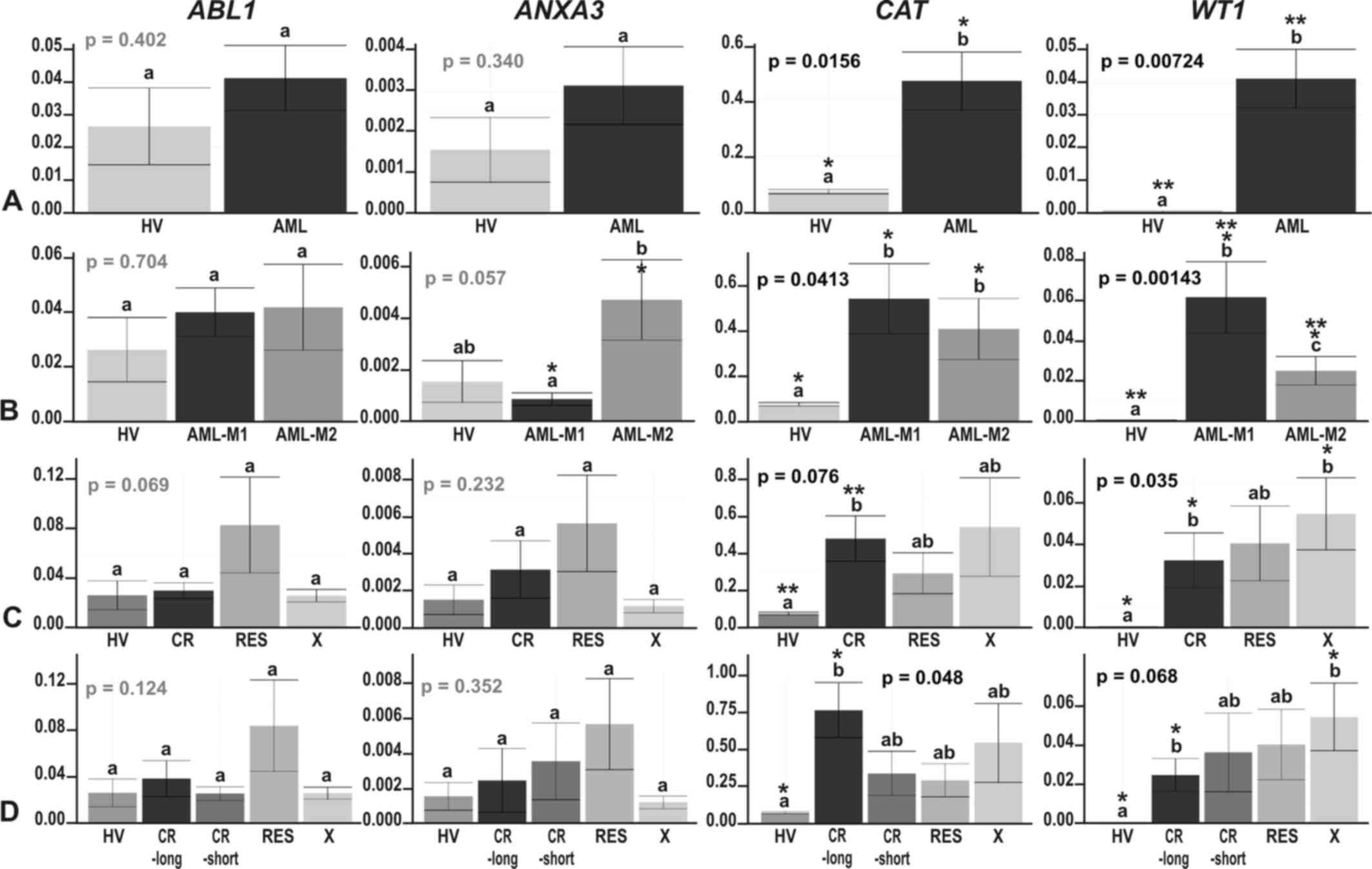
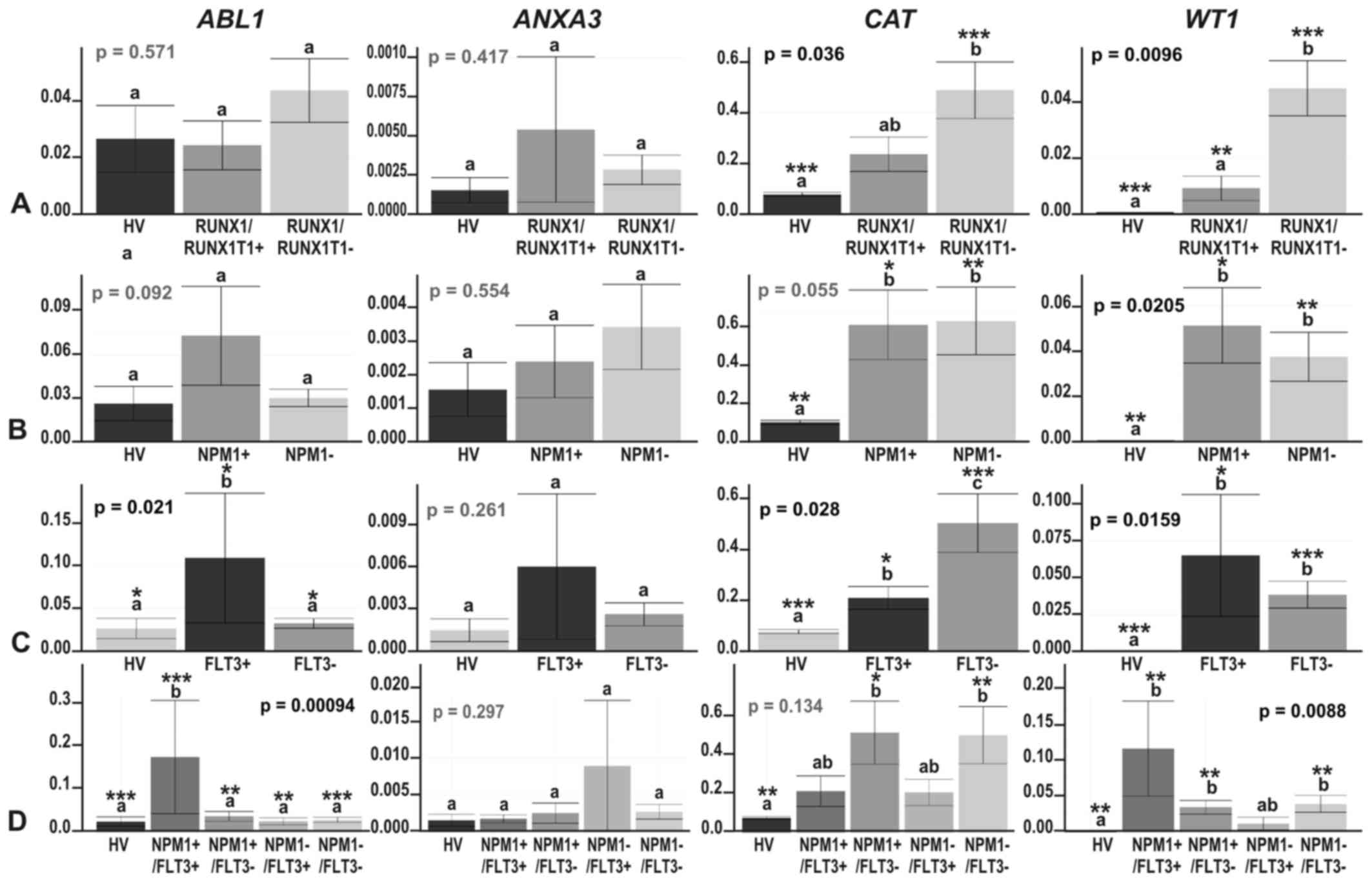
|
1
|
Estey E and Döhner H: Acute myeloid
leukaemia. Lancet. 368:1894–1907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Döhner H, Estey EH, Amadori S, Appelbaum
FR, Büchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson
RA, et al European LeukemiaNet: Diagnosis and management of acute
myeloid leukemia in adults: Recommendations from an international
expert panel, on behalf of the European LeukemiaNet. Blood.
115:453–474. 2010. View Article : Google Scholar
|
|
3
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposed revised
criteria for the classification of acute myeloid leukemia. A report
of the French-American-British Cooperative Group. Ann Intern Med.
103:620–625. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A, et al: The 2008 revision of the
World Health Organization (WHO) classification of myeloid neoplasms
and acute leukemia: Rationale and important changes. Blood.
114:937–951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Döhner H: Implication of the molecular
characterization of acute myeloid leukemia. Hematology Am Soc
Hematol Educ Program Washington, DC: pp. 412–419. 2007
|
|
6
|
Grimwade D and Hills RK: Independent
prognostic factors for AML outcome. Hematology Am Soc Hematol Educ
Program Washington, DC: pp. 385–395. 2009
|
|
7
|
Falini B: Acute myeloid leukemia with
mutated nucleophosmin (NPM1): Molecular, pathological, and clinical
features. Cancer Treat Res. 145:149–168. 2010. View Article : Google Scholar
|
|
8
|
Nakao M, Yokota S, Iwai T, Kaneko H,
Horiike S, Kashima K, Sonoda Y, Fujimoto T and Misawa S: Internal
tandem duplication of the flt3 gene found in acute myeloid
leukemia. Leukemia. 10:1911–1918. 1996.PubMed/NCBI
|
|
9
|
Lin LI, Chen CY, Lin DT, Tsay W, Tang JL,
Yeh YC, Shen HL, Su FH, Yao M, Huang SY, et al: Characterization of
CEBPA mutations in acute myeloid leukemia: Most patients with CEBPA
mutations have biallelic mutations and show a distinct
immunophenotype of the leukemic cells. Clin Cancer Res.
11:1372–1379. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Care RS, Valk PJ, Goodeve AC, Abu-Duhier
FM, Geertsma-Kleinekoort WM, Wilson GA, Gari MA, Peake IR,
Löwenberg B and Reilly JT: Incidence and prognosis of c-KIT and
FLT3 mutations in core binding factor (CBF) acute myeloid
leukaemias. Br J Haematol. 121:775–777. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tyner JW, Erickson H, Deininger MWN,
Willis SG, Eide CA, Levine RL, Heinrich MC, Gattermann N, Gilliland
DG, Druker BJ, et al: High-throughput sequencing screen reveals
novel, transforming RAS mutations in myeloid leukemia patients.
Blood. 113:1749–1755. 2009. View Article : Google Scholar :
|
|
12
|
Delhommeau F, Dupont S, Della Valle V,
James C, Trannoy S, Massé A, Kosmider O, Le Couedic JP, Robert F,
Alberdi A, et al: Mutation in TET2 in myeloid cancers. N Engl J
Med. 360:2289–2301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ley TJ, Ding L, Walter MJ, McLellan MD,
Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et
al: DNMT3A mutations in acute myeloid leukemia. N Engl J Med.
363:2424–2433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aref S, Kamel Areida S, Abdel Aaal MF,
Adam OM, El-Ghonemy MS, El-Baiomy MA and Zeid TA: Prevalence and
clinical effect of IDH1 and IDH2 mutations among cytogenetically
normal acute myeloid leukemia patients. Clin Lymphoma Myeloma Leuk.
15:550–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mrózek K, Marcucci G, Paschka P, Whitman
SP and Bloomfield CD: Clinical relevance of mutations and
gene-expression changes in adult acute myeloid leukemia with normal
cytogenetics: Are we ready for a prognostically prioritized
molecular classification? Blood. 109:431–448. 2007. View Article : Google Scholar
|
|
16
|
Bacher U, Kohlmann A and Haferlach T: Gene
expression profiling for diagnosis and therapy in acute leukaemia
and other haematologic malignancies. Cancer Treat Rev. 36:637–646.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Riva L, Luzi L and Pelicci PG: Genomics of
acute myeloid leukemia: The next generation. Front Oncol. 2:402012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kohlmann A, Grossmann V, Nadarajah N and
Haferlach T: Next-generation sequencing - feasibility and
practicality in haematology. Br J Haematol. 160:736–753. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Golub TR, Slonim DK, Tamayo P, Huard C,
Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri
MA, et al: Molecular classification of cancer: Class discovery and
class prediction by gene expression monitoring. Science.
286:531–537. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alizadeh A, Eisen M, Davis RE, Ma C, Sabet
H, Tran T, Powell JI, Yang L, Marti GE, Moore DT, et al: The
lymphochip: A specialized cDNA microarray for the genomic-scale
analysis of gene expression in normal and malignant lymphocytes.
Cold Spring Harb Symp Quant Biol. 64:71–78. 1999. View Article : Google Scholar
|
|
21
|
Bullinger L, Döhner K, Bair E, Fröhling S,
Schlenk RF, Tibshirani R, Döhner H and Pollack JR: Use of
gene-expression profiling to identify prognostic subclasses in
adult acute myeloid leukemia. N Engl J Med. 350:1605–1616. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Valk PJ, Verhaak RG, Beijen MA, Erpelinck
CAJ, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM,
Beverloo HB, Moorhouse MJ, van der Spek PJ, Löwenberg B, et al:
Prognostically useful gene-expression profiles in acute myeloid
leukemia. N Engl J Med. 350:1617–1628. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bacher U, Kohlmann A, Haferlach C and
Haferlach T: Gene expression profiling in acute myeloid leukaemia
(AML). Best Pract Res Clin Haematol. 22:169–180. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Alcalay M, Tiacci E, Bergomas R, Bigerna
B, Venturini E, Minardi SP, Meani N, Diverio D, Bernard L, Tizzoni
L, et al: Acute myeloid leukemia bearing cytoplasmic nucleophosmin
(NPMc+ AML) shows a distinct gene expression profile
characterized by up-regulation of genes involved in stem-cell
maintenance. Blood. 106:899–902. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang L, Zhou K, Yang Y, Shang Z, Wang J,
Wang D, Wang N, Xu D and Zhou J: FLT3-ITD-associated
gene-expression signatures in NPM1-mutated cytogenetically normal
acute myeloid leukemia. Int J Hematol. 96:234–240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Radmacher MD, Marcucci G, Ruppert AS,
Mrózek K, Whitman SP, Vardiman JW, Paschka P, Vukosavljevic T,
Baldus CD, Kolitz JE, et al Cancer and Leukemia Group B:
Independent confirmation of a prognostic gene-expression signature
in adult acute myeloid leukemia with a normal karyotype: A Cancer
and Leukemia Group B study. Blood. 108:1677–1683. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wilson CS, Davidson GS, Martin SB, Andries
E, Potter J, Harvey R, Ar K, Xu Y, Kopecky KJ, Ankerst DP, et al:
Gene expression profiling of adult acute myeloid leukemia
identifies novel biologic clusters for risk classification and
outcome prediction. Blood. 108:685–696. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marcinkowska-Swojak M, Handschuh L,
Wojciechowski P, Goralski M, Tomaszewski K, Kazmierczak M,
Lewandowski K, Komarnicki M, Blazewicz J, Figlerowicz M, et al:
Simultaneous detection of mutations and copy number variation of
NPM1 in the acute myeloid leukemia using multiplex
ligation-dependent probe amplification. Mutat Res. 786:14–26. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oshlack A, Emslie D, Corcoran LM and Smyth
GK: Normalization of boutique two-color microarrays with a high
proportion of differentially expressed probes. Genome Biol.
8:R22007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Uszczyńska B, Zyprych-Walczak J, Handschuh
L, Szabelska A, Kaźmierczak M, Woronowicz W, Kozłowski P, Sikorski
MM, Komarnicki M, Siatkowski I, et al: Analysis of boutique arrays:
A universal method for the selection of the optimal data
normalization procedure. Int J Mol Med. 32:668–684. 2013.
View Article : Google Scholar
|
|
31
|
Wenne R, Handschuh L, Pocwierz-Kotus A,
Urbaniak R, Formanowicz P, Całkiewicz J, Brzozowska K, Figlerowicz
M, Węgrzyn G and Wróbel B: The application of microarray technology
to the identification of Tc1-like element sequences in fish
genomes. Mar Biol Res. 7:466–477. 2011. View Article : Google Scholar
|
|
32
|
Zmieńko A, Guzowska-Nowowiejska M,
Urbaniak R, Pląder W, Formanowicz P and Figlerowicz M: A tiling
microarray for global analysis of chloroplast genome expression in
cucumber and other plants. Plant Methods. 7:292011. View Article : Google Scholar
|
|
33
|
Luczak M, Kaźmierczak M, Handschuh L,
Lewandowski K, Komarnicki M and Figlerowicz M: Comparative proteome
analysis of acute myeloid leukemia with and without maturation. J
Proteomics. 75:5734–5748. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kaźmierczak M, Luczak M, Lewandowski K,
Handschuh L, Czyż A, Jarmuż M, Gniot M, Michalak M, Figlerowicz M
and Komarnicki M: Esterase D and gamma 1 actin level might predict
results of induction therapy in patients with acute myeloid
leukemia without and with maturation. Med Oncol. 30:7252013.
View Article : Google Scholar
|
|
35
|
Ley TJ, Miller C, Ding L, Raphael BJ,
Mungall AJ, Robertson A, Hoadley K, Triche TJ Jr, Laird PW, Baty
JD, et al Cancer Genome Atlas Research Network: Genomic and
epigenomic landscapes of adult de novo acute myeloid leukemia. N
Engl J Med. 368:2059–2074. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Smyth GK: Limma: linear models for
microarray data. Bioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Springer; New York, NY: pp. 397–420. 2005,
View Article : Google Scholar
|
|
37
|
Benjamini Y and Hochberg Y: Controlling
the False Discovery Rate: A Practical and Powerful Approach to
Multiple Testing. J R Stat Soc B. 57:289–300. 1995.
|
|
38
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
39
|
Bergmann L, Miething C, Maurer U, Brieger
J, Karakas T, Weidmann E and Hoelzer D: High levels of Wilms’ tumor
gene (wt1) mRNA in acute myeloid leukemias are associated with a
worse long-term outcome. Blood. 90:1217–1225. 1997.PubMed/NCBI
|
|
40
|
Lyu X, Xin Y, Mi R, Ding J, Wang X, Hu J,
Fan R, Wei X, Song Y and Zhao RY: Overexpression of Wilms tumor 1
gene as a negative prognostic indicator in acute myeloid leukemia.
PLoS One. 9:e924702014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wouters BJ, Löwenberg B and Delwel R: A
decade of genome-wide gene expression profiling in acute myeloid
leukemia: Flashback and prospects. Blood. 113:291–298. 2009.
View Article : Google Scholar :
|
|
42
|
Haferlach T, Kohlmann A, Schnittger S,
Dugas M, Hiddemann W, Kern W and Schoch C: Global approach to the
diagnosis of leukemia using gene expression profiling. Blood.
106:1189–1198. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Payton JE, Grieselhuber NR, Chang L-W,
Murakami M, Geiss GK, Link DC, Nagarajan R, Watson MA and Ley TJ:
High throughput digital quantification of mRNA abundance in primary
human acute myeloid leukemia samples. J Clin Invest. 119:1714–1726.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gutiérrez NC, López-Pérez R, Hernández JM,
Isidro I, González B, Delgado M, Fermiñán E, García JL, Vázquez L,
González M, et al: Gene expression profile reveals deregulation of
genes with relevant functions in the different subclasses of acute
myeloid leukemia. Leukemia. 19:402–409. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Andersson A, Edén P, Lindgren D, Nilsson
J, Lassen C, Heldrup J, Fontes M, Borg A, Mitelman F, Johansson B,
et al: Gene expression profiling of leukemic cell lines reveals
conserved molecular signatures among subtypes with specific genetic
aberrations. Leukemia. 19:1042–1050. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Park MH, Cho SA, Yoo KH, Yang MH, Ahn JY,
Lee HS, Lee KE, Mun YC, Cho DH, Seong CM, et al: Gene expression
profile related to prognosis of acute myeloid leukemia. Oncol Rep.
18:1395–1402. 2007.PubMed/NCBI
|
|
47
|
Tominaga-Sato S, Tsushima H, Ando K,
Itonaga H, Imaizumi Y, Imanishi D, Iwanaga M, Taguchi J, Fukushima
T, Yoshida S, et al: Expression of myeloperoxidase and gene
mutations in AML patients with normal karyotype: Double CEBPA
mutations are associated with high percentage of MPO positivity in
leukemic blasts. Int J Hematol. 94:81–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cristóbal I, Garcia-Orti L, Cirauqui C,
Cortes-Lavaud X, García-Sánchez MA, Calasanz MJ and Odero MD:
Overexpression of SET is a recurrent event associated with poor
outcome and contributes to protein phosphatase 2A inhibition in
acute myeloid leukemia. Haematologica. 97:543–550. 2012. View Article : Google Scholar :
|
|
49
|
Artero-Castro A, Castellvi J, García A,
Hernández J, Ramón y Cajal S and Lleonart ME: Expression of the
ribosomal proteins Rplp0, Rplp1, and Rplp2 in gynecologic tumors.
Hum Pathol. 42:194–203. 2011. View Article : Google Scholar
|
|
50
|
Tsai ST, Chien IH, Shen WH, Kuo YZ, Jin
YT, Wong TY, Hsiao JR, Wang HP, Shih NY and Wu LW: ENO1, a
potential prognostic head and neck cancer marker, promotes
transformation partly via chemokine CCL20 induction. Eur J Cancer.
46:1712–1723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rowlands DC, Williams A, Jones NA, Guest
SS, Reynolds GM, Barber PC and Brown G: Stathmin expression is a
feature of proliferating cells of most, if not all, cell lineages.
Lab Invest. 72:100–113. 1995.PubMed/NCBI
|
|
52
|
Roos G, Brattsand G, Landberg G, Marklund
U and Gullberg M: Expression of oncoprotein 18 in human leukemias
and lymphomas. Leukemia. 7:1538–1546. 1993.PubMed/NCBI
|
|
53
|
Johnson WE, Jones NA, Rowlands DC,
Williams A, Guest SS and Brown G: Down-regulation but not
phosphorylation of stathmin is associated with induction of HL60
cell growth arrest and differentiation by physiological agents.
FEBS Lett. 364:309–313. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Matushansky I, Radparvar F and Skoultchi
AI: Reprogramming leukemic cells to terminal differentiation by
inhibiting specific cyclin-dependent kinases in G1. Proc Natl Acad
Sci USA. 97:14317–14322. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Müller A, Lange K, Gaiser T, Hofmann M,
Bartels H, Feller AC and Merz H: Expression of angiopoietin-1 and
its receptor TEK in hematopoietic cells from patients with myeloid
leukemia. Leuk Res. 26:163–168. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tagliafico E, Tenedini E, Manfredini R,
Grande A, Ferrari F, Roncaglia E, Bicciato S, Zini R, Salati S,
Bianchi E, et al: Identification of a molecular signature
predictive of sensitivity to differentiation induction in acute
myeloid leukemia. Leukemia. 20:1751–1758. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Qian Z, Fernald AA, Godley LA, Larson RA
and Le Beau MM: Expression profiling of CD34+ hematopoietic
stem/progenitor cells reveals distinct subtypes of therapy-related
acute myeloid leukemia. Proc Natl Acad Sci USA. 99:14925–14930.
2002. View Article : Google Scholar
|
|
58
|
Shimamura R, Kudo J, Kondo H, Dohmen K,
Gondo H, Okamura S, Ishibashi H and Niho Y: Expression of the
thymosin beta 4 gene during differentiation of hematopoietic cells.
Blood. 76:977–984. 1990.PubMed/NCBI
|
|
59
|
Schwanhäusser B, Wolf J, Selbach M and
Busse D: Synthesis and degradation jointly determine the
responsiveness of the cellular proteome. BioEssays. 35:597–601.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Vogel C and Marcotte EM: Insights into the
regulation of protein abundance from proteomic and transcriptomic
analyses. Nat Rev Genet. 13:227–232. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen G, Gharib TG, Huang CC, Taylor JM,
Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB,
Hanash SM, et al: Discordant protein and mRNA expression in lung
adenocarcinomas. Mol Cell Proteomics. 1:304–313. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jansen R, Greenbaum D and Gerstein M:
Relating whole-genome expression data with protein-protein
interactions. Genome Res. 12:37–46. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Miwa H, Beran M and Saunders GF:
Expression of the Wilms’ tumor gene (WT1) in human leukemias.
Leukemia. 6:405–409. 1992.PubMed/NCBI
|
|
64
|
El-Rifai W, Moskaluk CA, Abdrabbo MK,
Harper J, Yoshida C, Riggins GJ, Frierson HF Jr and Powell SM:
Gastric cancers overexpress S100A calcium-binding proteins. Cancer
Res. 62:6823–6826. 2002.PubMed/NCBI
|
|
65
|
Su YJ, Xu F, Yu JP, Yue DS, Ren XB and
Wang CL: Up-regulation of the expression of S100A8 and S100A9 in
lung adenocarcinoma and its correlation with inflammation and other
clinical features. Chin Med J (Engl). 123:2215–2220. 2010.
|
|
66
|
Whitman SP, Maharry K, Radmacher MD,
Becker H, Mrózek K, Margeson D, Holland KB, Wu YZ, Schwind S,
Metzeler KH, et al: FLT3 internal tandem duplication associates
with adverse outcome and gene- and microRNA-expression signatures
in patients 60 years of age or older with primary cytogenetically
normal acute myeloid leukemia: A Cancer and Leukemia Group B study.
Blood. 116:3622–3626. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kim HS, Lee TB and Choi CH:
Down-regulation of catalase gene expression in the
doxorubicin-resistant AML subline AML-2/DX100. Biochem Biophys Res
Commun. 281:109–114. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bergmann L, Maurer U and Weidmann E: Wilms
tumor gene expression in acute myeloid leukemias. Leuk Lymphoma.
25:435–443. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lossos IS, Czerwinski DK, Wechser MA and
Levy R: Optimization of quantitative real-time RT-PCR parameters
for the study of lymphoid malignancies. Leukemia. 17:789–795. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hussein S, Michael P, Brabant D, Omri A,
Narain R, Passi K, Ramana CV, Parrillo JE, Kumar A, Parissenti A,
et al: Characterization of human septic sera induced gene
expression modulation in human myocytes. Int J Clin Exp Med.
2:131–148. 2009.PubMed/NCBI
|
|
71
|
Kreuzer KA, Saborowski A, Lupberger J,
Appelt C, Na IK, le Coutre P and Schmidt CA: Fluorescent
5′-exonuclease assay for the absolute quantification of Wilms’
tumour gene (WT1) mRNA: Implications for monitoring human
leukaemias. Br J Haematol. 114:313–318. 2001. View Article : Google Scholar : PubMed/NCBI
|