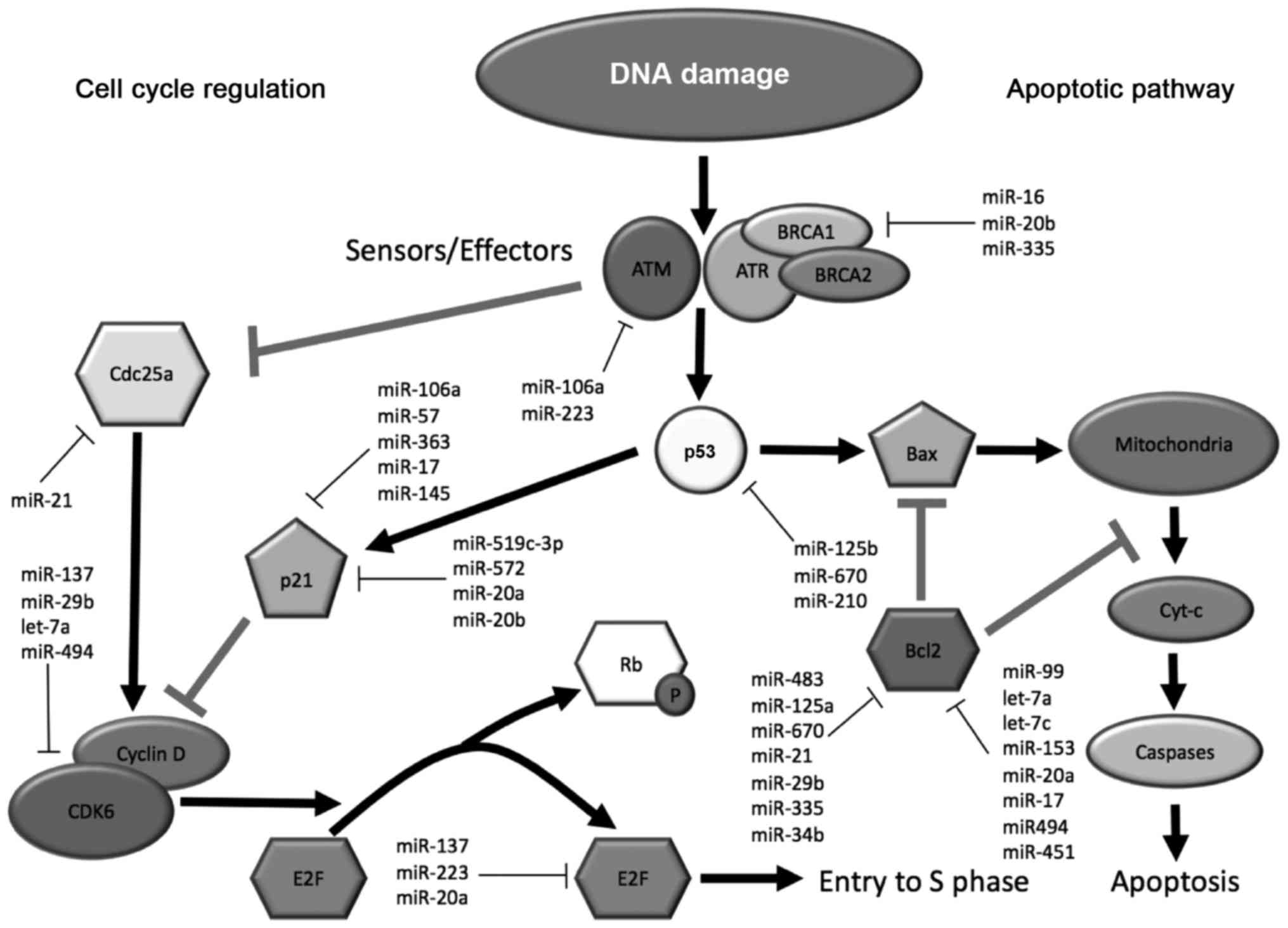
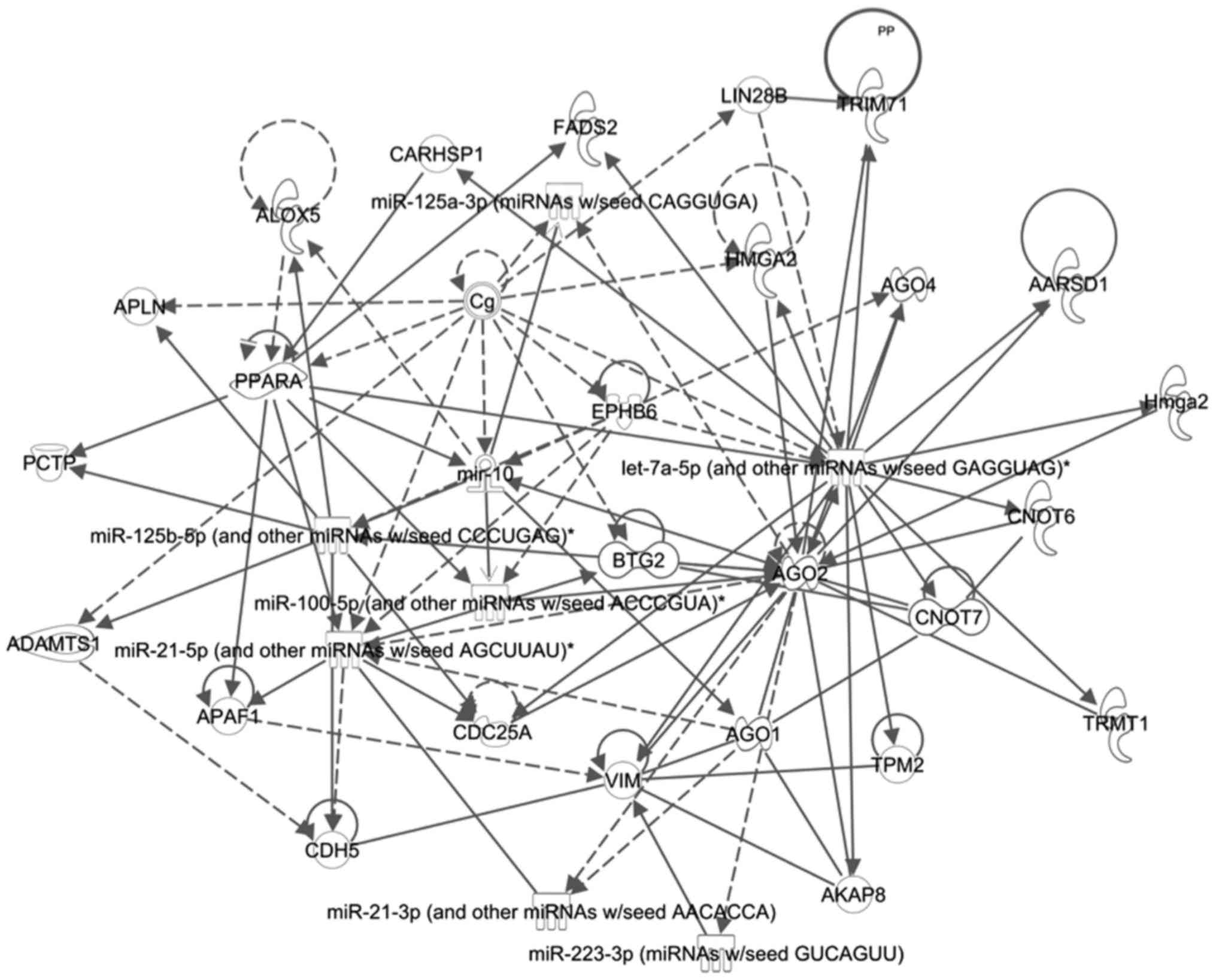
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sauer R, Liersch T, Merkel S, Fietkau R,
Hohenberger W, Hess C, Becker H, Raab HR, Villanueva MT, Witzigmann
H, et al: Preoperative versus postoperative chemoradiotherapy for
locally advanced rectal cancer: Results of the German
CAO/ARO/AIO-94 randomized phase III trial after a median follow-up
of 11 years. J Clin Oncol. 30:1926–1933. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roh MS, Colangelo LH, O'Connell MJ,
Yothers G, Deutsch M, Allegra CJ, Kahlenberg MS, Baez-Diaz L,
Ursiny CS, Petrelli NJ, et al: Preoperative multimodality therapy
improves disease-free survival in patients with carcinoma of the
rectum: NSABP R-03. J Clin Oncol. 27:5124–5130. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luna-Pérez P, Rodríguez-Ramírez S,
Hernández-Pacheco F, Gutiérrez De La Barrera M, Fernández R and
Labastida S: Anal sphincter preservation in locally advanced low
rectal adenocarcinoma after preoperative chemoradiation therapy and
coloanal anastomosis. J Surg Oncol. 82:3–9. 2003. View Article : Google Scholar
|
|
5
|
Hiotis SP, Weber SM, Cohen AM, Minsky BD,
Paty PB, Guillem JG, Wagman R, Saltz LB and Wong WD: Assessing the
predictive value of clinical complete response to neoadjuvant
therapy for rectal cancer: An analysis of 488 patients. J Am Coll
Surg. 194:131–135; discussion 135–136. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Habr-Gama A, de Souza PM, Ribeiro U Jr,
Nadalin W, Gansl R, Sousa AH Jr, Campos FG and Gama-Rodrigues J:
Low rectal cancer: Impact of radiation and chemotherapy on surgical
treatment. Dis Colon Rectum. 41:1087–1096. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Medich D, McGinty J, Parda D, Karlovits S,
Davis C, Caushaj P and Lembersky B: Preoperative chemoradiotherapy
and radical surgery for locally advanced distal rectal
adenocarcinoma: Pathologic findings and clinical implications. Dis
Colon Rectum. 44:1123–1128. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grann A, Minsky BD, Cohen AM, Saltz L,
Guillem JG, Paty PB, Kelsen DP, Kemeny N, Ilson D and Bass-Loeb J:
Preliminary results of preoperative 5-fluorouracil, low-dose
leucovorin, and concurrent radiation therapy for clinically
resectable T3 rectal cancer. Dis Colon Rectum. 40:515–522. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Habr-Gama A, Gama-Rodrigues J, São Julião
GP, Proscurshim I, Sabbagh C, Lynn PB and Perez RO: Local
recurrence after complete clinical response and watch and wait in
rectal cancer after neoadjuvant chemoradiation: Impact of salvage
therapy on local disease control. Int J Radiat Oncol Biol Phys.
88:822–828. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Habr-Gama A, Perez RO, Nadalin W, Sabbaga
J, Ribeiro U Jr, Silva e Sousa AH Jr, Campos FG, Kiss DR and
Gama-Rodrigues J: Operative versus nonoperative treatment for stage
0 distal rectal cancer following chemoradiation therapy: Long-term
results. Ann Surg. 240:711–717; discussion 717–718. 2004.PubMed/NCBI
|
|
11
|
Renehan AG, Malcomson L, Emsley R, Gollins
S, Maw A, Myint AS, Rooney PS, Susnerwala S, Blower A, Saunders MP,
et al: Watch-and-wait approach versus surgical resection after
chemoradiotherapy for patients with rectal cancer (the OnCoRe
project): A propensity-score matched cohort analysis. Lancet Oncol.
17:174–183. 2016. View Article : Google Scholar
|
|
12
|
Smith JD, Ruby JA, Goodman KA, Saltz LB,
Guillem JG, Weiser MR, Temple LK, Nash GM and Paty PB: Nonoperative
management of rectal cancer with complete clinical response after
neoadjuvant therapy. Ann Surg. 256:965–972. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Janjan NA, Khoo VS, Abbruzzese J, Pazdur
R, Dubrow R, Cleary KR, Allen PK, Lynch PM, Glober G, Wolff R, et
al: Tumor downstaging and sphincter preservation with preoperative
chemoradiation in locally advanced rectal cancer: The M. D.
Anderson Cancer Center experience. Int J Radiat Oncol Biol Phys.
44:1027–1038. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lièvre A, Bachet JB, Le Corre D, Boige V,
Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M, et al:
KRAS mutation status is predictive of response to cetuximab therapy
in colorectal cancer. Cancer Res. 66:3992–3995. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luna-Pérez P, Segura J, Alvarado I,
Labastida S, Santiago-Payán H and Quintero A: Specific c-K-ras gene
mutations as a tumor-response marker in locally advanced rectal
cancer treated with preoperative chemoradiotherapy. Ann Surg Oncol.
7:727–731. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duldulao MP, Lee W, Nelson RA, Li W, Chen
Z, Kim J and Garcia-Aguilar J: Mutations in specific codons of the
KRAS oncogene are associated with variable resistance to
neoadjuvant chemoradiation therapy in patients with rectal
adenocarcinoma. Ann Surg Oncol. 20:2166–2171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Davies JM, Trembath D, Deal AM, Funkhouser
WK, Calvo BF, Finnegan T, Weck KE, Tepper JE and O'Neil BH:
Phospho-ERK and AKT status, but not KRAS mutation status, are
associated with outcomes in rectal cancer treated with
chemoradiotherapy. Radiat Oncol. 6:1142011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Clancy C, Burke JP and Coffey JC: KRAS
mutation does not predict the efficacy of neo-adjuvant
chemoradiotherapy in rectal cancer: A systematic review and
meta-analysis. Surg Oncol. 22:105–111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krishnan S and Chang GJ: KRAS mutations
and rectal cancer response to chemoradiation: Are we closer to
personalization of therapy? Ann Surg Oncol. 20:3359–3362. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ghadimi BM, Grade M, Difilippantonio MJ,
Varma S, Simon R, Montagna C, Füzesi L, Langer C, Becker H, Liersch
T, et al: Effectiveness of gene expression profiling for response
prediction of rectal adenocarcinomas to preoperative
chemoradiotherapy. J Clin Oncol. 23:1826–1838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Watanabe T, Komuro Y, Kiyomatsu T,
Kanazawa T, Kazama Y, Tanaka J, Tanaka T, Yamamoto Y, Shirane M,
Muto T, et al: Prediction of sensitivity of rectal cancer cells in
response to preoperative radiotherapy by DNA microarray analysis of
gene expression profiles. Cancer Res. 66:3370–3374. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Agostini M, Zangrando A, Pastrello C,
D'Angelo E, Romano G, Giovannoni R, Giordan M, Maretto I, Bedin C,
Zanon C, et al: A functional biological network centered on XRCC3:
A new possible marker of chemoradiotherapy resistance in rectal
cancer patients. Cancer Biol Ther. 16:1160–1171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Conde-Muíño R, Cuadros M, Zambudio N,
Segura-Jiménez I, Cano C and Palma P: Predictive biomarkers to
chemoradiation in locally advanced rectal cancer. BioMed Res Int.
2015:9214352015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Molinari C, Casadio V, Foca F, Zingaretti
C, Giannini M, Avanzolini A, Lucci E, Saragoni L, Passardi A,
Amadori D, et al: Gene methylation in rectal cancer: Predictive
marker of response to chemoradiotherapy? J Cell Physiol.
228:2343–2349. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang G, Li Z, Zhao Q, Zhu Y, Zhao C, Li X,
Ma Z, Li X and Zhang Y: LincRNA-p21 enhances the sensitivity of
radiotherapy for human colorectal cancer by targeting the
Wnt/β-catenin signaling pathway. Oncol Rep. 31:1839–1845.
2014.PubMed/NCBI
|
|
26
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: A new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
D'Angelo E, Vicentini C, Agostini M, Kiss
A, Baffa R, Scarpa A and Fassan M: MicroRNAs as tools and effectors
for patient treatment in gastrointestinal carcinogenesis. Curr Drug
Targets. 16:383–392. 2015. View Article : Google Scholar
|
|
30
|
Suárez J, Vera R, Balén E, Gómez M, Arias
F, Lera JM, Herrera J and Zazpe C: Pathologic response assessed by
Mandard grade is a better prognostic factor than down staging for
disease-free survival after preoperative radiochemotherapy for
advanced rectal cancer. Colorectal Dis. 10:563–568. 2008.
View Article : Google Scholar
|
|
31
|
Drebber U, Lay M, Wedemeyer I, Vallböhmer
D, Bollschweiler E, Brabender J, Mönig SP, Hölscher AH, Dienes HP
and Odenthal M: Altered levels of the onco-microRNA 21 and the
tumor-supressor microRNAs 143 and 145 in advanced rectal cancer
indicate successful neoadjuvant chemoradiotherapy. Int J Oncol.
39:409–415. 2011.PubMed/NCBI
|
|
32
|
Akao Y, Nakagawa Y and Naoe T: MicroRNAs
143 and 145 are possible common onco-microRNAs in human cancers.
Oncol Rep. 16:845–850. 2006.PubMed/NCBI
|
|
33
|
Bandrés E, Cubedo E, Agirre X, Malumbres
R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, et
al: Identification by real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lopes-Ramos CM, Habr-Gama A, Quevedo BS,
Felício NM, Bettoni F, Koyama FC, Asprino PF, Galante PA,
Gama-Rodrigues J, Camargo AA, et al: Overexpression of miR-21–5p as
a predictive marker for complete tumor regression to neoadjuvant
chemoradiotherapy in rectal cancer patients. BMC Med Genomics.
7:682014. View Article : Google Scholar
|
|
36
|
Caramés C, Cristóbal I, Moreno V, del
Puerto L, Moreno I, Rodriguez M, Marín JP, Correa AV, Hernández R,
Zenzola V, et al: MicroRNA-21 predicts response to preoperative
chemoradiotherapy in locally advanced rectal cancer. Int J
Colorectal Dis. 30:899–906. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
D'Angelo E, Fassan M, Maretto I,
Pucciarelli S, Zanon C, Digito M, Rugge M, Nitti D and Agostini M:
Serum miR-125b is a non-invasive predictive biomarker of the
pre-operative chemoradiotherapy responsiveness in patients with
rectal adenocarcinoma. Oncotarget. 7:28647–28657. 2016.PubMed/NCBI
|
|
38
|
Svoboda M, Sana J, Fabian P, Kocakova I,
Gombosova J, Nekvindova J, Radova L, Vyzula R and Slaby O: MicroRNA
expression profile associated with response to neoadjuvant
chemoradiotherapy in locally advanced rectal cancer patients.
Radiat Oncol. 7:1952012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hotchi M, Shimada M, Kurita N, Iwata T,
Sato H, Morimoto S, Yoshikawa K, Higashijima J and Miyatani T:
microRNA expression is able to predict response to
chemoradiotherapy in rectal cancer. Mol Clin Oncol. 1:137–142.
2013.PubMed/NCBI
|
|
40
|
Nakao T, Iwata T, Hotchi M, Yoshikawa K,
Higashijima J, Nishi M, Takasu C, Eto S, Teraoku H and Shimada M:
Prediction of response to preoperative chemoradiotherapy and
establishment of individualized therapy in advanced rectal cancer.
Oncol Rep. 34:1961–1967. 2015.PubMed/NCBI
|
|
41
|
Millino C, Maretto I, Pacchioni B, Digito
M, De Paoli A, Canzonieri V, D'Angelo E, Agostini M, Rizzolio F,
Giordano A, et al: Gene and microRNA expression are predictive of
tumor response in rectal adenocarcinoma patients treated with
preoperative chemoradiotherapy. J Cell Physiol. 232:426–435. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Della Vittoria Scarpati G, Falcetta F,
Carlomagno C, Ubezio P, Marchini S, De Stefano A, Singh VK,
D'Incalci M, De Placido S and Pepe S: A specific miRNA signature
correlates with complete pathological response to neoadjuvant
chemoradiotherapy in locally advanced rectal cancer. Int J Radiat
Oncol Biol Phys. 83:1113–1119. 2012. View Article : Google Scholar
|
|
43
|
Bandres E, Arias F, Guerrero D, Lopez I,
Gonzalez-Huarriz M, Gomez Dorronsoro ML, Montes M, Monzon F, Torrea
N and Pedro Armendariz P: Association between a specific miRNA
signature and pathological response to neoadjuvant
chemoradiotherapy (CRT) in locally advanced rectal cancer (LARC)
patients. J Clin Oncol. 30:e140572012.
|
|
44
|
Kheirelseid EA, Miller N, Chang KH, Curran
C, Hennessey E, Sheehan M, Newell J, Lemetre C, Balls G and Kerin
MJ: miRNA expressions in rectal cancer as predictors of response to
neoadjuvant chemoradiation therapy. Int J Colorectal Dis.
28:247–260. 2013. View Article : Google Scholar
|
|
45
|
Svoboda M, Izakovicova Holla L, Sefr R,
Vrtkova I, Kocakova I, Tichy B and Dvorak J: Micro-RNAs miR125b and
miR137 are frequently upregulated in response to capecitabine
chemoradiotherapy of rectal cancer. Int J Oncol. 33:541–547.
2008.PubMed/NCBI
|
|
46
|
Deng J, Lei W, Fu JC, Zhang L, Li JH and
Xiong JP: Targeting miR-21 enhances the sensitivity of human colon
cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys
Res Commun. 443:789–795. 2014. View Article : Google Scholar
|
|
47
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar
|
|
48
|
Mima K, Nishihara R, Yang J, Dou R, Masugi
Y, Shi Y, da Silva A, Cao Y, Song M, Nowak J, et al: MicroRNA MIR21
(miR-21) and PTGS2 expression in colorectal cancer and patient
survival. Clin Cancer Res. 22:3841–3848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chang KH, Miller N, Kheirelseid EA,
Ingoldsby H, Hennessy E, Curran CE, Curran S, Smith MJ, Regan M,
McAnena OJ, et al: MicroRNA-21 and PDCD4 expression in colorectal
cancer. Eur J Surg Oncol. 37:597–603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fassan M, Pizzi M, Giacomelli L, Mescoli
C, Ludwig K, Pucciarelli S and Rugge M: PDCD4 nuclear loss
inversely correlates with miR-21 levels in colon carcinogenesis.
Virchows Arch. 458:413–419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Allgayer H: Pdcd4, a colon cancer
prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol.
73:185–191. 2010. View Article : Google Scholar
|
|
52
|
Li T, Leong MH, Harms B, Kennedy G and
Chen L: MicroRNA-21 as a potential colon and rectal cancer
biomarker. World J Gastroenterol. 19:5615–5621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chang KH, Miller N, Kheirelseid EA,
Lemetre C, Ball GR, Smith MJ, Regan M, McAnena OJ and Kerin MJ:
MicroRNA signature analysis in colorectal cancer: Identification of
expression profiles in stage II tumors associated with aggressive
disease. Int J Colorectal Dis. 26:1415–1422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dou X, Wang RB, Meng XJ, Yan HJ, Jiang SM,
Zhu KL, Xu XQ, Chen D, Song XR and Mu DB: PDCD4 as a predictor of
sensitivity to neoadjuvant chemoradiotherapy in locally advanced
rectal cancer patients. Asian Pac J Cancer Prev. 15:825–830. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kohwi-Shigematsu T, Poterlowicz K,
Ordinario E, Han HJ, Botchkarev VA and Kohwi Y: Genome organizing
function of SATB1 in tumor progression. Semin Cancer Biol.
23:72–79. 2013. View Article : Google Scholar
|
|
56
|
Kowalczyk AE, Krazinski BE, Godlewski J,
Grzegrzolka J, Kiewisz J, Kwiatkowski P, Sliwinska-Jewsiewicka A,
Dziegiel P and Kmiec Z: SATB1 is downregulated in clear cell renal
cell carcinoma and correlates with miR-21–5p overexpression and
poor prognosis. Cancer Genomics Proteomics. 13:209–217.
2016.PubMed/NCBI
|
|
57
|
Wang P, Zou F, Zhang X, Li H, Dulak A,
Tomko RJ Jr, Lazo JS, Wang Z, Zhang L and Yu J: microRNA-21
negatively regulates Cdc25A and cell cycle progression in colon
cancer cells. Cancer Res. 69:8157–8165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Salendo J, Spitzner M, Kramer F, Zhang X,
Jo P, Wolff HA, Kitz J, Kaulfuß S, Beißbarth T, Dobbelstein M, et
al: Identification of a microRNA expression signature for
chemoradiosensitivity of colorectal cancer cells, involving
miRNAs-320a, -224, -132 and let7g. Radiother Oncol. 108:451–457.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sklar MD: The ras oncogenes increase the
intrinsic resistance of NIH 3T3 cells to ionizing radiation.
Science. 239:645–647. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Weidhaas JB, Eisenmann DM, Holub JM and
Nallur SV: A conserved RAS/mitogen-activated protein kinase pathway
regulates DNA damage-induced cell death postirradiation in
Radelegans. Cancer Res. 66:10434–10438. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tong Z, Liu N, Lin L, Guo X, Yang D and
Zhang Q: miR-125a-5p inhibits cell proliferation and induces
apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1.
Biomed Pharmacother. 75:129–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xie B, Ding Q, Han H and Wu D: miRCancer:
A microRNA-cancer association database constructed by text mining
on literature. Bioinformatics. 29:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Nishida N, Yokobori T, Mimori K, Sudo T,
Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H and Mori M: MicroRNA
miR-125b is a prognostic marker in human colorectal cancer. Int J
Oncol. 38:1437–1443. 2011.PubMed/NCBI
|
|
65
|
Banzhaf-Strathmann J and Edbauer D: Good
guy or bad guy: The opposing roles of microRNA 125b in cancer. Cell
Commun Signal. 12:302014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mueller AC, Sun D and Dutta A: The miR-99
family regulates the DNA damage response through its target SNF2H.
Oncogene. 32:1164–1172. 2013. View Article : Google Scholar
|
|
67
|
Sun J, Chen Z, Tan X, Zhou F, Tan F, Gao
Y, Sun N, Xu X, Shao K and He J: MicroRNA-99a/100 promotes
apoptosis by targeting mTOR in human esophageal squamous cell
carcinoma. Med Oncol. 30:4112013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Xu K, Liu P and Wei W: mTOR signaling in
tumorigenesis. Biochim Biophys Acta. 1846:638–654. 2014.PubMed/NCBI
|
|
69
|
Tokunaga C, Yoshino K and Yonezawa K: mTOR
integrates amino acid- and energy-sensing pathways. Biochem Biophys
Res Commun. 313:443–446. 2004. View Article : Google Scholar
|
|
70
|
Hay N and Sonenberg N: Upstream and
downstream of mTOR. Genes Dev. 18:1926–1945. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chen D, Chen Z, Jin Y, Dragas D, Zhang L,
Adjei BS, Wang A, Dai Y and Zhou X: MicroRNA-99 family members
suppress Homeobox A1 expression in epithelial cells. PLoS One.
8:e806252013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I,
Wang C, Liu X and Zhou X: Downregulation of the microRNA-99 family
members in head and neck squamous cell carcinoma. Oral Oncol.
48:686–691. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang Y, Yu J, Liu H, Ma W, Yan L, Wang J
and Li G: Novel epigenetic CREB-miR-630 signaling axis regulates
radiosensitivity in colorectal cancer. PLoS One. 10:e01338702015.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sugatani T and Hruska KA: MicroRNA-223 is
a key factor in osteoclast differentiation. J Cell Biochem.
101:996–999. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Fazi F, Rosa A, Fatica A, Gelmetti V, De
Marchis ML, Nervi C and Bozzoni I: A minicircuitry comprised of
microRNA-223 and transcription factors NFI-A and C/EBPalpha
regulates human granulopoiesis. Cell. 123:819–831. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wong QW, Lung RW, Law PT, Lai PB, Chan KY,
To KF and Wong N: MicroRNA-223 is commonly repressed in
hepatocellular carcinoma and potentiates expression of Stathmin1.
Gastroenterology. 135:257–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Rubin CI and Atweh GF: The role of
stathmin in the regulation of the cell cycle. J Cell Biochem.
93:242–250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ghosh R, Gu G, Tillman E, Yuan J, Wang Y,
Fazli L, Rennie PS and Kasper S: Increased expression and
differential phosphorylation of stathmin may promote prostate
cancer progression. Prostate. 67:1038–1052. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Saal LH, Johansson P, Holm K,
Gruvberger-Saal SK, She QB, Maurer M, Koujak S, Ferrando AA,
Malmström P, Memeo L, et al: Poor prognosis in carcinoma is
associated with a gene expression signature of aberrant PTEN tumor
suppressor pathway activity. Proc Natl Acad Sci USA. 104:7564–7569.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Alli E, Yang JM, Ford JM and Hait WN:
Reversal of stathmin-mediated resistance to paclitaxel and
vinblastine in human breast carcinoma cells. Mol Pharmacol.
71:1233–1240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Völler D, Linck L, Bruckmann A, Hauptmann
J, Deutzmann R, Meister G and Bosserhoff AK: Argonaute family
protein expression in normal tissue and cancer entities. PLoS One.
11:e01611652016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sexl V, Diehl JA, Sherr CJ, Ashmun R,
Beach D and Roussel MF: A rate limiting function of cdc25A for S
phase entry inversely correlates with tyrosine dephosphorylation of
Cdk2. Oncogene. 18:573–582. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Shen T and Huang S: The role of Cdc25A in
the regulation of cell proliferation and apoptosis. Anticancer
Agents Med Chem. 12:631–639. 2012. View Article : Google Scholar : PubMed/NCBI
|