | 45 Gy capecitabine
+ oxaliplatin | Mandard regression
grade | 373 | miScript assay | Upregulated
Complete response: Signature: miR-1183, miR-483-5p, miR-622,
miR-125a-3p,
miR-1224-5p, miR-188-5p, miR-1471, miR-671-5p, miR-1909,
miR-630,
miR-75
Downregulated
Complete response: Signature: miR-1274b, miR-720 |
|
| Svoboda et
al (38) | 20 | 50.4 Gy
capecitabine/5-FU | Mandard regression
grade | n/a | TaqMan MicroRNA
assay (TLDA) | Upregulated
Poor response: miR-215, miR-190b, miR-29b-2
Downregulated
Poor response: let7ea, miR-196b,
miR-450a, miR-450b-5p, miR-99a |
|
| Bandres et
al (43) | 61 | 50.4 Gy
capecitabine | Mandard regression
grade | 667 | TaqMan MicroRNA
assay (TLDA) | Upregulated
Complete response: Signature: miR-21, miR-99, miR-125ba, miR-125bl, let-7ca, miR-490
Downregulated
No response: Signature: miR-21, miR-125a-3p |
|
| Kheirelseid et
al (44) | 12 | Not specified | Mandard regression
grade | n/a | TaqMan MicroRNA
assay (TLDA) | Upregulated
Complete response: Signature: miR-16, miR-590-5p, miR-153
Partial response: Signature: 519c-3p, miR-561 |
|
| Lopes-Ramos et
al (35) | 43 | 50.4-54 Gy
5-FU | Dworak's regression
grade; pelvic MRI; proctoscopy;
CEA | n/a | SOLiD Total RNA-Seq
kit | Upregulated
Complete response: miR-21-5p |
|
| Hotchi et al
(39) | 43 | 40 Gy/20 fractions
S-l | RECIST
Histopathological examination for pathologic downstaging | 821 | Agilent Human miRNA
microarray v2.0 | Upregulated (RECIST)
Response: miR-223
Downregulated
(RECIST)
Response: miR-20b, miR-92, let-7aa, miR-20a, miR-17, miR-106a
Upregulated
(Histopatholopic patholopic)
Response: miR-223, miR-142-3p, miR-223, miR-630, miR-126 |
|
| Carames et
al (36) | 76 | Radiation dose
unspecified capecitabine/5-FU | Ryan classification
of downgrading | 1 | TaqMan MicroRNA
assay | Upregulated
Poor response: miR-21a |
|
| D'Angelo et
al (37) | 38 | 50.4 Gy
capecitabine/5-FU | Mandard regression
grade | 866 | Agilent SurePrint
Technology (Rel 12.0 v3) | Upregulated
Poor response: miR-125b |
|
| Millino et
al (41) | 59 | 50.4 Gy
capecitabine/5-FU | Mandard regression
grade | 866 | Agilent SurePrint
Technology (Rel 12.0 v3) | Upregulated
Complete response: miR-572, miR-939, miR-538, miR-1260, miR-575,
miR-210, miR-150, miR-324-5p, miR-638, miR-572
Poor response: miR-630a,
miR-210 |
|
| Nakao et al
(40) | 59 | 40 Gy
tegafur/gimeracil/oteracil | RECIST | n/a | Agilent Human miRNA
microarray v3.0 | Upregulated
Response: miR-19-3p, miR-866-3p, mR-923, miR-494, miR-513a-5p,
miR-513b, miR-154, miR-379, miR-223, miR-1542-5p, miR-144,
miR-363, miR-31, miR-1290, miR-382, miR-193a-5p, miR-451, miR-335,
miR-486-5p, miR-1246, miR-34b, miR-144 |
Summary of patient studies identifying a
single pre-therapeutic miRNA associated with response
A number of studies have identified relationships
between single miRs and pathological response to chemoradiation.
Typically, these studies have measured the relationship between the
pre-therapeutic levels of certain miRs and the pathological
response to therapy. The first of these studies measured the
relationship between miR-145, a miR known to downregulate IRS-1
expression and cellular proliferation and treatment response
(24,31). Higher pre-therapeutic miR-145
expression levels correlated with chemoradiosensitivity and more
pathological tumor down-staging. Other studies have shown that
miR-145 levels are often decreased in colorectal cancer, further
supporting its role as a tumor suppressor (32–34).
Another study by Ramos et al (35) found pre-treatment miR-21-5p
expression to be upregulated in patients who demonstrated an
improved pathological response to chemoradiation. This result,
however, was contradicted by a study by Carames et al
(36) which found that increased
miR-21 correlates with worse pathologic response to therapy. A
study by D'Angelo et al (37) similarly identified that
upregulation of miR-125b correlates with a worse pathological
response to therapy. This study is particularly interesting, as it
found that high expression levels of both tissue and serum miR-125b
correlated with a poor response to therapy. This study supports the
potential for serum-based miRNA analysis as a less invasive and
cost-effective biomarker compared to tissue-based analysis and
offers the potential for serial monitoring.
Summary of patient studies identifying
multiple pre-therapeutic miRNAs associated with response
Other studies have identified multiple miRNAs whose
individual pre-therapeutic levels correlate with pathological
response to chemoradiation. A study by Svoboda et al
(38) examining 20 rectal cancer
patients detected eight miRNAs that differed in expression between
responders and non-responders. Five miRNAs (miR-196b, 450a, 450b-5p
and 99a) were elevated in responders while three different miRNAs
(miR-215, 190b and 29b-2) were elevated in non-responders. A study
by Hotchi et al (39) was
unique in that it used three separate methods of measuring response
to chemoradiation: RECIST (Response Evaluation Criteria in Solid
Tumors), histopathological analysis (tumor regression grade) and
clinical tumor downstaging. Each of these methods detected distinct
miRNAs associated with response to therapy. Using the RECIST method
to evaluate response, miR-223 was found to be elevated in
responders, while miR-20b, miR-92, let-7a, miR-20a, miR-17 and
miR-106a were decreased in responders. Histopathological
examination with tumor regression grade revealed miR-142-3p and
miR-223 to be elevated in responders. Clinical tumor downstaging
showed elevated levels of miR-223, miR-630 and miR-126 to correlate
with improved response to therapy. Of these, miR-223 was the only
miR found to be elevated in responders via all three responder
classification methods. A study by Nakao et al (40) validated these results by
demonstrating that elevated pre-therapeutic miR-223 levels
predicted for a pCR. In addition, many other miRs correlated with a
complete response, and these can be referenced in Table I. A more recent study by Millino
et al (41) found a large
number of miRs to be upregulated in complete responders (Table I) They also detected two miRs to be
significantly upregulated in non-responders: miR-630 and miR-210.
Notably, this finding for miR-630 contradicts studies by Hotchi
et al (39) and Della
Vittoria Scarpati et al (42).
Summary of patient studies predicting
clinical response
As mentioned, pathologic staging appears to
correlate better with outcomes than clinical staging and is likely
to serve as a better endpoint for development of molecular
signatures given that clinical response may be more subjective and
that investigators use different methods to assess clinical
response (e.g. MRI, PET scan and endoscopic biopsies). However,
clinical staging evaluation has the advantage of not requiring a
thorough pathologic evaluation of the resected surgical specimen.
In addition to the study by Hotchi et al (39), two other studies used
clinical/imaging indicators with or without pathological
downstaging to measure response to therapy. Lopez-Ramos et
al (35) assessed clinical
response by biopsy, rectal examination, pelvic MRI, proctoscopy and
CEA levels. miR-21-5p upregulation prior to therapy was found to
predict better clinical and pathologic responses to therapy. Nakao
et al (40) was the only
study that did not combine clinical downstaging with pathologic
downstaging. In the present review, associations between miRs were
based purely on RECIST response, and many miRs were upregulated and
associated with complete imaging response, including miR-223, which
was identified in the study by Hotchi et al (39) (Table
I).
Summary of patient studies that have
developed miRNA signatures
Since single molecular aberrations are often
unlikely to reliably predict response across a large number of
patients, investigators have attempted to develop signatures by
incorporating multiple miRs. In doing so, it is hoped that
biomolecular signatures will exhibit improved predictive power
compared to single miR biomarkers. Bandres et al (43) examined the expression profile of
667 miRNAs in 85 rectal cancer patients. They found a signature
consisting of miR-21, miR-99, miR-125b, miR-125b1, miR-let-7c and
miR-490 upregulation that was associated with pCR. Conversely, a
signature incorporating miR-21 and 125a-3p downregulation was
associated with no response to treatment.
Another study by Della Vittoria Scarpati et
al (42) identified many
different miRNAs that correlated with treatment response and used
these miRs to develop a signature that best correlated with pCR.
They identified 13 miRNAs (Table
I) that were differentially expressed between complete
responders versus incomplete responders. The miRNAs with the
strongest predictive value for treatment response were miR-630 and
miR-622. A study by Kheirelseid et al (44) reported a unique signature
consisting of miR-16, miR-590-5 and miR-153 that, when upregulated,
predicted for pCR. The authors also identified a signature
comprised of miR-519-3p and miR-561 that could predict a better
treatment response. Further efforts are warranted to investigate
the utility of miR expression signatures in predicting clinical
outcomes and validate them across multiple clinical datasets.
Patient studies comparing miRNA levels
before and after therapy
All of the previously mentioned studies used
pre-therapeutic miRNA levels to predict response to therapy.
Notably, only one study by Svoboda et al (45) measured changes in miRNA expression
levels after therapy, and how this difference could predict
response to therapy. The authors found that two miRs (miR-125b and
miR-137) increased in expression during treatment (from tumor
biopsy tissue obtained 2 weeks into chemoradiation) and correlated
with a poor response to therapy. Overall, there is a lack of
studies utilizing this methodologic approach and further work is
warranted to explore how the expression of miRNA biomarkers change
during and after therapy. These studies could provide a better
understanding of how tumor tissue responds to chemoradiation while
simultaneously identifying molecular pathways that could mediate
resistance (particularly by assessing miRNAs in recurrent or
persistent disease).
3. Summary of individual miRs and in
vitro studies
Despite the large number of studies, a significant
confounding factor is that there has been minimal overlap amongst
the miRNAs identified as being predictive of treatment response.
This may be due to different tumor down-staging criteria,
histopathologic regression grading systems, treatment methods
and/or patient characteristics (i.e. cancer stage, grade,
perineural invasion and lymphovascular space invasion). However,
some miRNAs were identified in multiple studies, and the discussion
that follows is centered on many of these, along with some of the
in vitro evidence that assists to characterize their
mechanisms of action in determining response to CRT. We summarize
the major findings of individual miRNAs identified in these rectal
cancer studies in Table II.
 | Table IISpecific miRs that were common among
studies: effect on radiation sensitivity and relevant targets. |
Table II
Specific miRs that were common among
studies: effect on radiation sensitivity and relevant targets.
| miRNA | Effect on
radiation | Relevant
target(s) | Function of
target(s) | Study/Authors |
|---|
| miR-21 |
Radioresistance | SABT1: recruits
chromatin remodeling and epigenetic modifying proteins | Regulation of gene
expression at G1/S checkpoint | Kohwi-Shigematsu
et al (55)
Kowalzyk et al (56)
Mima et al (48) |
|
Radiosensitivity | PDCD4: a protein
which helps induce apoptosis | Apoptosis | |
| Let-7 family |
Radiosensitivity | RAS: a GTPase in
the MAPK pathway (identified in other cell lines) | Regulation of
growth, transcription, and translation | Johnson et
al (59)
Weidhass et al (61) |
| miR-125a-5p |
Radiosensitivity | Bcl2 family:
anti-apoptotic proteins | Anti-apoptosis | Tong et al
(62)
Xie et al (63) |
| miR-125b |
Radioresistance | p53: regulates cell
division and apoptosis | DNA repair
induction, G1/S checkpoint | Banzhaf-Strathmann
et al (65) |
| miR-99 |
Radiosensitivity | SNF2H/SMARCA5:
chromatin remodeling factor implicated in DNA repair mTOR:
integrates signaling pathways to promote cellular growth and
survival HOXA1: transcription factor and proto-oncogene that
regulates anti-apoptotic | DNA repair,
cellular proliferation, survival, apoptosis | Xu et al
(68)
Tokunaga et al (69)
Hay et al (70) |
| miR-630 |
Radiosensitivity | BCL2CL2, TP53RK:
proteins that prevent apoptosis | Anti-apoptosis | Zhang et al
(73) |
| miR-223 |
Radiosensitivity | STMN1: contributes
to mitotic spindles | Exit from
mitosis | Sugatani et
al (74)
Fazi et al (75)
Wong et al (76)
Rubin et al (77)
Ghosh et al (78)
Saal et al (79)
Alli et al (80) |
miR-21
miR-21 is significant as it is the most prolific
miRNA in patient studies predicting response to CRT. It was found
to have a significant correlation with response to therapy in 3 of
the 12 studies examined for this analysis (35,36,43),
and had a correlation approaching significance in one other study
(36). In addition, its molecular
targets and oncogene and tumor suppressor properties are well
documented in preclinical studies (46–54).
Its role in predicting patient response to CRT, however, remains
controversial. Two of the above patient studies found upregulation
of miR-21 to be correlated with a complete response to CRT
(35,43). An in vitro study by
Lopes-Ramos et al (35)
showed that miR-21-5p upregulation induces radiosensitization by
inhibiting SATB1 expression. SATB1 is a gene regulator that is
associated with poor outcomes in rectal cancer (55). This inverse relationship between
miR-21-5p and SATB1 has also been confirmed in other cancer types
(56). In addition, another in
vitro study with colon cancer cells found that miR-21 inhibits
cdc25a levels, therefore, arresting the cell cycle at the G1/S
checkpoint and preventing tumor growth (57).
A potential radiosensitizing property for miR-21 has
been contradicted by other studies, however. Carames et al
(36) found that patients having
upregulated miR-21 experienced worse response to CRT, postulating
that miR-21 conferred radioresistance in these patients. This
result has been replicated in an in vitro study by Deng
et al (46) which
demonstrated that inhibiting miR-21 can increase the sensitivity of
CRC cells to CRT. Mechanistically, a link between miR-21 and PDCDR,
a programmed cell-death protein, has been identified. PDCD4 helps
induce apoptosis, and therefore leads to cellular death. Dou et
al showed that PDCD4 inhibition rendered rectal cancer cells
less likely to commit to apoptosis after radiation therapy, thereby
decreasing the sensitivity of cancer cells to radiation therapy
(52). Inhibition of PDCDR by
miR-21 has also been shown in several other in vitro studies
(47,50,51,53).
These seemingly contradictory results may be due, in
part, to miR-21 affecting different gene targets under different
cellular circumstances. While targeting SATB1 may induce
radiosensitivity, targeting PDCD4 may lead to radioresistance.
Context dependencies whereby a gene, RNA transcript, protein, or
other molecules have both oncogenic and tumor suppressor roles have
been identified for many other molecules, and are similarly
possible for miRNAs.
Let-7 family
The Let-7 family of miRNAs was implicated in three
of the above studies (38,39,43).
Svoboda et al (38) found
that let-7e downregulation was associated with a poor response to
chemoradiation. Bandres et al (43) report similar results, with let-7c
upregulation correlating with complete response. Let-7's role in
radiation sensitivity has been extensively studied in vitro,
although only one study focused on rectal cancer cells. Salendo
et al (58) found that
let-7g, in addition to other miRs, promotes increased
radiosensitivity in rectal cancer cells, congruent with the Svoboda
(38) and Bandres (43) studies. The Salendo study (58) also quantified pre-treatment
expression levels of miR-let-7g in rectal cancer biopsy samples and
found that higher levels of let-7g were associated with improved
disease-free survival.
The possible mechanisms for let-7's radiosensitizing
properties can be elucidated via studies in other cancer cell
lines. The major target appears to be RAS (59). RAS is a protein in the EGFR/MAPK
pathway that has been implicated in diminishing the effectiveness
of radiation in multiple cancer types (60,61).
Its role in promoting radioresistance appears to be mediated by DNA
repair mechanisms, thereby correcting radiation-induced DNA damage
and preventing subsequent cell death (61). Let-7 can silence the RAS gene,
therefore eliminating this protection and increasing cancer cell
susceptibility to radiation therapy. Another study found let-7 to
be a master regulator of cell division, possibly affecting more
than 30 genes involved in mitosis (59). While very interesting and
hypothesis-generating, these studies should be extrapolated to
rectal cancer with caution.
A study by Hotchi et al (39) found radioresistance properties
associated with a let-7 family member. It showed that let-7a was
one of many miRs whose downregulation actually correlated with a
complete response. However, this correlation was only seen in one
of the three downstaging methods used, and let-7a appears to be the
only member of the let-7 family to be associated with
radioresistance.
miR-125 family
The miR-125 family was identified in four of the ten
patient studies (37,42,43,45).
Two of these found upregulated miR-125a-5p levels to be associated
with a complete response to therapy. Della Vittoria Scarpati et
al (42) found elevated
miR-125a-3p to be 1 of 11 elevated miRs implicated in a signature
that correlated with complete response to CRT, while Bandres et
al (43) confirmed this result
by finding downregulation of miR-125a-5p to be associated with no
response to therapy. Cellular and human tissue studies further
confirm these results (62).
A study by Tong et al (62) investigated the cellular targets of
miR-125a-5p. In concordance with the two patient studies, they
found miR-125a-5p to be a tumor suppressor in colon cancer,
inhibiting cell proliferation and growth. Furthermore, they found
the anti-apoptotic genes BCL2, BCL2L12 and Mcl-1 to be targets of
miR-125a-5p. Increased miR-125a-5p levels decreased the expression
of these anti-apoptotic genes, while overexpression of these
anti-apoptotic genes overcame the tumor suppressive effect of
miR-125a-5p. Additional support for a tumor suppressive role for
miR-125a-5p was provided by a study showing miR-125a levels to be
decreased in colorectal cancer (63).
These results are distinct from those for miR-125b.
miR-125b has been consistently shown to be upregulated in
colorectal cancer and correlated with poor prognosis (64). Svoboda et al (45) found elevated miR-125b levels to
correlate with a poor response to therapy. This result is supported
by a study by D'Angelo et al (37) which found elevated miR-125b levels
to correlate with a poor response to chemoradiation. These apparent
oncogenic properties of miR-125b were further confirmed by a study
by Banzhaf-Strathmann et al (65) identifying the targets of miR-125b
to be the apoptosis-associated gene BAK1, as well as cell cycle
proteins Puma, cyclin C, Cdc25c and p53. The only contradictory
study was published by Bandres et al (43) who reported upregulated miR-125b to
be part of a miR signature that correlated with complete
response.
miR-99
miR-99 was identified in two of the patient studies.
Svoboda et al (38) found
downregulated miR-99 levels to correlate with a poor response to
therapy, and this result was further corroborated by Bandres et
al (43) who found that high
miR-99 levels correlated with a complete response to therapy. The
data may suggest that miR-99 has a radiosensitizing effect. In
vitro studies in other cancer cell lines have identified
plausible targets for miR-99 that may explain its association with
radiosensitization/response. In one study, miR-99 family miRNAs
were identified in a screen for miRNAs that correlate with
radiosensitivity. They were found to target SNF2H/SMARCA5 (a
SWI/SNF chromatin remodeling factor), reduce BRCA1 localization to
sites of DNA damage, and reduce the efficiency of multiple types of
double-strand break repair (homologous recombination and
non-homologous end-joining) (66).
Another study by Sun et al (67) in esophageal cancer cells found that
miR-99 induces apoptosis by inhibiting mTOR. mTOR has been
identified as an important protein in oncogenesis, as its
overexpression and dysregulation leads to uncontrolled
proliferation and survival (68).
Its functions in cellular growth and proliferation include
integration of nutrient sensing pathways and mitochondrial activity
(69). mTOR receives extensive
signaling input from many upstream cell signaling pathways
regulating growth, including insulin and IGF-1 (70). In addition, another study showed
that miR-99 family miRNAs target homeobox A1 (HOXA1), a
proto-oncogene, and Bcl2 and then reduced proliferation, migration
and enhanced apoptosis (69,71).
Consistent with these functions, miR-99 has been shown to be
downregulated in human cancers, including prostate, head and neck
and esophageal cancer, consistent with tumor suppressive functions
(72). Thus, higher miR-99 appears
to be associated with improved response.
miR-630
miR-630 was identified in three of the patient
studies (39,41,42).
Two of these, Hotchi et al (39) and Della Vittoria Scarpati et
al (42) found that
upregulated miR-630 correlated with a better response to
chemoradiation. Additional support for this result was provided by
an in vitro study by Zhang et al (73) who found that miR-630 induces
apoptosis in cancer cells after radiation therapy. Subsequent
mechanistic investigations identified that the targets of miR-630
are BCL2L2 and TP53RK, two proteins which prevent apoptosis.
However, these results were contradicted by the most recent study
by D'Angelo et al (37),
which found that miR-630 upregulation correlates with a poor
response to CRT.
miR-223
miR-223 was identified in two of the studies, and in
both cases, its upregulation increased response to CRT. In Hotch
et al (39), the evidence
for miR-223 radiosensitizing effect was especially strong, as it
was the only miR to be consistently associated with an increased
response in all three methods of response assessment. Nakao et
al (40) provided supporting
evidence of this, as miR-223 was one of many miRs associated with
an increase in response to CRT. No in vitro studies have
been performed to examine the mechanism of action of miR-223 in
rectal cancer cells specifically, but studies from other cell lines
propose a role in modulating cell differentiation and proliferation
(74,75). In hepatocellular carcinoma, STMN1
has been identified as a target of miR-223 which is responsible for
its tumor suppressor effect (76).
STMN1 is a microtubule regulator which promotes depolymerization of
tubulin and is important for forming the mitotic spindle.
Inhibition of STMN1 leads to cell accumulation in the G2/M phase,
unable to exit mitosis (77). In
addition, overexpression of STMN1 has been correlated with poor
treatment response in other tumor types (78–80).
Therefore, the tumor suppressive effects of miR-223 may be mediated
by silencing of STMN1, thereby preventing cancer cells from
proliferating.
4. IPA Molecular Network Analysis
An Ingenuity Pathway Analysis (IPA) was carried out
to find potential links between some of the miRs found in the
present review. miR-223, miR-21, miR-125a, miR-125b, miR-630,
miR-99 and the let-7 family were analyzed. The IPA analysis
searched the literature for upstream and downstream targets of
these miRs, and then tried to connect them into a single possible
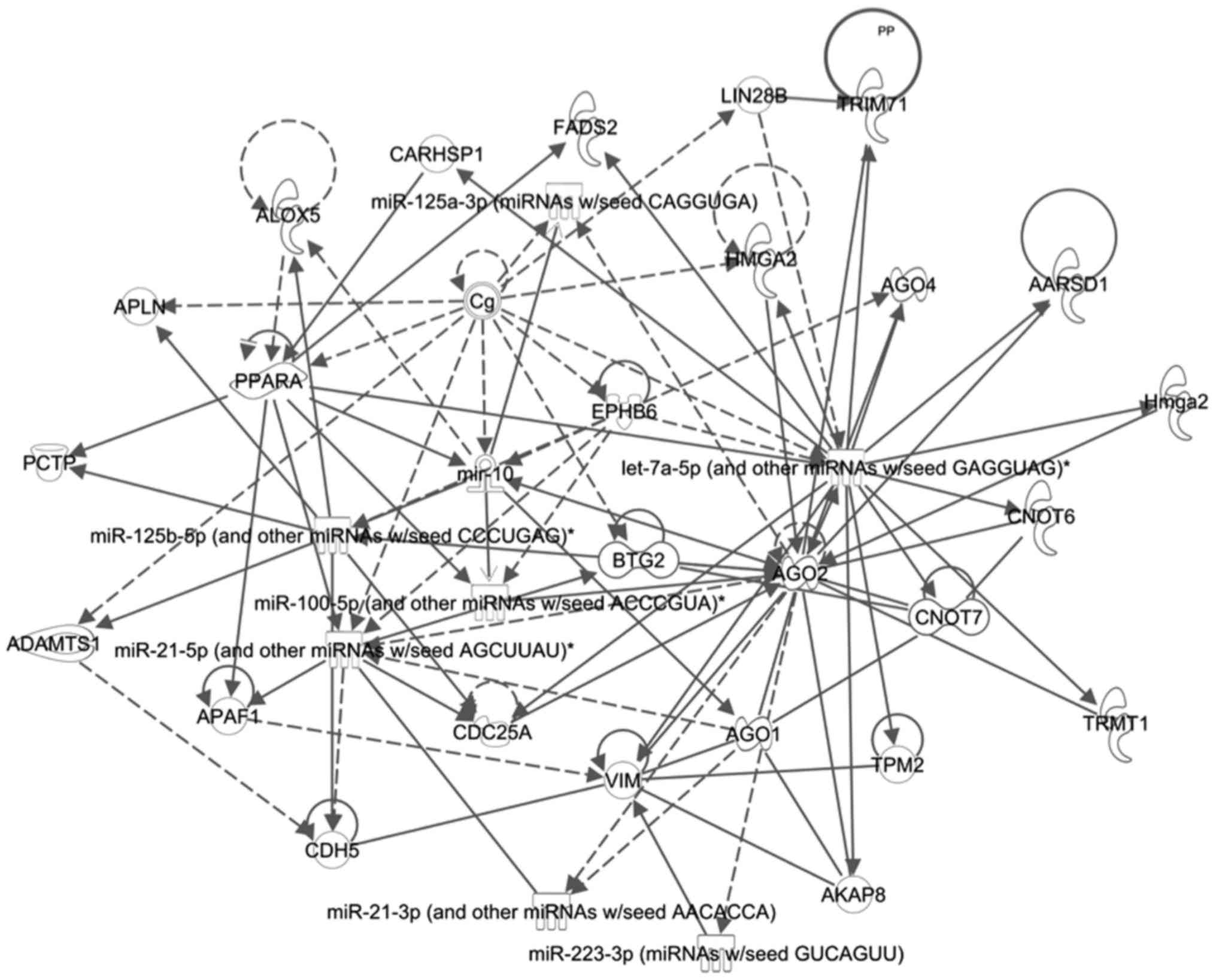
network. A network (Fig. 2) was
identified containing six of the seven miRs, with an IPA
correlation score of 17. One of the main molecules in this network
was AGO2, which has been shown to interact with many of the miRs.
AGO2 is a member of the argonaute family of proteins which guide
miRs to their targets for silencing (81). One important target of this pathway
is CDC25A. CDC25A has been identified as an oncogene, encoding for
cdc25a, which is required to transition from the G1 to S of mitosis
(82,83). These results suggest that some of
identified miRs may work together to modulate the ability of tumor
cells to progress through the cell cycle, and therefore ultimately
modify their radiosensitivity.
5. Conclusions/Future directions
There are promising results with the use of miRNAs
as biomarkers to predict response in rectal cancer after CRT.
However, as shown above, there is no consensus among studies with
regards to the individual miRs or miR signatures that predict pCR
or cCR. This may be due to many different confounding factors. One
is that some studies used different chemotherapy regimens with
distinct agents (e.g. fluorouracil, capecitabine, platinum agent
and S-1) and/or doses that may induce different miR responses. In
addition, the different types of tissue fixation methods that were
used (paraffin-embedded versus frozen) might impact results as
well. Another potential confounding factor is that the studies used
different methodologies to evaluate expression of miRNAs as shown
in Table I, certainly leading to
variability in quantification. Furthermore, different staging
techniques and different endpoints (e.g. clinical response versus
pathologic response) may have contributed to some inconsistencies
between studies. Finally, performing large-scale molecular testing
can be subject to type I error from multiple comparisons
testing.
Future studies should attempt to develop and
validate consistent miR signatures that correlate with pathological
and/or clinical complete responses, and be cognizant of the
endpoint that is being used to develop the signature. For example,
if a signature is designed to best select patients for avoiding
surgery after neoadjuvant CRT, the quality of the signature should
be based on its ability to predict pCR. Conversely, if a signature
is being developed to predict a poor response after CRT, then the
signature should reliably predict which patients will have lymph
node positive disease or poor tumor regression grade (e.g. Mandard
TRG ≥4) after standard CRT in efforts to potentially intensify the
neoadjuvant regimen. Finally, if the clinical objective is to
potentially alter post-operative (adjuvant) treatment, the
signature should be able to predict local or distant recurrence.
Further research should also be focused on developing more
predictive signatures using less invasive tests (e.g. urine/serum
miRNAs). In addition, other clinical outcomes beyond primary tumor
response (e.g. survival, cause-specific survival, colostomy-free
survival, local failure, regional failure and distant failure)
should ultimately be correlated with miRNA expression. Such
predictive biomarkers could then be used to identify patients with
a high probability of pCR/cCR from chemoradiation, or low
probability of response. Ideally, those patients identified as high
likelihood of responding could be initially spared the morbidity
and quality of life issues associated with total mesorectal
excision, particularly for distal tumors. Lastly, miRNA analysis
has the potential to identify pathways conferring radiation or
chemoradiation resistance, by comparing pre-therapeutic and
post-therapeutic samples (i.e. at the time of surgery). These
analyses hold promise for identifying novel molecular pathways for
targeting in combination with radiation or chemoradiation, in an
effort to further improve upon current cCR and pCR rates.
Acknowledgments
The present review was supported by the following
grants: the Award Number Grant KL2TR001068 from the National Center
for Advancing Translational Sciences, and NIH R01 CA198128 (to
T.W.). The content is solely the responsibility of the authors and
does not necessarily represent the official views of the National
Center for Advancing Translational Sciences or the National
Institutes of Health.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sauer R, Liersch T, Merkel S, Fietkau R,
Hohenberger W, Hess C, Becker H, Raab HR, Villanueva MT, Witzigmann
H, et al: Preoperative versus postoperative chemoradiotherapy for
locally advanced rectal cancer: Results of the German
CAO/ARO/AIO-94 randomized phase III trial after a median follow-up
of 11 years. J Clin Oncol. 30:1926–1933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roh MS, Colangelo LH, O'Connell MJ,
Yothers G, Deutsch M, Allegra CJ, Kahlenberg MS, Baez-Diaz L,
Ursiny CS, Petrelli NJ, et al: Preoperative multimodality therapy
improves disease-free survival in patients with carcinoma of the
rectum: NSABP R-03. J Clin Oncol. 27:5124–5130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luna-Pérez P, Rodríguez-Ramírez S,
Hernández-Pacheco F, Gutiérrez De La Barrera M, Fernández R and
Labastida S: Anal sphincter preservation in locally advanced low
rectal adenocarcinoma after preoperative chemoradiation therapy and
coloanal anastomosis. J Surg Oncol. 82:3–9. 2003. View Article : Google Scholar
|
|
5
|
Hiotis SP, Weber SM, Cohen AM, Minsky BD,
Paty PB, Guillem JG, Wagman R, Saltz LB and Wong WD: Assessing the
predictive value of clinical complete response to neoadjuvant
therapy for rectal cancer: An analysis of 488 patients. J Am Coll
Surg. 194:131–135; discussion 135–136. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Habr-Gama A, de Souza PM, Ribeiro U Jr,
Nadalin W, Gansl R, Sousa AH Jr, Campos FG and Gama-Rodrigues J:
Low rectal cancer: Impact of radiation and chemotherapy on surgical
treatment. Dis Colon Rectum. 41:1087–1096. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Medich D, McGinty J, Parda D, Karlovits S,
Davis C, Caushaj P and Lembersky B: Preoperative chemoradiotherapy
and radical surgery for locally advanced distal rectal
adenocarcinoma: Pathologic findings and clinical implications. Dis
Colon Rectum. 44:1123–1128. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grann A, Minsky BD, Cohen AM, Saltz L,
Guillem JG, Paty PB, Kelsen DP, Kemeny N, Ilson D and Bass-Loeb J:
Preliminary results of preoperative 5-fluorouracil, low-dose
leucovorin, and concurrent radiation therapy for clinically
resectable T3 rectal cancer. Dis Colon Rectum. 40:515–522. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Habr-Gama A, Gama-Rodrigues J, São Julião
GP, Proscurshim I, Sabbagh C, Lynn PB and Perez RO: Local
recurrence after complete clinical response and watch and wait in
rectal cancer after neoadjuvant chemoradiation: Impact of salvage
therapy on local disease control. Int J Radiat Oncol Biol Phys.
88:822–828. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Habr-Gama A, Perez RO, Nadalin W, Sabbaga
J, Ribeiro U Jr, Silva e Sousa AH Jr, Campos FG, Kiss DR and
Gama-Rodrigues J: Operative versus nonoperative treatment for stage
0 distal rectal cancer following chemoradiation therapy: Long-term
results. Ann Surg. 240:711–717; discussion 717–718. 2004.PubMed/NCBI
|
|
11
|
Renehan AG, Malcomson L, Emsley R, Gollins
S, Maw A, Myint AS, Rooney PS, Susnerwala S, Blower A, Saunders MP,
et al: Watch-and-wait approach versus surgical resection after
chemoradiotherapy for patients with rectal cancer (the OnCoRe
project): A propensity-score matched cohort analysis. Lancet Oncol.
17:174–183. 2016. View Article : Google Scholar
|
|
12
|
Smith JD, Ruby JA, Goodman KA, Saltz LB,
Guillem JG, Weiser MR, Temple LK, Nash GM and Paty PB: Nonoperative
management of rectal cancer with complete clinical response after
neoadjuvant therapy. Ann Surg. 256:965–972. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Janjan NA, Khoo VS, Abbruzzese J, Pazdur
R, Dubrow R, Cleary KR, Allen PK, Lynch PM, Glober G, Wolff R, et
al: Tumor downstaging and sphincter preservation with preoperative
chemoradiation in locally advanced rectal cancer: The M. D.
Anderson Cancer Center experience. Int J Radiat Oncol Biol Phys.
44:1027–1038. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lièvre A, Bachet JB, Le Corre D, Boige V,
Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, et al:
KRAS mutation status is predictive of response to cetuximab therapy
in colorectal cancer. Cancer Res. 66:3992–3995. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luna-Pérez P, Segura J, Alvarado I,
Labastida S, Santiago-Payán H and Quintero A: Specific c-K-ras gene
mutations as a tumor-response marker in locally advanced rectal
cancer treated with preoperative chemoradiotherapy. Ann Surg Oncol.
7:727–731. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duldulao MP, Lee W, Nelson RA, Li W, Chen
Z, Kim J and Garcia-Aguilar J: Mutations in specific codons of the
KRAS oncogene are associated with variable resistance to
neoadjuvant chemoradiation therapy in patients with rectal
adenocarcinoma. Ann Surg Oncol. 20:2166–2171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Davies JM, Trembath D, Deal AM, Funkhouser
WK, Calvo BF, Finnegan T, Weck KE, Tepper JE and O'Neil BH:
Phospho-ERK and AKT status, but not KRAS mutation status, are
associated with outcomes in rectal cancer treated with
chemoradiotherapy. Radiat Oncol. 6:1142011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Clancy C, Burke JP and Coffey JC: KRAS
mutation does not predict the efficacy of neo-adjuvant
chemoradiotherapy in rectal cancer: A systematic review and
meta-analysis. Surg Oncol. 22:105–111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krishnan S and Chang GJ: KRAS mutations
and rectal cancer response to chemoradiation: Are we closer to
personalization of therapy? Ann Surg Oncol. 20:3359–3362. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ghadimi BM, Grade M, Difilippantonio MJ,
Varma S, Simon R, Montagna C, Füzesi L, Langer C, Becker H, Liersch
T, et al: Effectiveness of gene expression profiling for response
prediction of rectal adenocarcinomas to preoperative
chemoradiotherapy. J Clin Oncol. 23:1826–1838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Watanabe T, Komuro Y, Kiyomatsu T,
Kanazawa T, Kazama Y, Tanaka J, Tanaka T, Yamamoto Y, Shirane M,
Muto T, et al: Prediction of sensitivity of rectal cancer cells in
response to preoperative radiotherapy by DNA microarray analysis of
gene expression profiles. Cancer Res. 66:3370–3374. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Agostini M, Zangrando A, Pastrello C,
D'Angelo E, Romano G, Giovannoni R, Giordan M, Maretto I, Bedin C,
Zanon C, et al: A functional biological network centered on XRCC3:
A new possible marker of chemoradiotherapy resistance in rectal
cancer patients. Cancer Biol Ther. 16:1160–1171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Conde-Muíño R, Cuadros M, Zambudio N,
Segura-Jiménez I, Cano C and Palma P: Predictive biomarkers to
chemoradiation in locally advanced rectal cancer. BioMed Res Int.
2015:9214352015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Molinari C, Casadio V, Foca F, Zingaretti
C, Giannini M, Avanzolini A, Lucci E, Saragoni L, Passardi A,
Amadori D, et al: Gene methylation in rectal cancer: Predictive
marker of response to chemoradiotherapy? J Cell Physiol.
228:2343–2349. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang G, Li Z, Zhao Q, Zhu Y, Zhao C, Li X,
Ma Z, Li X and Zhang Y: LincRNA-p21 enhances the sensitivity of
radiotherapy for human colorectal cancer by targeting the
Wnt/β-catenin signaling pathway. Oncol Rep. 31:1839–1845.
2014.PubMed/NCBI
|
|
26
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: A new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
D'Angelo E, Vicentini C, Agostini M, Kiss
A, Baffa R, Scarpa A and Fassan M: MicroRNAs as tools and effectors
for patient treatment in gastrointestinal carcinogenesis. Curr Drug
Targets. 16:383–392. 2015. View Article : Google Scholar
|
|
30
|
Suárez J, Vera R, Balén E, Gómez M, Arias
F, Lera JM, Herrera J and Zazpe C: Pathologic response assessed by
Mandard grade is a better prognostic factor than down staging for
disease-free survival after preoperative radiochemotherapy for
advanced rectal cancer. Colorectal Dis. 10:563–568. 2008.
View Article : Google Scholar
|
|
31
|
Drebber U, Lay M, Wedemeyer I, Vallböhmer
D, Bollschweiler E, Brabender J, Mönig SP, Hölscher AH, Dienes HP
and Odenthal M: Altered levels of the onco-microRNA 21 and the
tumor-supressor microRNAs 143 and 145 in advanced rectal cancer
indicate successful neoadjuvant chemoradiotherapy. Int J Oncol.
39:409–415. 2011.PubMed/NCBI
|
|
32
|
Akao Y, Nakagawa Y and Naoe T: MicroRNAs
143 and 145 are possible common onco-microRNAs in human cancers.
Oncol Rep. 16:845–850. 2006.PubMed/NCBI
|
|
33
|
Bandrés E, Cubedo E, Agirre X, Malumbres
R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, et
al: Identification by real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lopes-Ramos CM, Habr-Gama A, Quevedo BS,
Felício NM, Bettoni F, Koyama FC, Asprino PF, Galante PA,
Gama-Rodrigues J, Camargo AA, et al: Overexpression of miR-21–5p as
a predictive marker for complete tumor regression to neoadjuvant
chemoradiotherapy in rectal cancer patients. BMC Med Genomics.
7:682014. View Article : Google Scholar
|
|
36
|
Caramés C, Cristóbal I, Moreno V, del
Puerto L, Moreno I, Rodriguez M, Marín JP, Correa AV, Hernández R,
Zenzola V, et al: MicroRNA-21 predicts response to preoperative
chemoradiotherapy in locally advanced rectal cancer. Int J
Colorectal Dis. 30:899–906. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
D'Angelo E, Fassan M, Maretto I,
Pucciarelli S, Zanon C, Digito M, Rugge M, Nitti D and Agostini M:
Serum miR-125b is a non-invasive predictive biomarker of the
pre-operative chemoradiotherapy responsiveness in patients with
rectal adenocarcinoma. Oncotarget. 7:28647–28657. 2016.PubMed/NCBI
|
|
38
|
Svoboda M, Sana J, Fabian P, Kocakova I,
Gombosova J, Nekvindova J, Radova L, Vyzula R and Slaby O: MicroRNA
expression profile associated with response to neoadjuvant
chemoradiotherapy in locally advanced rectal cancer patients.
Radiat Oncol. 7:1952012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hotchi M, Shimada M, Kurita N, Iwata T,
Sato H, Morimoto S, Yoshikawa K, Higashijima J and Miyatani T:
microRNA expression is able to predict response to
chemoradiotherapy in rectal cancer. Mol Clin Oncol. 1:137–142.
2013.PubMed/NCBI
|
|
40
|
Nakao T, Iwata T, Hotchi M, Yoshikawa K,
Higashijima J, Nishi M, Takasu C, Eto S, Teraoku H and Shimada M:
Prediction of response to preoperative chemoradiotherapy and
establishment of individualized therapy in advanced rectal cancer.
Oncol Rep. 34:1961–1967. 2015.PubMed/NCBI
|
|
41
|
Millino C, Maretto I, Pacchioni B, Digito
M, De Paoli A, Canzonieri V, D'Angelo E, Agostini M, Rizzolio F,
Giordano A, et al: Gene and microRNA expression are predictive of
tumor response in rectal adenocarcinoma patients treated with
preoperative chemoradiotherapy. J Cell Physiol. 232:426–435. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Della Vittoria Scarpati G, Falcetta F,
Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK,
D'Incalci M, De Placido S and Pepe S: A specific miRNA signature
correlates with complete pathological response to neoadjuvant
chemoradiotherapy in locally advanced rectal cancer. Int J Radiat
Oncol Biol Phys. 83:1113–1119. 2012. View Article : Google Scholar
|
|
43
|
Bandres E, Arias F, Guerrero D, Lopez I,
Gonzalez-Huarriz M, Gomez Dorronsoro ML, Montes M, Monzon F, Torrea
N and Pedro Armendariz P: Association between a specific miRNA
signature and pathological response to neoadjuvant
chemoradiotherapy (CRT) in locally advanced rectal cancer (LARC)
patients. J Clin Oncol. 30:e140572012.
|
|
44
|
Kheirelseid EA, Miller N, Chang KH, Curran
C, Hennessey E, Sheehan M, Newell J, Lemetre C, Balls G and Kerin
MJ: miRNA expressions in rectal cancer as predictors of response to
neoadjuvant chemoradiation therapy. Int J Colorectal Dis.
28:247–260. 2013. View Article : Google Scholar
|
|
45
|
Svoboda M, Izakovicova Holla L, Sefr R,
Vrtkova I, Kocakova I, Tichy B and Dvorak J: Micro-RNAs miR125b and
miR137 are frequently upregulated in response to capecitabine
chemoradiotherapy of rectal cancer. Int J Oncol. 33:541–547.
2008.PubMed/NCBI
|
|
46
|
Deng J, Lei W, Fu JC, Zhang L, Li JH and
Xiong JP: Targeting miR-21 enhances the sensitivity of human colon
cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys
Res Commun. 443:789–795. 2014. View Article : Google Scholar
|
|
47
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
48
|
Mima K, Nishihara R, Yang J, Dou R, Masugi
Y, Shi Y, da Silva A, Cao Y, Song M, Nowak J, et al: MicroRNA MIR21
(miR-21) and PTGS2 expression in colorectal cancer and patient
survival. Clin Cancer Res. 22:3841–3848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chang KH, Miller N, Kheirelseid EA,
Ingoldsby H, Hennessy E, Curran CE, Curran S, Smith MJ, Regan M,
McAnena OJ, et al: MicroRNA-21 and PDCD4 expression in colorectal
cancer. Eur J Surg Oncol. 37:597–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fassan M, Pizzi M, Giacomelli L, Mescoli
C, Ludwig K, Pucciarelli S and Rugge M: PDCD4 nuclear loss
inversely correlates with miR-21 levels in colon carcinogenesis.
Virchows Arch. 458:413–419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Allgayer H: Pdcd4, a colon cancer
prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol.
73:185–191. 2010. View Article : Google Scholar
|
|
52
|
Li T, Leong MH, Harms B, Kennedy G and
Chen L: MicroRNA-21 as a potential colon and rectal cancer
biomarker. World J Gastroenterol. 19:5615–5621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chang KH, Miller N, Kheirelseid EA,
Lemetre C, Ball GR, Smith MJ, Regan M, McAnena OJ and Kerin MJ:
MicroRNA signature analysis in colorectal cancer: Identification of
expression profiles in stage II tumors associated with aggressive
disease. Int J Colorectal Dis. 26:1415–1422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dou X, Wang RB, Meng XJ, Yan HJ, Jiang SM,
Zhu KL, Xu XQ, Chen D, Song XR and Mu DB: PDCD4 as a predictor of
sensitivity to neoadjuvant chemoradiotherapy in locally advanced
rectal cancer patients. Asian Pac J Cancer Prev. 15:825–830. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kohwi-Shigematsu T, Poterlowicz K,
Ordinario E, Han HJ, Botchkarev VA and Kohwi Y: Genome organizing
function of SATB1 in tumor progression. Semin Cancer Biol.
23:72–79. 2013. View Article : Google Scholar
|
|
56
|
Kowalczyk AE, Krazinski BE, Godlewski J,
Grzegrzolka J, Kiewisz J, Kwiatkowski P, Sliwinska-Jewsiewicka A,
Dziegiel P and Kmiec Z: SATB1 is downregulated in clear cell renal
cell carcinoma and correlates with miR-21–5p overexpression and
poor prognosis. Cancer Genomics Proteomics. 13:209–217.
2016.PubMed/NCBI
|
|
57
|
Wang P, Zou F, Zhang X, Li H, Dulak A,
Tomko RJ Jr, Lazo JS, Wang Z, Zhang L and Yu J: microRNA-21
negatively regulates Cdc25A and cell cycle progression in colon
cancer cells. Cancer Res. 69:8157–8165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Salendo J, Spitzner M, Kramer F, Zhang X,
Jo P, Wolff HA, Kitz J, Kaulfuß S, Beißbarth T, Dobbelstein M, et
al: Identification of a microRNA expression signature for
chemoradiosensitivity of colorectal cancer cells, involving
miRNAs-320a, -224, -132 and let7g. Radiother Oncol. 108:451–457.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sklar MD: The ras oncogenes increase the
intrinsic resistance of NIH 3T3 cells to ionizing radiation.
Science. 239:645–647. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Weidhaas JB, Eisenmann DM, Holub JM and
Nallur SV: A conserved RAS/mitogen-activated protein kinase pathway
regulates DNA damage-induced cell death postirradiation in
Radelegans. Cancer Res. 66:10434–10438. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tong Z, Liu N, Lin L, Guo X, Yang D and
Zhang Q: miR-125a-5p inhibits cell proliferation and induces
apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1.
Biomed Pharmacother. 75:129–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xie B, Ding Q, Han H and Wu D: miRCancer:
A microRNA-cancer association database constructed by text mining
on literature. Bioinformatics. 29:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Nishida N, Yokobori T, Mimori K, Sudo T,
Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H and Mori M: MicroRNA
miR-125b is a prognostic marker in human colorectal cancer. Int J
Oncol. 38:1437–1443. 2011.PubMed/NCBI
|
|
65
|
Banzhaf-Strathmann J and Edbauer D: Good
guy or bad guy: The opposing roles of microRNA 125b in cancer. Cell
Commun Signal. 12:302014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mueller AC, Sun D and Dutta A: The miR-99
family regulates the DNA damage response through its target SNF2H.
Oncogene. 32:1164–1172. 2013. View Article : Google Scholar
|
|
67
|
Sun J, Chen Z, Tan X, Zhou F, Tan F, Gao
Y, Sun N, Xu X, Shao K and He J: MicroRNA-99a/100 promotes
apoptosis by targeting mTOR in human esophageal squamous cell
carcinoma. Med Oncol. 30:4112013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xu K, Liu P and Wei W: mTOR signaling in
tumorigenesis. Biochim Biophys Acta. 1846:638–654. 2014.PubMed/NCBI
|
|
69
|
Tokunaga C, Yoshino K and Yonezawa K: mTOR
integrates amino acid- and energy-sensing pathways. Biochem Biophys
Res Commun. 313:443–446. 2004. View Article : Google Scholar
|
|
70
|
Hay N and Sonenberg N: Upstream and
downstream of mTOR. Genes Dev. 18:1926–1945. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen D, Chen Z, Jin Y, Dragas D, Zhang L,
Adjei BS, Wang A, Dai Y and Zhou X: MicroRNA-99 family members
suppress Homeobox A1 expression in epithelial cells. PLoS One.
8:e806252013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I,
Wang C, Liu X and Zhou X: Downregulation of the microRNA-99 family
members in head and neck squamous cell carcinoma. Oral Oncol.
48:686–691. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang Y, Yu J, Liu H, Ma W, Yan L, Wang J
and Li G: Novel epigenetic CREB-miR-630 signaling axis regulates
radiosensitivity in colorectal cancer. PLoS One. 10:e01338702015.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sugatani T and Hruska KA: MicroRNA-223 is
a key factor in osteoclast differentiation. J Cell Biochem.
101:996–999. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fazi F, Rosa A, Fatica A, Gelmetti V, De
Marchis ML, Nervi C and Bozzoni I: A minicircuitry comprised of
microRNA-223 and transcription factors NFI-A and C/EBPalpha
regulates human granulopoiesis. Cell. 123:819–831. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wong QW, Lung RW, Law PT, Lai PB, Chan KY,
To KF and Wong N: MicroRNA-223 is commonly repressed in
hepatocellular carcinoma and potentiates expression of Stathmin1.
Gastroenterology. 135:257–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Rubin CI and Atweh GF: The role of
stathmin in the regulation of the cell cycle. J Cell Biochem.
93:242–250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ghosh R, Gu G, Tillman E, Yuan J, Wang Y,
Fazli L, Rennie PS and Kasper S: Increased expression and
differential phosphorylation of stathmin may promote prostate
cancer progression. Prostate. 67:1038–1052. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Saal LH, Johansson P, Holm K,
Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA,
Malmström P, Memeo L, et al: Poor prognosis in carcinoma is
associated with a gene expression signature of aberrant PTEN tumor
suppressor pathway activity. Proc Natl Acad Sci USA. 104:7564–7569.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Alli E, Yang JM, Ford JM and Hait WN:
Reversal of stathmin-mediated resistance to paclitaxel and
vinblastine in human breast carcinoma cells. Mol Pharmacol.
71:1233–1240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Völler D, Linck L, Bruckmann A, Hauptmann
J, Deutzmann R, Meister G and Bosserhoff AK: Argonaute family
protein expression in normal tissue and cancer entities. PLoS One.
11:e01611652016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sexl V, Diehl JA, Sherr CJ, Ashmun R,
Beach D and Roussel MF: A rate limiting function of cdc25A for S
phase entry inversely correlates with tyrosine dephosphorylation of
Cdk2. Oncogene. 18:573–582. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Shen T and Huang S: The role of Cdc25A in
the regulation of cell proliferation and apoptosis. Anticancer
Agents Med Chem. 12:631–639. 2012. View Article : Google Scholar : PubMed/NCBI
|