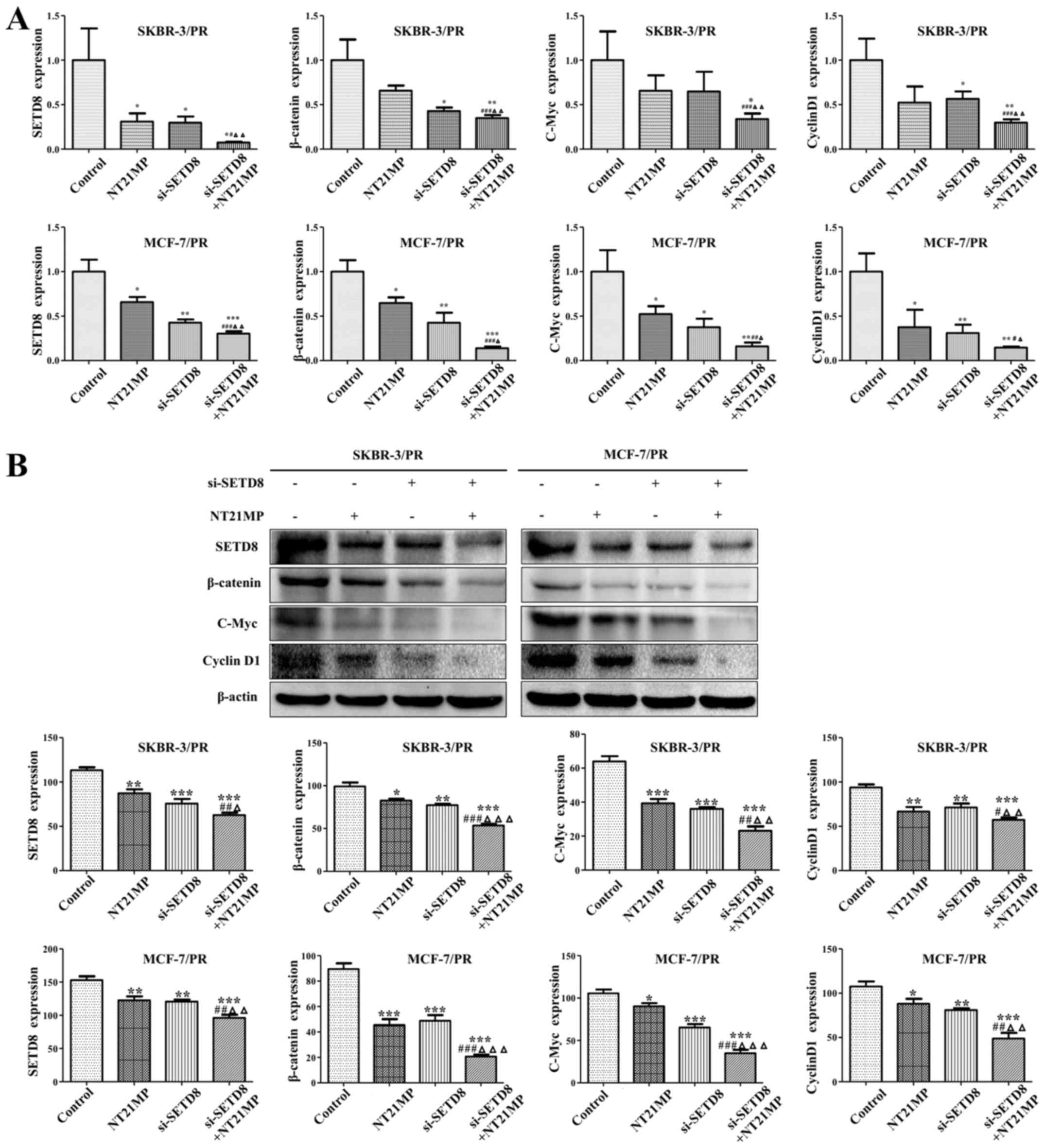
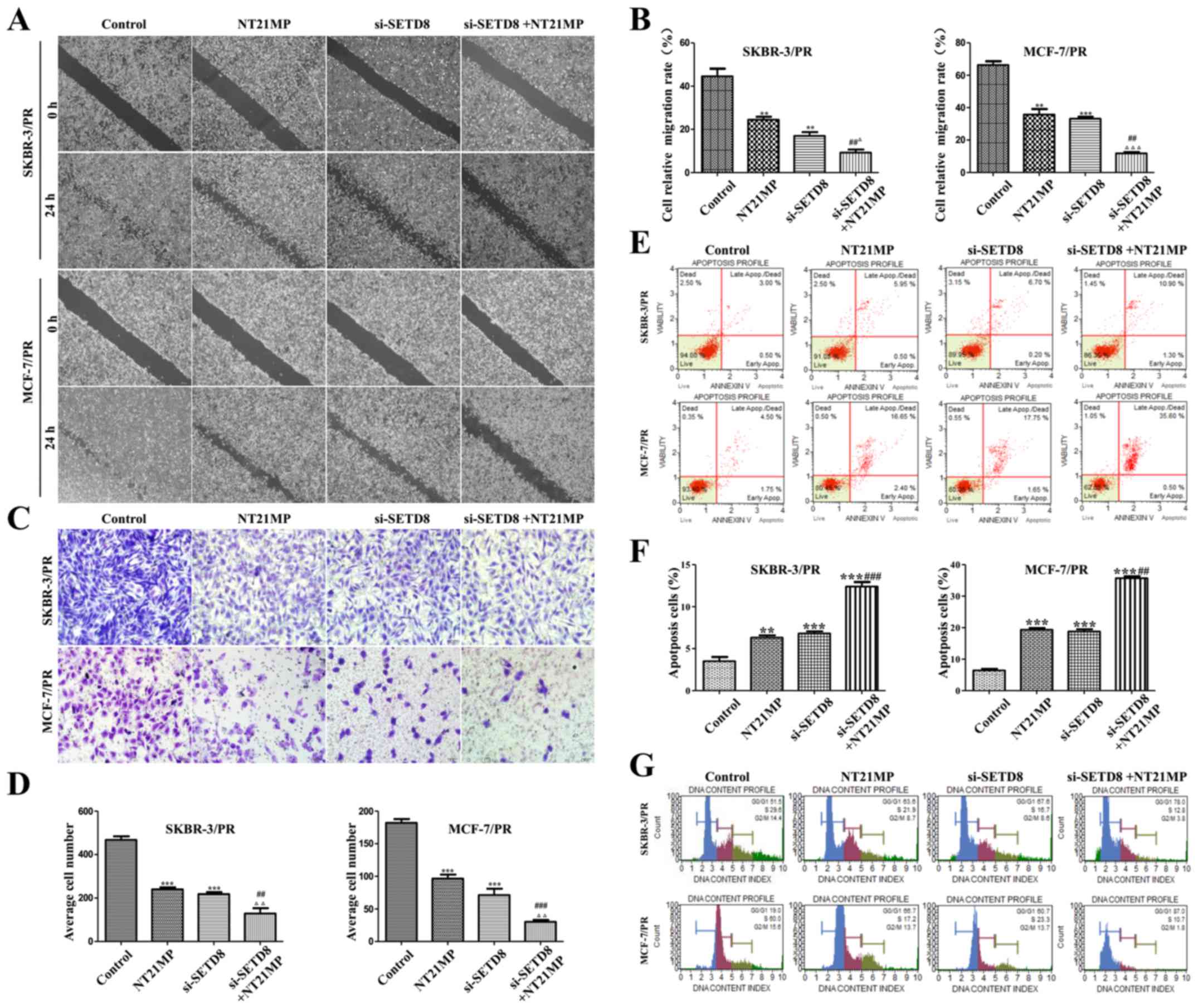
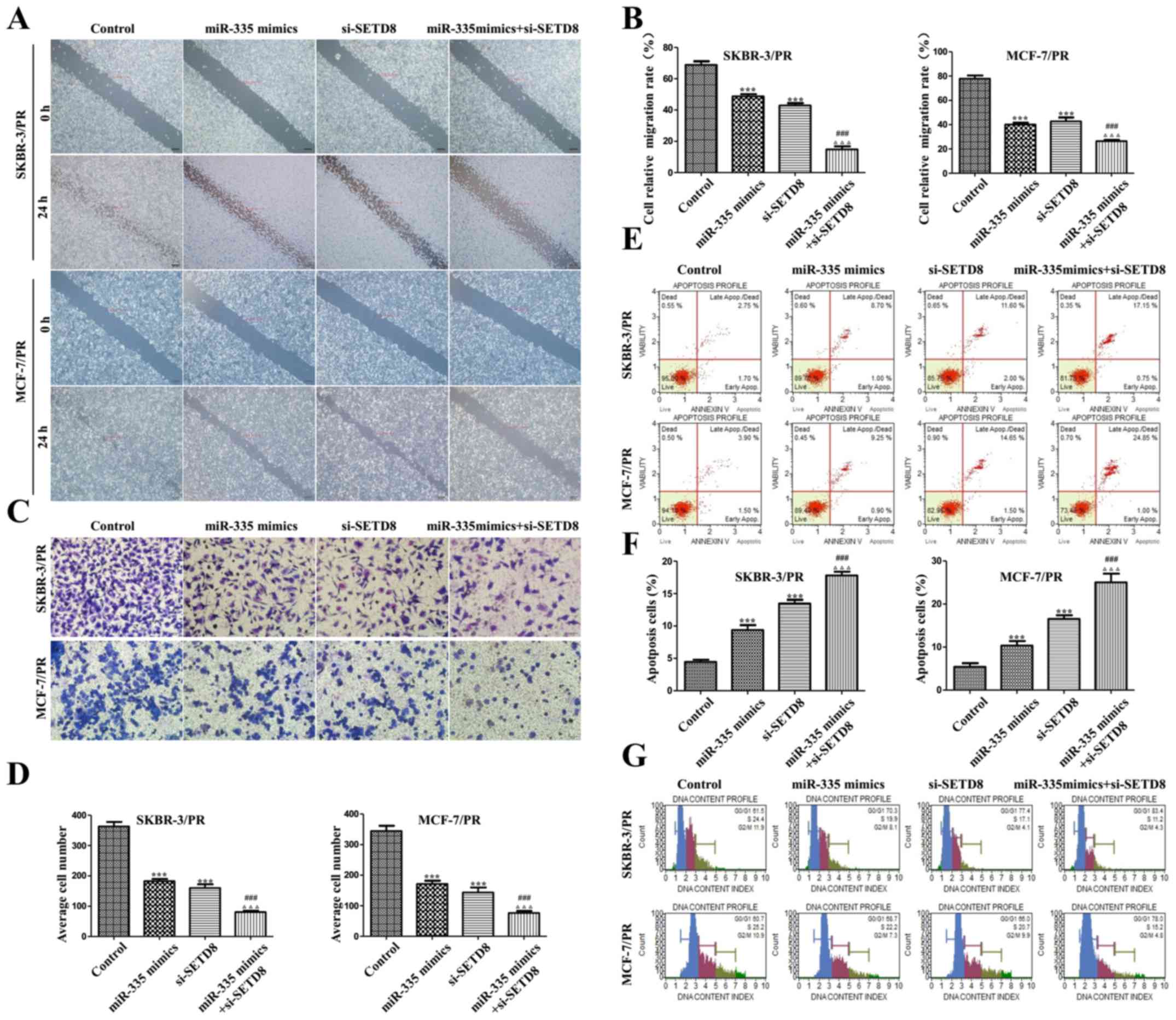
|
1
|
Rebecca L, Siegel MPH, Kimberly D and
Miller MPH: Issue Information. CA Cancer J Clin. 67:1–85. 2017.
View Article : Google Scholar
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zardavas D, Baselga J and Piccart M:
Emerging targeted agents in metastatic breast cancer. Nat Rev Clin
Oncol. 10:191–210. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang X, Jin L, Cao P, Cao K, Huang C, Luo
Y, Ma J, Shen S, Tan M, Li X, et al: MicroRNA-16 sensitizes breast
cancer cells to paclitaxel through suppression of IKBKB expression.
Oncotarget. 7:23668–23683. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Armat M, Oghabi Bakhshaiesh T, Sabzichi M,
Shanehbandi D, Sharifi S, Molavi O, Mohammadian J, Saeid Hejazi M
and Samadi N: The role of Six1 signaling in paclitaxel-dependent
apoptosis in MCF-7 cell line. Bosn J Basic Med Sci. 16:28–34.
2016.PubMed/NCBI
|
|
6
|
Mirnezami AH, Pickard K, Zhang L, Primrose
JN and Packhamå G: MicroRNAs: Key players in carcinogenesis and
novel therapeutic targets. Eur J Surg Oncol. 35:339–347. 2009.
View Article : Google Scholar
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Verghese ET, Hanby AM, Speirs V and Hughes
TA: Small is beautiful: microRNAs and breast cancer-where are we
now? J Pathol. 215:214–221. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu Y, Qiu Y, Yagüe E, Ji W, Liu J and
Zhang J: miRNA-205 targets VEGFA and FGF2 and regulates resistance
to chemotherapeutics in breast cancer. Cell Death Dis. 7:e22912016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu J, Zhou Q and Tan S: Targeting miRNAs
associated with surface expression of death receptors to modulate
TRAIL resistance in breast cancer. Cancer Lett. 383:154–160. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Raza U, Saatci Ö, Uhlmann S, Ansari SA,
Eyüpoğlu E, Yurdusev E, Mutlu M, Ersan PG, Altundağ MK, Zhang JD,
et al: The miR-644a/CTBP1/p53 axis suppresses drug resistance by
simultaneous inhibition of cell survival and epithelial-mesenchymal
transition in breast cancer. Oncotarget. 7:49859–49877. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu X, Lv YG, Yan CY, Yi J and Ling R:
Enforced expression of hsa-miR-125a-3p in breast cancer cells
potentiates docetaxel sensitivity via modulation of BRCA1
signaling. Biochem Biophys Res Commun. 479:893–900. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu Q, Gong JP, Li J, Zhong SL, Chen WX,
Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH, et al: Down-regulation of
miRNA-452 is associated with adriamycin-resistance in breast cancer
cells. Asian Pac J Cancer Prev. 15:5137–5142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Zhong S, Xu Y, Yu D, Ma T, Chen
L, Zhao Y, Chen X, Yang S, Wu Y, et al: MicroRNA-3646 contributes
to docetaxel resistance in human breast cancer cells by
GSK-3β/β-catenin signaling pathway. PLoS One. 11:e01531942016.
View Article : Google Scholar
|
|
16
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17–5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie
T, Zhang J, Peng C, Lin Y and Chen J: MicroRNA-25 regulates
chemoresistance-associated autophagy in breast cancer cells, a
process modulated by the natural autophagy inducer
isoliquiritigenin. Oncotarget. 5:7013–7026. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen H, Wang D, Li L, Yang S, Chen X, Zhou
S, Zhong S, Zhao J and Tang J: MiR-222 promotes drug-resistance of
breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1
pathway. Gene. 596:110–118. 2017. View Article : Google Scholar
|
|
19
|
Ward A, Balwierz A, Zhang JD, Küblbeck M,
Pawitan Y, Hielscher T, Wiemann S and Sahin Ö: Re-expression of
microRNA-375 reverses both tamoxifen resistance and accompanying
EMT-like properties in breast cancer. Oncogene. 32:1173–1182. 2013.
View Article : Google Scholar
|
|
20
|
Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang
B, Min L and Liu W: miRNA-200c increases the sensitivity of breast
cancer cells to doxorubicin through the suppression of
E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 7:1579–1584.
2013.PubMed/NCBI
|
|
21
|
Shen H, Li L, Yang S, Wang D, Zhong S,
Zhao J and Tang J: MicroRNA-29a contributes to drug-resistance of
breast cancer cells to adriamycin through PTEN/AKT/GSK3β signaling
pathway. Gene. 593:84–90. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gao M, Miao L, Liu M, Li C, Yu C, Yan H,
Yin Y, Wang Y, Qi X and Ren J: miR-145 sensitizes breast cancer to
doxorubicin by targeting multidrug resistance-associated protein-1.
Oncotarget. 7:59714–59726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang L, He D, Yang D, Chen Z, Pan Q, Mao
A, Cai Y, Li X, Xing H, Shi M, et al: MiR-489 regulates
chemoresistance in breast cancer via epithelial mesenchymal
transition pathway. FEBS Lett. 588:2009–2015. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Wang YW, Xing AY, Xiang S, Shi DB,
Liu L, Li YX and Gao P: Suppression of SPIN1-mediated PI3K-Akt
pathway by miR-489 increases chemosensitivity in breast cancer. J
Pathol. 239:459–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim Y, Kim H, Park D and Jeoung D: miR-335
targets SIAH2 and confers sensitivity to anti-cancer drugs by
increasing the expression of HDAC3. Mol Cells. 38:562–572. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang R, Lei Y, Hu B, Yang J, Fang S, Wang
Q, Li M and Guo L: WW domain binding protein 5 induces multidrug
resistance of small cell lung cancer under the regulation of
miR-335 through the Hippo pathway. Br J Cancer. 115:243–251. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang QL, Li CH, Ding YX, Chen CJ, Zhang J
and Wang H: Inhibitory effect of polypeptide to inhibit CXCR4 on
metastasis of breast cancer cell line. CTM. 20:89–92. 2008.
|
|
29
|
Yang QL, Ding YX, Chen CJ, Tang J, Zhang J
and Yang ZF: Suppression of murine breast cancer metastasis by
selective inhibition of CXCR4 by synthetic polypeptide derived from
viral macrophage inflammatory protein II. Chin Sci Bull.
55:2152–2159. 2010. View Article : Google Scholar
|
|
30
|
Yang Q, Huang J, Wu Q, Cai Y, Zhu L, Lu X,
Chen S, Chen C and Wang Z: Acquisition of epithelial-mesenchymal
transition is associated with Skp2 expression in
paclitaxel-resistant breast cancer cells. Br J Cancer.
110:1958–1967. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen S, Zhu L, Huang J, Cai Y, Lu X, Yang
Q, Wu Q, Chen C and Wang Z: Arsenic trioxide targets miR-125b in
glioma cells. Curr Pharm Des. 20:5354–5361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu Q, Wang R, Yang Q, Hou X, Chen S, Hou
Y, Chen C, Yang Y, Miele L, Sarkar FH, et al: Chemoresistance to
gemcitabine in hepatoma cells induces epithelial-mesenchymal
transition and involves activation of PDGF-D pathway. Oncotarget.
4:1999–2009. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Heidary MF, Mahmoodzadeh Hosseini H,
Mehdizadeh Aghdam E, Nourani MR, Ranjbar R, Mirnejad R and Imani
Fooladi AA: Overexpression of metastatic related microRNAs, mir-335
and mir-10b, by Staphylococcal enterotoxin B in the metastatic
breast cancer cell line. Adv Pharm Bull. 5:255–259. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang R, Dick M, Marme F, Schneeweiss A,
Langheinz A, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, Varon
R, et al: Genetic variants within miR-126 and miR-335 are not
associated with breast cancer risk. Breast Cancer Res Treat.
127:549–554. 2011. View Article : Google Scholar
|
|
35
|
Meng Y, Zou Q, Liu T, Cai X, Huang Y and
Pan J: microRNA-335 inhibits proliferation, cell-cycle progression,
colony formation, and invasion via targeting PAX6 in breast cancer
cells. Mol Med Rep. 11:379–385. 2015.
|
|
36
|
Gao Y, Zeng F, Wu JY, Li HY, Fan JJ, Mai
L, Zhang J, Ma DM, Li Y and Song FZ: MiR-335 inhibits migration of
breast cancer cells through targeting oncoprotein c-Met. Tumour
Biol. 36:2875–2883. 2015. View Article : Google Scholar
|
|
37
|
Negrini M and Calin GA: Breast cancer
metastasis: A microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tavazoie SF, Alarcón C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massagué J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Png KJ, Yoshida M, Zhang XH, Shu W, Lee H,
Rimner A, Chan TA, Comen E, Andrade VP, Kim SW, et al: MicroRNA-335
inhibits tumor reinitiation and is silenced through genetic and
epigenetic mechanisms in human breast cancer. Genes Dev.
25:226–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Erturk E, Cecener G, Egeli U, Tunca B,
Tezcan G, Gokgoz S, Tolunay S and Tasdelen I: Expression status of
let-7a and miR-335 among breast tumors in patients with and without
germ-line BRCA mutations. Mol Cell Biochem. 395:77–88. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nishioka K, Rice JC, Sarma K,
Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P,
Tempst P, Steward R, et al: PR-Set7 is a nucleosome-specific
methyltransferase that modifies lysine 20 of histone H4 and is
associated with silent chromatin. Mol Cell. 9:1201–1213. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fang J, Feng Q, Ketel CS, Wang H, Cao R,
Xia L, Erdjument-Bromage H, Tempst P, Simon JA and Zhang Y:
Purification and functional characterization of SET8, a nucleosomal
histone H4-lysine 20-specific methyltransferase. Curr Biol.
12:1086–1099. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Beck DB, Oda H, Shen SS and Reinberg D:
PR-Set7 and H4K20me1: At the crossroads of genome integrity, cell
cycle, chromosome condensation, and transcription. Genes Dev.
26:325–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
DeVilbiss AW, Sanalkumar R, Hall BD,
Katsumura KR, de Andrade IF and Bresnick EH: Epigenetic
determinants of erythropoiesis: Role of the histone
methyltransferase SetD8 in promoting erythroid cell maturation and
survival. Mol Cell Biol. 35:2073–2087. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Malik J, Getman M and Steiner LA: Histone
methyltransferase Setd8 represses Gata2 expression and regulates
erythroid maturation. Mol Cell Biol. 35:2059–2072. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang J, Hou W, Chai M, Zhao H, Jia J, Sun
X, Zhao B and Wang R: MicroRNA-127-3p inhibits proliferation and
invasion by targeting SETD8 in human osteosarcoma cells. Biochem
Biophys Res Commun. 469:1006–1011. 2016. View Article : Google Scholar
|
|
47
|
Shi XL, Guo ZJ, Wang XL, Liu XL and Shi
GF: SET8 expression is associated with overall survival in gastric
cancer. Genet Mol Res. 14:15609–15615. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Song F, Zheng H, Liu B, Wei S, Dai H,
Zhang L, Calin GA, Hao X, Wei Q, Zhang W, et al: An miR-502-binding
site single-nucleotide polymorphism in the 3′-untranslated region
of the SET8 gene is associated with early age of breast cancer
onset. Clin Cancer Res. 15:6292–6300. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu N, Huangyang P, Yang X, Han X, Yan R,
Jia H, Shang Y and Sun L: microRNA-7 suppresses the invasive
potential of breast cancer cells and sensitizes cells to DNA
damages by targeting histone methyltransferase SET8. J Biol Chem.
288:19633–19642. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang F, Sun L, Li Q, Han X, Lei L, Zhang H
and Shang Y: SET8 promotes epithelial-mesenchymal transition and
confers TWIST dual transcriptional activities. EMBO J. 31:110–123.
2012. View Article : Google Scholar
|
|
51
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Schotta G: H4K20 monomethylation faces the
WNT. Proc Natl Acad Sci USA. 108:3097–3098. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li Z, Nie F, Wang S and Li L: Histone H4
Lys 20 monomethylation by histone methylase SET8 mediates Wnt
target gene activation. Proc Natl Acad Sci USA. 108:3116–3123.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ke X, Xing B, Yu B, Yu X, Majnik A, Cohen
S, Lane R and Joss-Moore L: IUGR disrupts the
PPARγ-Setd8-H4K20me(1) and Wnt signaling pathways in the juvenile
rat hippocampus. Int J Dev Neurosci. 38:59–67. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tamamori-Adachi M, Ito H,
Sumrejkanchanakij P, Adachi S, Hiroe M, Shimizu M, Kawauchi J,
Sunamori M, Marumo F, Kitajima S, et al: Critical role of cyclin D1
nuclear import in cardiomyocyte proliferation. Circ Res.
92:e12–e19. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Boonmuen N, Thongon N, Chairoungdua A,
Suksen K, Pompimon W, Tuchinda P, Reutrakul V and Piyachaturawat P:
5-Acetyl goniothalamin suppresses proliferation of breast cancer
cells via Wnt/β-catenin signaling. Eur J Pharmacol. 791:455–464.
2016. View Article : Google Scholar : PubMed/NCBI
|