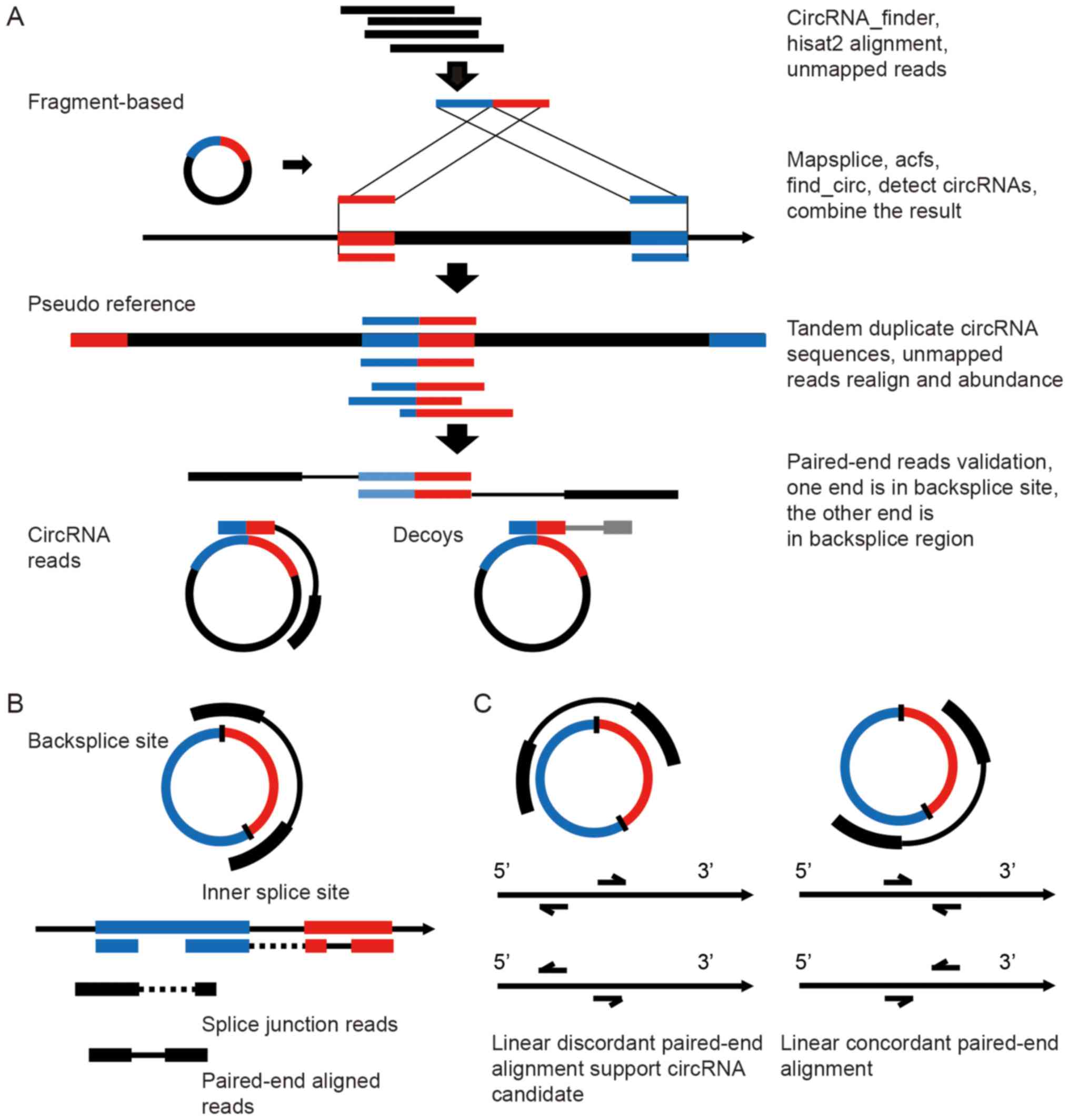
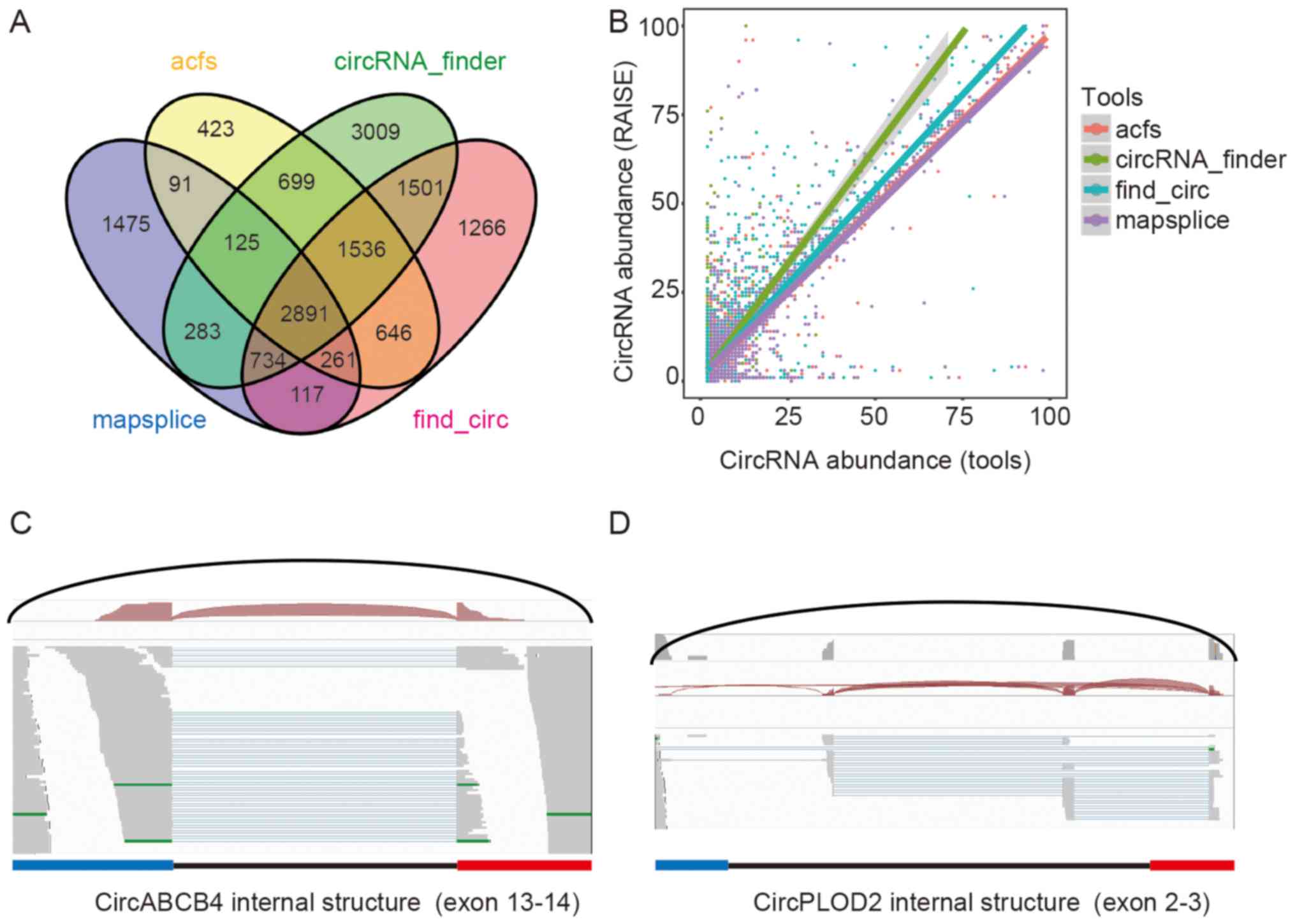
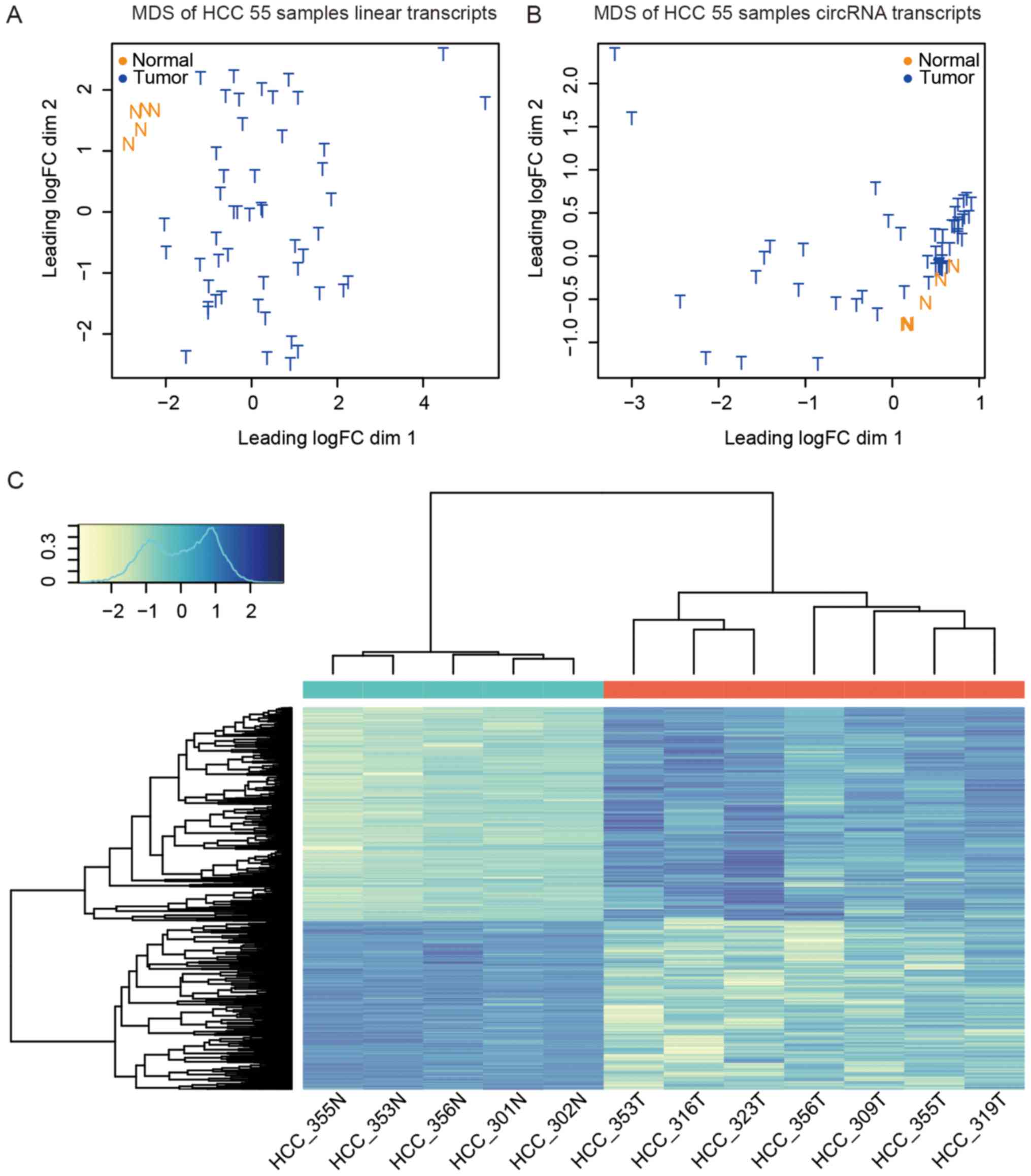
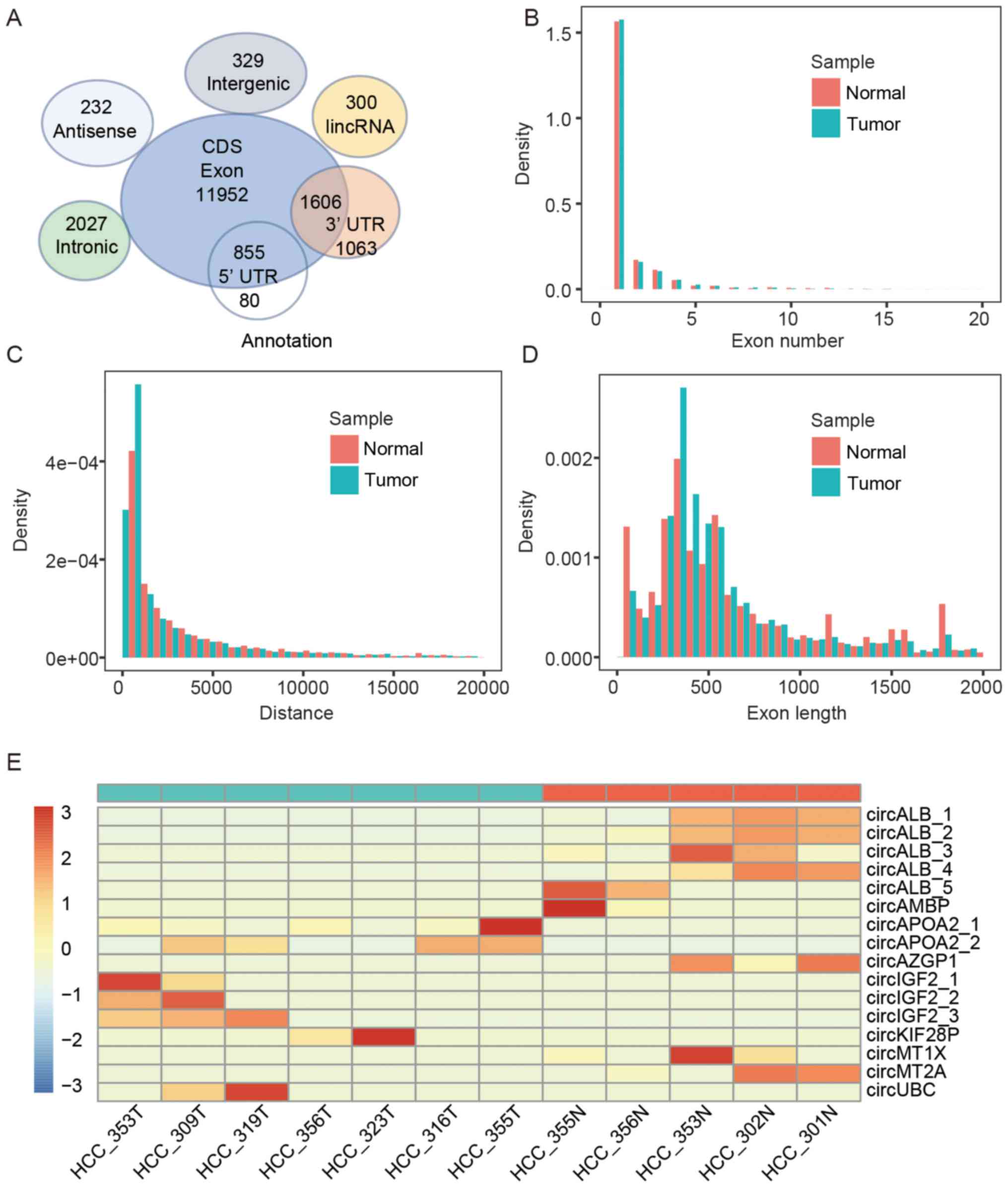
|
1
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One. 7(e30733):
e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
3
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C, et al: Analysis of intron sequences reveals hallmarks
of circular RNA biogenesis in animals. Cell Rep. 10:170–177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kelly S, Greenman C, Cook PR and
Papantonis A: Exon skipping is correlated with exon
circularization. J Mol Biol. 427:2414–2417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Barrett SP, Wang PL and Salzman J:
Circular RNA biogenesis can proceed through an exon-containing
lariat precursor. eLife. 4:e075402015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Starke S, Jost I, Rossbach O, Schneider T,
Schreiner S, Hung LH and Bindereif A: Exon circularization requires
canonical splice signals. Cell Rep. 10:103–111. 2015. View Article : Google Scholar
|
|
13
|
Szabo L, Morey R, Palpant NJ, Wang PL,
Afari N, Jiang C, Parast MM, Murry CE, Laurent LC and Salzman J:
Statistically based splicing detection reveals neural enrichment
and tissue-specific induction of circular RNA during human fetal
development. Genome Biol. 16:1262015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahmed I, Karedath T, Andrews SS, Al-Azwani
IK, Mohamoud YA, Querleu D, Rafii A and Malek JA: Altered
expression pattern of circular RNAs in primary and metastatic sites
of epithelial ovarian carcinoma. Oncotarget. 7:36366–36381. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang K, Singh D, Zeng Z, Coleman SJ, Huang
Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al:
MapSplice: Accurate mapping of RNA-seq reads for splice junction
discovery. Nucleic Acids Res. 38:e1782010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hoffmann S, Otto C, Kurtz S, Sharma CM,
Khaitovich P, Vogel J, Stadler PF and Hackermüller J: Fast mapping
of short sequences with mismatches, insertions and deletions using
index structures. PLOS Comput Biol. 5:e10005022009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Westholm JO, Miura P, Olson S, Shenker S,
Joseph B, Sanfilippo P, Celniker SE, Graveley BR and Lai EC:
Genome-wide analysis of drosophila circular RNAs reveals their
structural and sequence properties and age-dependent neural
accumulation. Cell Rep. 9:1966–1980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao Y, Wang J and Zhao F: CIRI: An
efficient and unbiased algorithm for de novo circular RNA
identification. Genome Biol. 16:42015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
You X and Conrad TO: Acfs: Accurate
circRNA identification and quantification from RNA-Seq data. Sci
Rep. 6:388202016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng J, Metge F and Dieterich C: Specific
identification and quantification of circular RNAs from sequencing
data. Bioinformatics. 32:1094–1096. 2016. View Article : Google Scholar
|
|
22
|
Szabo L and Salzman J: Detecting circular
RNAs: Bioinformatic and experimental challenges. Nat Rev Genet.
17:679–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rybak-Wolf A, Stottmeister C, Glažar P,
Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss
R, et al: Circular RNAs in the mammalian brain are highly abundant,
conserved, and dynamically expressed. Mol Cell. 58:870–885. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Venø MT, Hansen TB, Venø ST, Clausen BH,
Grebing M, Finsen B, Holm IE and Kjems J: Spatiotemporal regulation
of circular RNA expression during porcine embryonic brain
development. Genome Biol. 16:2452015. View Article : Google Scholar
|
|
26
|
Tan WL, Lim BT, Anene-Nzelu CG,
Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD,
et al: A landscape of circular RNA expression in the human heart.
Cardiovasc Res. 113:298–309. 2017.PubMed/NCBI
|
|
27
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alnasir J and Shanahan HP: Investigation
into the annotation of protocol sequencing steps in the sequence
read archive. Gigascience. 4:232015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Birney E, Stamatoyannopoulos JA, Dutta A,
Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis
ET, Thurman RE, et al: Children's Hospital Oakland Research
Institute: Identification and analysis of functional elements in 1%
of the human genome by the ENCODE pilot project. Nature.
447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dong H, Zhang L, Qian Z, Zhu X, Zhu G,
Chen Y, Xie X, Ye Q, Zang J, Ren Z, et al: Identification of
HBV-MLL4 integration and its molecular basis in Chinese
hepatocellular carcinoma. PLoS One. 10:e01231752015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kornienko AE, Dotter CP, Guenzl PM,
Gisslinger H, Gisslinger B, Cleary C, Kralovics R, Pauler FM and
Barlow DP: Long non-coding RNAs display higher natural expression
variation than protein-coding genes in healthy humans. Genome Biol.
17:142016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Scheckel C, Drapeau E, Frias MA, Park CY,
Fak J, Zucker-Scharff I, Kou Y, Haroutunian V, Ma'ayan A, Buxbaum
JD, et al: Regulatory consequences of neuronal ELAV-like protein
binding to coding and non-coding RNAs in human brain. eLife.
5:52016. View Article : Google Scholar
|
|
33
|
Liu SJ, Nowakowski TJ, Pollen AA, Lui JH,
Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz
AA, et al: Single-cell analysis of long non-coding RNAs in the
developing human neocortex. Genome Biol. 17:672016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hansen TB, Venø MT, Damgaard CK and Kjems
J: Comparison of circular RNA prediction tools. Nucleic Acids Res.
44:e582016. View Article : Google Scholar :
|
|
35
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Quinlan AR and Hall IM: BEDTools: A
flexible suite of utilities for comparing genomic features.
Bioinformatics. 26:841–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kroll KW, Mokaram NE, Pelletier AR,
Frankhouser DE, Westphal MS, Stump PA, Stump CL, Bundschuh R,
Blachly JS and Yan P: Quality control for RNA-Seq (QuaCRS): An
integrated quality control pipeline. Cancer Inform. 13(Suppl 3):
7–14. 2014.PubMed/NCBI
|
|
40
|
Chen C, Khaleel SS, Huang H and Wu CH:
Software for pre-processing Illumina next-generation sequencing
short read sequences. Source Code Biol Med. 9:82014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
Project Data Processing Subgroup: The Sequence Alignment/Map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Anders S, Pyl PT and Huber W: HTSeq - a
Python framework to work with high-throughput sequencing data.
Bioinformatics. 31:166–169. 2015. View Article : Google Scholar
|
|
44
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
46
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar
|
|
47
|
Kadota PO, Hajjiri Z, Finn PW and Perkins
DL: Precision subtypes of T cell-mediated rejection identified by
molecular profiles. Front Immunol. 6:5362015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Alhasan AA, Izuogu OG, Al-Balool HH, Steyn
JS, Evans A, Colzani M, Ghevaert C, Mountford JC, Marenah L,
Elliott DJ, et al: Circular RNA enrichment in platelets is a
signature of transcriptome degradation. Blood. 127:e1–e11. 2016.
View Article : Google Scholar :
|
|
54
|
Thomas LF and Sætrom P: Circular RNAs are
depleted of polymorphisms at microRNA binding sites.
Bioinformatics. 30:2243–2246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gao Y, Wang J, Zheng Y, Zhang J, Chen S
and Zhao F: Comprehensive identification of internal structure and
alternative splicing events in circular RNAs. Nat Commun.
7:120602016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Caiment F, Gaj S, Claessen S and Kleinjans
J: High-throughput data integration of RNA-miRNA-circRNA reveals
novel insights into mechanisms of benzo[a]pyrene-induced
carcinogenicity. Nucleic Acids Res. 43:2525–2534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ghosal S, Das S, Sen R, Basak P and
Chakrabarti J: Circ2Traits: A comprehensive database for circular
RNA potentially associated with disease and traits. Front Genet.
4:2832013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44D:D209–D215. 2016. View Article : Google Scholar
|
|
60
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar :
|
|
61
|
Huang M, Zhong Z, Lv M, Shu J, Tian Q and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs and circRNAs with associated co-expression and ceRNA
networks in bladder carcinoma. Oncotarget. 7:47186–47200. 2016.
View Article : Google Scholar : PubMed/NCBI
|