Introduction
Oral squamous cell carcinomas (OSCC) is one of the
most common types of cancer, and its incidence is increasing
worldwide (1). In spite of
considerable advances being made in diagnostics and treatment, the
5-year survival rate for patients with OSCC has not improved during
the past few decades and remains <50% (2). Although much scientific research has
indicated that local recurrence and nodal metastasis are the major
causes of mortality in patients with OSCC, the precise molecular
mechanisms remain unclear (3–5).
Therefore, in order to improve the diagnosis and management of
patients with OSCC, it is important to identify effective
diagnostic biomarkers and therapeutic targets.
The mammalian genome encodes large numbers of
non-coding transcripts that have structural, regulatory or unknown
functions (6,7). Long non-coding RNAs (lncRNAs), a
class of transcripts >200 nucleotides in length, have been shown
to play significant regulatory roles in epigenetic modulation, as
well as transcriptional and post-transcriptional in recent years
(8,9). Increasing evidence has indicated that
lncRNAs participate in the biological processes of cell
proliferation, differentiation, apoptosis and cancer metastasis
during cancer development and progression (10–12).
However, whether such distinct functions of lncRNAs are involved in
the development of OSCC remains unknown. H19 a 2.3 kb of lncRNA
molecule which is one of the earliest identified imprinted genes
(13). Previous studies have
reported that H19 is aberrantly expressed in human cancers,
including hepatocellular carcinoma, as well as bladder and breast
cancers, and usually correlates with cancer progression, metastasis
and a poor prognosis, suggesting that H19 may be used as a
biomarker for the diagnosis of these types of cancer (14,15).
Moreover, H19 has been found to promote epithelial‑mesenchymal
transition (EMT) by antagonizing the activity of microRNAs (miRNAs
or miRs) as a competing endogenous RNA (ceRNA), and regulating
expression of their downstream genes in several type of cancer,
such as pancreatic and colorectal cancer (16,17).
Recently, H19 was found to be associated with the risk of OSCC in a
Chinese population (18). Zhang
et al also demonstrated that H19 played a crucial role in
the progression of tongue squamous cell carcinoma (TSCC) by
regulating the expression of β-catenin and glycogen synthase kinase
(GSK)-3β via enhancer of zeste homolog 2 (EZH2), indicating that
the inhibition of H19 may be a potential target for the treatment
of TSCC (19). However, the roles
and mechanism of H19 in the development and progression of OSCC
remain to be elucidated.
miRNAs are a class of endogenous small non-coding
RNAs, 20–25 nucleotides in length, which can suppress gene
expression by directly binding to the 3′-untranslated region
(3′-UTR) of target messenger RNAs (mRNAs) to induce mRNA decay or
translational repression (20).
Several deregulated miRNAs have also been reported in OSCC, and are
involved in OSCC cell growth, apoptosis, migration and invasion
(21,22). Xu et al found that miR‑138
was significantly downregulated in OSCC and the overexpression of
miR-138 inhibited cell proliferation OSCC in vitro and in
vivo (23). However, to date,
at least to the best of our knowledge, there are limited studies
available on the association between lncRNAs and miRNAs and their
role in the development of OSCC; thus, studies are required to shed
further insight into this matter.
In the present study, we first analyzed GEO datasets
to investigate aberrantly expressed lncRNAs, and found that H19 was
significantly upregulated both in OSCC tissues and cell lines.
Moreover, the knockdown of H19 inhibited OSCC proliferation and
invasion in vitro, and suppressed tumor growth in
vivo. Finally, H19 was found to play an oncogenic role in OSCC
cells by regulating EZH2 and by targeting miR‑138. These findings
provide a novel mechanism of the H19/miR‑138/EZH2 axis in OSCC, and
suggest that this axis may be a promising molecular therapeutic
target for OSCC.
Materials and methods
Cell lines and tissue samples
Six OSCC cell lines SCC-4 (Cat. no. CRL-1624),
SCC-15 (Cat. no. CRL-1623), CAL-27 (Cat. no. CRL-2095) [all from
American Type Culture Collection (ATCC), Manassas, VA, USA], HSC-3
(RCB1975), HSC-2 (RCB1945) (both from Riken Cell Bank, Tsukuba,
Japan) and Ca9-22 (JCRB0625) [from the Japanese Collection of
Research Bioresources (JCRB), Osaka, Japan] were used in this
study. All cells were cultured in Dulbecco's modified Eagle's
medium (DMEM). The medium was supplemented with 10% fetal bovine
serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 100 U/ml penicillin
and 100 μg/ml streptomycin at 37°C in a 5% CO2
atmosphere. A non-tumorigenic immortalized oral keratinocyte line
(HOK‑16B, generous gift from Dr No-Hee Park, University of
California, Los Angeles, CA, USA) was maintained in oral
keratinocyte medium, supplemented with 1% keratinocyte growth
factor plus epithelial growth factor mixture (Invitrogen, Carlsbad,
CA, USA). A total of 42 freshly frozen OSCC tissues, as well as 42
matched controls were obtained from the Department of Oral and
Maxillofacial Surgery, Shenzhen Hospital of Southern Medical
University, Shenzhen, China. None of the patients with OSCC had
received radiotherapy or chemotherapy prior to surgery. This study
protocol conformed to the Ethics Committee of Shenzhen Hospital of
Southern Medical University (Shenzhen, China). All human materials
were obtained with informed consent and approved by the Ethics
Committee of Shenzhen Hospital of Southern Medical University.
lncRNA expression profile data from
GEO
The micro-array data was downloaded from the open
GEO database (https://www.ncbi.nlm.nih.gov/geo/) and the GEO
accession number is GSE3524 (24).
These microarray expression data were analyzed by GEO2R
bioinformatics software (http://www.ncbi.nlm.nih.gov/geo/geo2r/), which can
analyze any GEO series. The adjusted P-values (adj. P-value) using
the Benjamini and Hochberg (BH) false discovery rate (FDR) method
by default were applied to correct for the occurrence of
false‑positive results. An adj. P<0.05 and a |logFC|≥1 were set
as the cut-off criteria. A heatmap of the 49 lncRNAs which the most
significant differences in expression was generated using the
online tool Morpheus (https://software.broadinsti-tute.org/morpheus/).
RNA extraction, reverse transcription and
quantitative RT-PCR
For lncRNA analysis, total RNA was isolated from the
cells and tissues using TRIzol reagent (Invitrogen) according to
the manufacturer's instructions and reverse transcribed using the
Superscript III first strand synthesis system (Life Technologies,
Carlsbad, CA, USA). For miRNA analysis, RNA was extracted from the
liver tissues using the miRNeasy mini kit (Qiagen, West Sussex, UK)
according to the manufacturer's instructions. The RNA was then
reverse transcribed into cDNA. Amplifications were carried out on
an ABI 7500 Real-Time PCR system (Life Technologies) using
SYBR-Green according to the manufacturer's instructions.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6 snRNA were
used as endogenous controls. Data were analyzed using 7500 software
v.2.0.1 (Applied Biosystems, Foster City, CA, USA), and calculated
using the 2−∆∆Cq method (25). All experiments were performed in
triplicate.
Cell transfection
The miR-138 mimic, miR-138 inhibitor, mimics
negative control (mimics NC) and inhibitor NC were purchased from
RiboBio Co., Ltd. (Guangzhou, China). A small interfering RNA
against H19 (si-H19) and si-Scramble were purchased from Santa Cruz
Biotechnology (Santa Cruz, CA, USA). Plasmid cDNA-H19 was
constructed by introducing a BamHI-EcoRI fragment
containing the H19 cDNA into the same site in the pcDNA3.1 plasmid
(Invitrogen). Cell transfections with si-H19, pcDNA-H19 and miR-138
mimic and inhibitor were performed as previously described
(26). The HSC-2 and Ca9-22 cells
were transfected with H19 siRNA, miR-138 mimic or miR-138 inhibitor
using the Lipofectamine® RNAiMAX kit (Thermo Fisher
Scientific, Waltham, MA, USA). The CAL-27 cells were transfected
with pcDNA-H19 or pcDNA-H19 plus miR-138 mimic using
Lipofectamine® 2000 (Life Technologies) according to the
manufacturer's instructions. The cells were harvested after 48 h
and used for further analysis.
Lentivirus production and infection
Lentiviral constructs carrying shRNA targeting H19
(Lv-shRNA), and an empty negative control vector (LV-GFP) were
obtained from GenePharma (Shanghai, China). The 293 cells
(CRL-1573, ATCC) were co-transfected with Lenti-Pac HIV Expression
Packaging Mix and the lentiviral vectors (or the control
lenti-virus vectors) using Lipofectamine 2000 (Life Technologies).
After 48 h, lentiviral particles in the supernatant were harvested
and ultra-centrifuged to concentrate the lentiviral particles.
Subsequently, the HSC-2 and Ca9-22 cells grown on 6-well plates
were transduced with the lentiviruses of 10 transduction units (TU)
per cell. The expression of H19 in the transduced cells was
examined by real-time PCR analysis.
Analysis of cell proliferation, apoptosis
and cell cycle progression
Cell proliferation was examined by MTT assay
according to the manufacturer's instructions (Roche Applied
Science, Basel, Switzerland). Briefly, approximately
1×103 cells were seeded in a 96-well culture plate for
24 h. The cells were then transfected with si-H19, si-Scramble,
miR-138 inhibitor or miR-control. At various time points, 0.5 mg/ml
MTT solution was added to each well. The absorbance was then
recorded at 490 nm on a Bio-Rad model 680 microplate reader,
(Bio-Rad Laboratories, Hercules, CA, USA). For the analysis of
apoptosis, the cells were stained with Annexin V-FITC and propidium
iodide (PI) (TACS Annexin V-FITC, Trevigen Inc., Gaithersburg, MD,
USA) and then analyzed with double‑label flow cytometry on a flow
cytometer (FACSCanto II; BD Biosciences, San Jose, CA, USA). For
cell cycle analysis, the cells were resuspended in PBS, stained
with PI containing RNase A for 30 min at 37°C, and analyzed by flow
cytometry. All the assays were conducted in triplicate.
In vivo tumor growth assay
All animal procedures were performed according to
national guidelines and approved by the Animal Care Ethics
Committee of Shenzhen Hospital of Southern Medical University.
Twenty female BALB/c nude mice (weighing 20±2 g; 4 weeks old,
Laboratory Animal Center of Shanghai, Academy of Science) were used
in this study. The HSC-2 and Ca9-22 cells transfected with Lv-shRNA
or Lv‑Control were injected into the left flanks of the nude mice
(2×106 cells/mouse). After 5 weeks, the mice were
sacrificed and tumor tissues were dissected. The tumor weight was
measured and the tumor volume was calculated according to the
following formula: (length x width2)/2. The largest
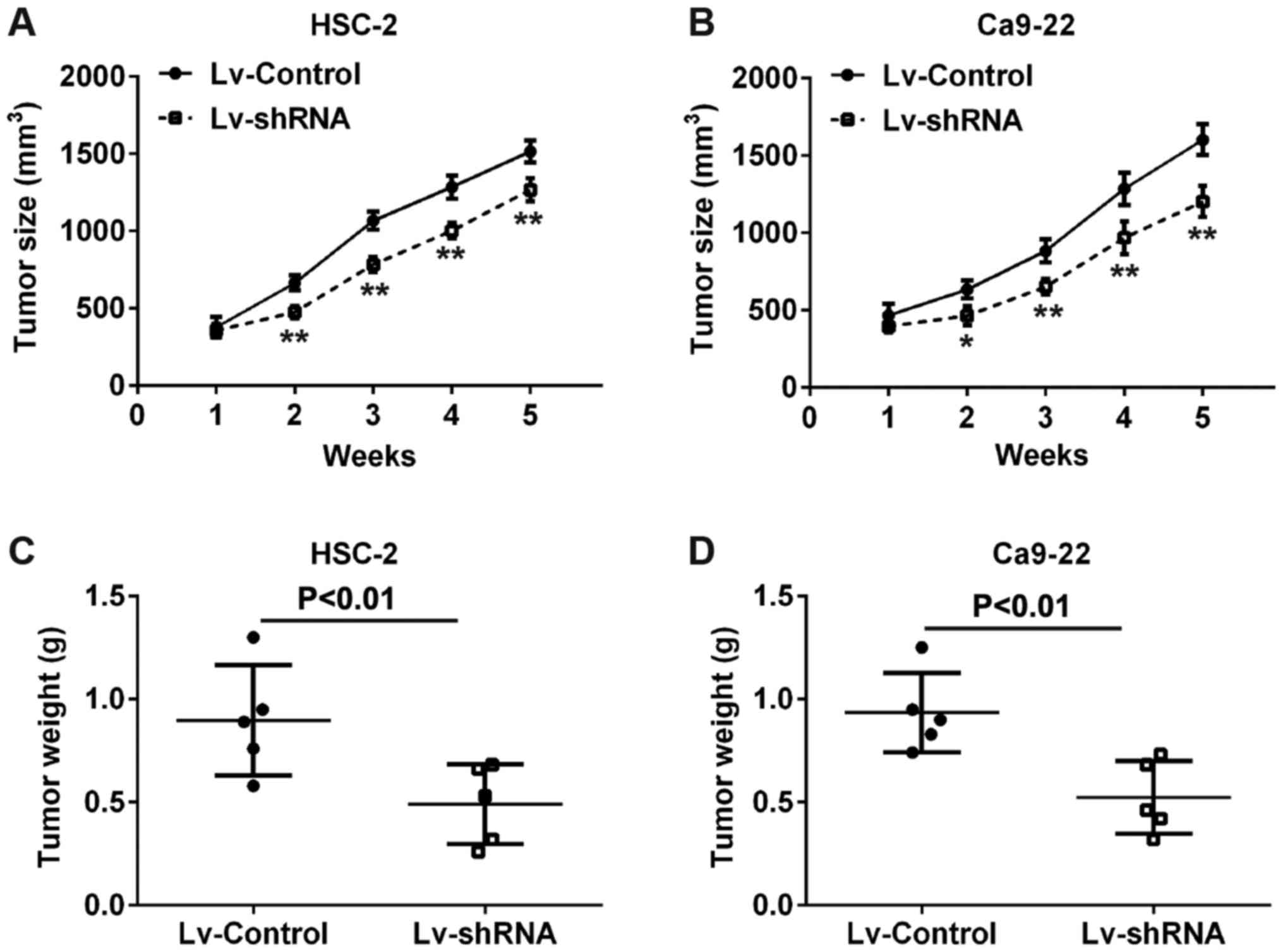
tumor size and volume in the mice of different groups was 1.30 g
and 1,500 mm3 (Lv-Control-transfected HSC-2 cells), 1.25
g and 1,700 mm3 (Lv-Control transfected Ca9-22 cells),
0.68 g and 1,150 mm3 (Lv-siRNA transfected HSC-2 cells),
0.73 g and 1,300 mm3 (Lv-siRNA transfected Ca9-22
cells).
Bioinformatics
In silico prediction of miRNA binding sites
within the H19 3′UTR was performed using TargetScan (www.targetscan.org) and PicTar (http://www.pictar.org).
Luciferase assays
The 3′-UTR of H19, with wild-type or mutant (Mut)
binding sites for miR-138, was amplified and cloned into the pGL3
vector (Promega, Madison, WI, USA) to generate the plasmid
pGL3-WT-H19-1 (wt-H9-1) or pGL3-WT-H19-2 (wt-H9-2). The putative
binding site of miR-138 in H19 was mutated by using a QuikChange
Site Directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA) to
synthetize mutant type pGL3‑mut‑H19‑1 (Mut‑H19‑1) or pGL3-mut-H19-2
vector (Mut-H19-2). For the luciferase reporter assay, the 293
cells were co-transfected with the luciferase reporter vectors and
miR-138 mimics or corresponding negative control (GenePharma) using
Lipofectamine 2000 reagent (Life Technologies). The pRL-TK plasmid
(Promega) was used as a normalizing control. After 48 h of
incubation, luciferase activity was analyzed using the
Dual‑Luciferase Reporter Assay System (Promega) according to the
manufacturer's instructions.
Wound healing assay
The cells were plated in 6-well plates and
transfected when cultured to 95% confluence. The cell layers were
then scratched using a 10 μl plastic pipette tip to produce
wounds. The wounds were photographed at 0 and 48 h under an
inverted phase contrast microscope (IX71; Olympus Corp., Tokyo,
Japan). Three random fields were marked and measured. All the
assays were carried out in triplicate.
Cell invasion assays
For invasion assays, a total of 3×104
HSC-2 and Ca9-22 cells in 150 μl serum-free medium at
post-transfection were seeded into the upper chamber (24‑well
insert, pore size 8 μm; Corning, NY, USA) pre‑coated with 30
μg/well Matrigel solution (BD Biosciences), and the lower
chambers were filled with 500 μl of 10% FBS medium.
Following incubation at 37°C for 48 h, the membranes were fixed
with 4% polyoxymethylene and stained with 0.1% crystal violet
(Sigma-Aldrich). Five pre-determined fields were counted under a
microscope (Olympus Corp., original magnification, ×200). All
assays were performed in triplicate.
Western blot analysis
Total proteins were extracted from cells using
radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma‑Aldrich)
and quantified with a Bicin Choninic Acid (BCA) protein assay kit
(Pierce, Rockford, IL, USA). A total of 40 μg of protein
were subjected to 10% SDS-PAGE, and subsequently transferred onto a
polyvinylidene difluoride membranes (Millipore, Billerica, MA,
USA). The blots were incubated with the primary antibodies specific
for cleaved caspase-3 (1:500; Cat. no. 9661), cleaved
poly(ADP-ribose) polymerase (PARP; 1:1,000; Cat. no. 5625), Bax
(1:1,000; Cat. no. 5023), EZH2 (1:1,000; Cat. no. 5246), zinc
finger E‑box‑binding homeobox 1 (ZEB1; 1:1,000; Cat. no. 3396),
E-cadherin (1:1,000; Cat. no. 14472), vimentin (1:1,000; Cat. no.
5741), N-cadherin (1:1,000; Cat. no. 13116) and β-actin (1:2,000;
Cat. no. 4970). All antibodies were obtained from Cell Signaling
Technology, Inc. (Danvers, MA, USA). Following 3 washes in TBST,
the membranes were incubated with corresponding horseradish
peroxidase (HRP)-conjugated secondary antibody (1:10,000; Santa
Cruz Biotechnology, Inc.) for 2 h at room temperature, and washed
with TBST 3 times. The protein bands were visualized by ECL
detection reagent (GE Healthcare Life Sciences, Piscataway, NJ,
USA). The intensity of the protein fragments was quantified with
the Quantity One software (4.5.0 basic; Bio-Rad).
Statistical analysis
Statistical analyses were performed with GraphPad
Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). All
data are presented as the means ± SD. Differences were analyzed by
a Student's t‑test between two groups or one-way analysis of
variance (ANOVA), followed by Tukey's multiple comparison tests
between multiple groups. Spearman's analysis was used in
correlation analysis. Survival analysis under the circumstance of
Kaplan-Meier method. A P-value <0.05 was considered to indicate
a statistically significant difference.
Results
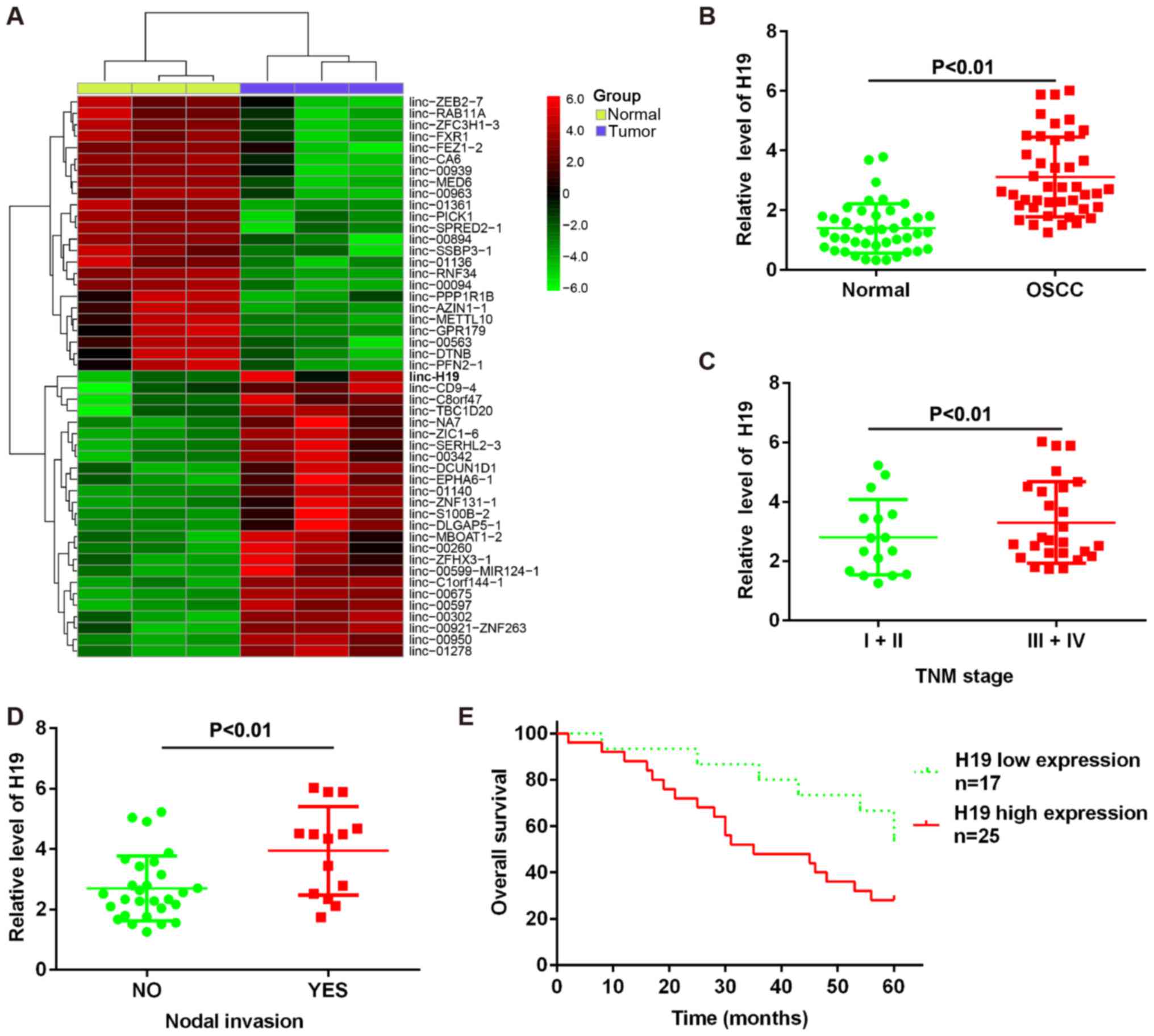
lncRNA H1 is overexpressed in OSCC
To identify the lncRNAs involved in the development
and progression of OSCC, a GSE dataset was obtained from the GEO
database under the accession number GSE3524 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3524).
Analysis of these data revealed that 24 lncRNAs were downregulated
and 25 miRNAs were upregulated in the tumor group, compared with
the normal group. Among the aberrantly expressed lncRNAs, lncRNA H1
was the most significantly upregulated lncRNA in GSE dataset
(Fig. 1A). To validate the
microarray analysis finding, we detected H19 expression in a cohort
of 42 paired tumor tissues and normal tissues. The detailed patient
clinical data are presented in Table
I. As shown in Fig. 1B, H19
expression was increased in tumor tissues compared with normal
tissues. These data suggested that H19 may be involved in the
process of carcinogenesis.
 | Table IClinicopathogical characteristics of
the study subjects. |
Table I
Clinicopathogical characteristics of
the study subjects.
| Characteristic | n (%) |
|---|
| Sex | |
| Male | 33 (78.57) |
| Female | 9 (21.43) |
| Age (years) | |
| ≥50 | 31 (73.81) |
| <50 | 11 (26.19) |
| Tobacco use | |
| Yes | 32 (76.19) |
| No | 10 (23.81) |
| Alcohol use | |
| Yes | 30 (71.43) |
| No | 12 (28.57) |
| Tumor site | |
| Tongue | 23 (54.76) |
| Floor of
mouth | 8 (19.05) |
| Alveolar | 6 (14.29) |
| Buccal mucosa | 4 (9.52) |
| Retromolar | 1 (2.38) |
| Tumor stage | |
| I–II | 16 (38.10) |
| III–IV | 26 (61.90) |
|
Differentiation | |
| Well and
moderate | 34 (80.95) |
| Poor | 8 (19.05) |
| Nodal invasion | |
| Negative | 28 (66.67) |
| Positive | 14 (33.33) |
To determine whether H19 expression was associated
with the grade of malignancy and nodal invasion in OSCC, we
examined the expression level of H19. We demonstrated that H19
expression was positively associated with the pathological grades
of OSCC and nodal invasion (Fig. 1C
and D). Based on the relative expression ratios of <0.5, the
42 clinical cases were divided into 2 groups as follows: the H19
low expression group (n=17) and the H19 high expression group
(n=25). We then assessed whether the expression of H19 correlated
with the post-operative survival time of patients with OSCC.
Kaplan-Meier survival analysis revealed that patients with a high
H19 expression had a poorer overall survival than those with a low
H19 expression (Fig. 1E). Taken
together, these findings indicate that H19 may be used as a
biomarker for the diagnosis and prognosis of OSCC.
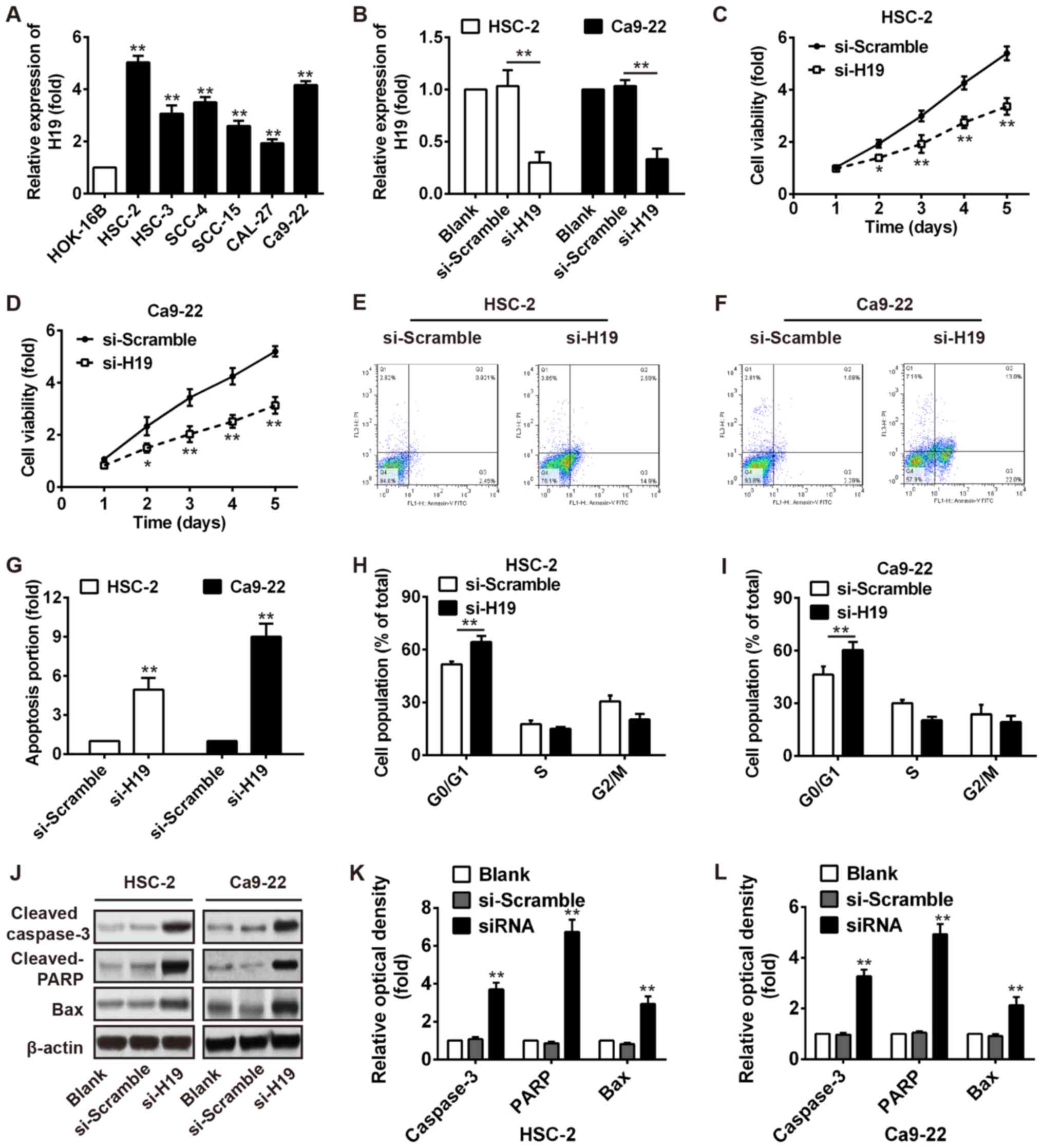
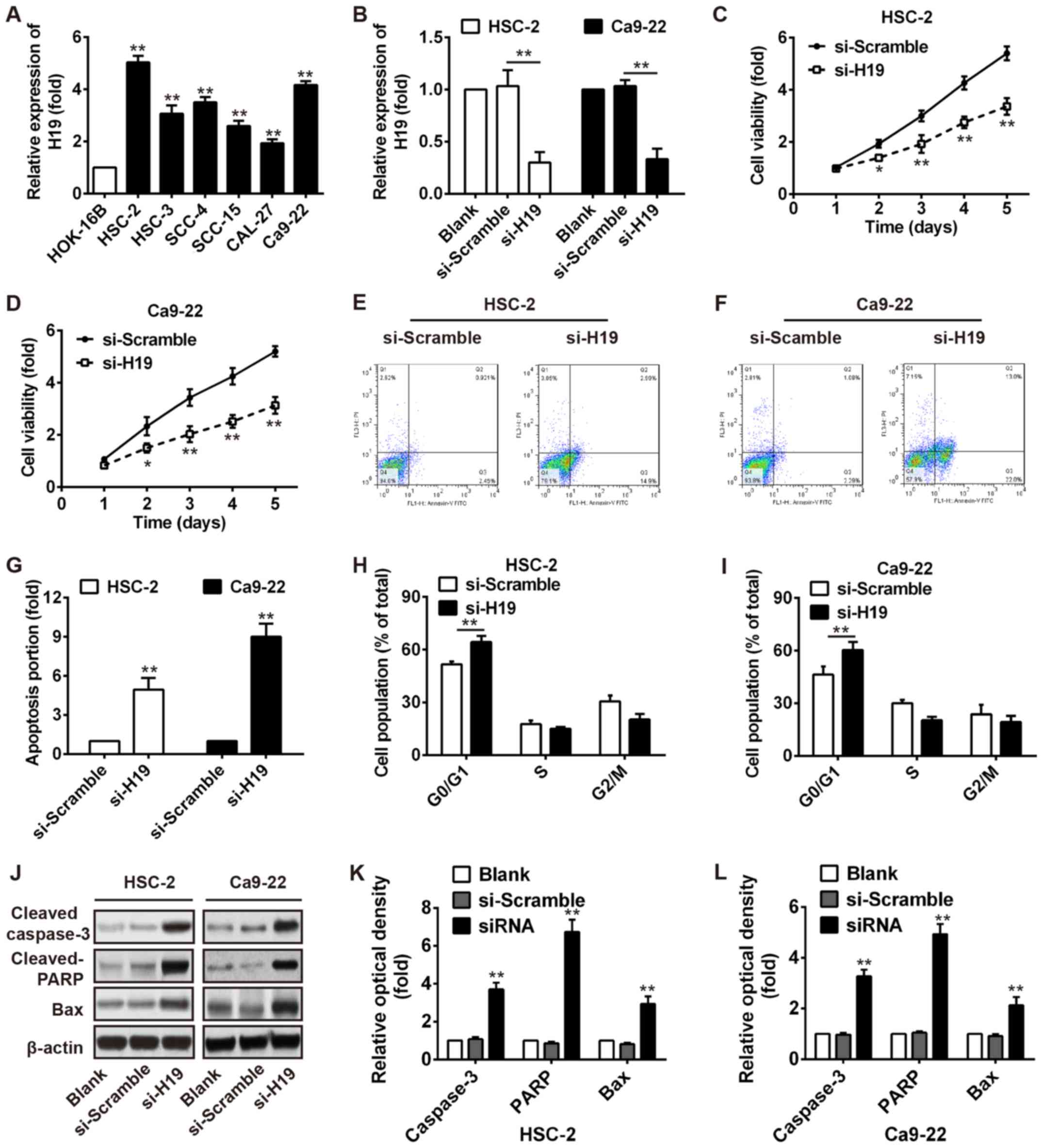
Knockdown of H19 inhibits the
proliferation, increases the G0/G1 phase population and induces the
apoptosis of OSCC cells
To examine the effect of H19 on cell growth, we
first measured the expression levels of H19 in 6 OSCC cell lines
(HSC-2, HSC-3, SCC-4, SCC-15, CAL-27 and Ca9-22) and a normal oral
mucosa cell line (HOK-16B). As shown in Fig. 2A, a higher expression of H19 was
observed in the OSCC cell lines compared with the normal oral
mucosa cell line, particularly in the HSC-2 and Ca9-22 cells.
Subsequently, the HSC-2 and Ca9-22 cells were transiently
transfected with si-H19 and the endogenous level of H19 was
effectively decreased (Fig. 2B).
As demonstrated by MTT assays, we found that H19 silencing
significantly decreased the viability of the HSC‑2 and Ca9‑22 cells
(Fig. 2C and D). Furthermore, the
proportion of the cell population undergoing apoptosis was
increased after knockdown of H19 in the HSC-2 and Ca9-22 cells
(Fig. 2E–G). Flow cytometry also
revealed a significant promotion of cells in the G0/G1 phase of the
cell cycle in the HSC-2 and Ca9-22 cells transfected with si-H19
(Fig. 2H and I). In addition,
western blot analysis revealed that the expression levels of
apoptosis-related proteins, including cleaved caspase-3 and cleaved
PARP and Bax were markedly increased after H19 knockdown (Fig. 2J–L). Our results thus revealed that
H19 knockdown inhibited several malignancy-related parameters of
OSCC in vitro.
Knockdown of H19 inhibits tumor growth in
vivo
To evaluate the functional roles of H19 in
vivo, we established a xenograft mouse mode in which Lv-shRNA
or Lv-Control-transfected HSC-2 and Ca9-22 cells were transplanted
into the flanks of BALB/c nude mice. Consistent with the results
obtained in vitro, after 5 weeks, tumor volumes in the mice
injected with cells from the si-H19 group were markedly smaller
compared with those in the mice injected with cells from the
Lv-Control (Fig. 3A and B).
Similarly, tumor weights in the mice injected with cells from the
Lv‑shRNA group were significantly lower compared with those in the
mice injected with cells from the Lv-Control group (Fig. 3C and D). These results indicate
that the knockdown of H19 expression inhibits tumor growth in
vivo.
Knockdown of H19 inhibits the migration
and invasion of OSCC cells
Based on the above-mentioned results in that the
expression of H19 was higher in metastatic tissues, we hypothesized
that H19 may be associated with the metastasis of OSCC. Thus, wound
healing and Transwell assays were performed to examine the effects
of H19 on OSCC cell metastasis. As shown in Fig. 4A, H19 silencing markedly inhibited
the migration of monolayer-cultured HSC-2 and Ca9-22 cells. In
addition, the numbers of invaded cells were markedly attenuated in
the cells in which H19 was knocked down compared with the control
cells (Fig. 4B). It is well known
that EMT plays a critical role in the invasion and metastasis of
OSCC cells (27). In this study,
we thus assessed the effect of H19 on the expression of EMT
related-genes. Our results demonstrated that H19 silencing
significantly decreased the expression levels of vimentin and
N-cadherin (mesenchymal markers), but increased the expression
levels of ZEB1 and E-cadherin (epithelial markers) (Fig. 4C). These data suggest that the
knockdown of H19 suppresses OSCC cell metastasis by inhibiting
EMT.
H19 directly targets miR-138 and inhibits
its level in OSCC cells
Recently, studies have confirmed that lncRNAs can
function as ceRNAs or molecular sponges that modulate miRNAs in
cancers (28,29). In this study, we thus performed
bioinformatics analysis using TargetScan and PicTar, and found that
miR-138 contains a binding site for H19. The predicted binding
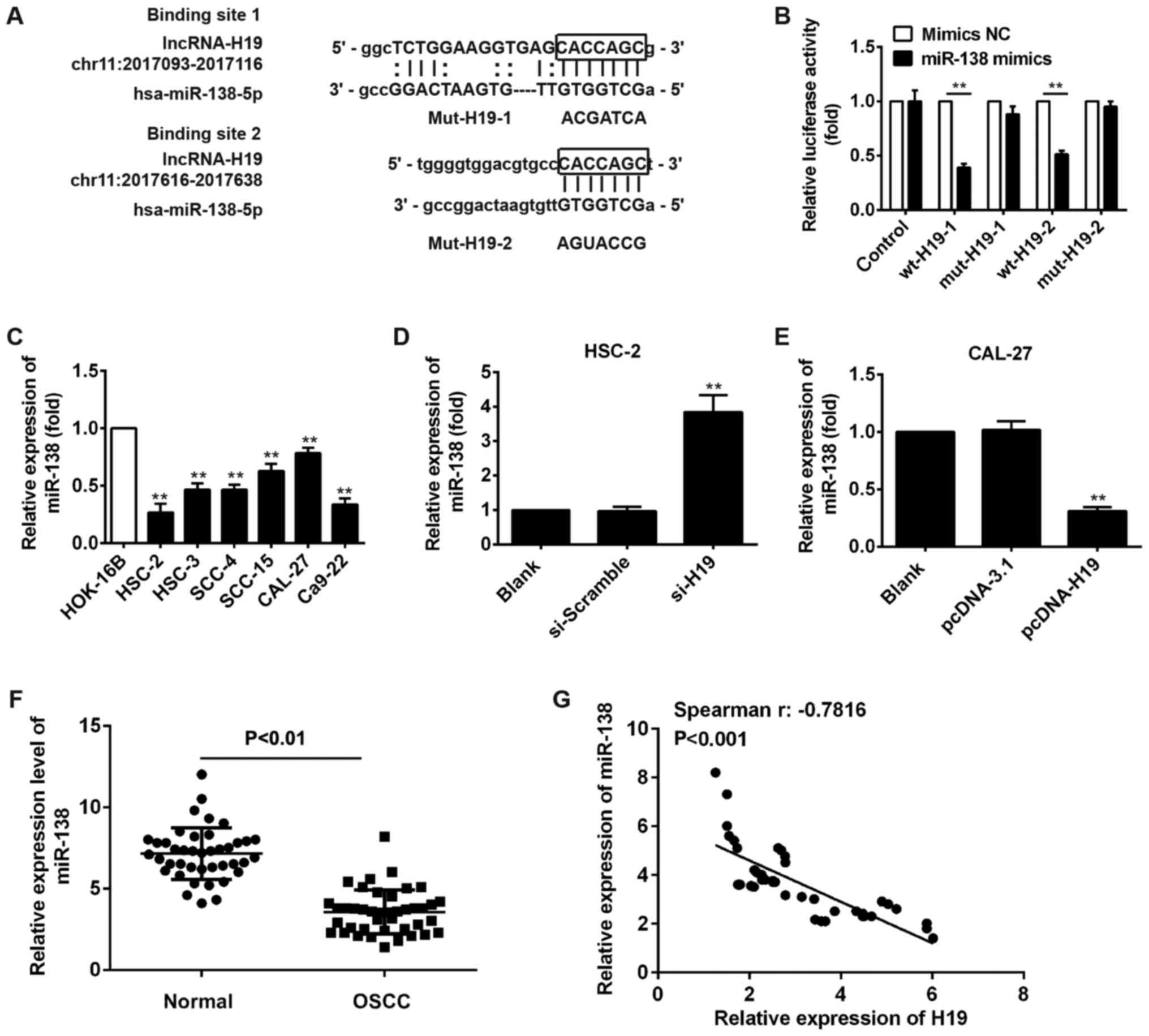
sites for H19 in the miR-138 sequence are illustrated in Fig. 5A. To examine whether H19 directly
targets miR-138, we conducted a luciferase assay. As shown in
Fig. 5B, luciferase activity was
significantly inhibited when wt‑H19‑1 or wt-H19-2 was
co-transfected with miR-138 mimics compared with that after mimic
NC co-transfection, whereas the inhibitory effect was abolished
when the H19 3′-UTR was mutated. This indicated that miR-138
probably interacted with H19.
It has been found that miR-138 functions as a tumor
suppressor in human OSCC (23).
Thus, we first measured miR-138 expression in 6 OSCC cell lines
(HSC-2, HSC-3, SCC-4, SCC-15, CAL-27 and Ca9-22) and a normal oral
mucosa cell line (HOK-16B). Consistent with the findings previous
of that study, the expression of miR-138 was also expressed at low
levels in the OSCC cell lines, particularly in the HSC-2 and Ca9-22
cells (Fig. 5C). To further
determine whether H19 affects OSCC cell proliferation and
metastasis by regulating miR-138 expression, we measured miR-138
expression after the silencing or overexpression of H19 in HSC-2
and CAL-27 cells, and the results of RT-qPCR indicated that miR-138
expression was upregulated after the knockdown of H19 in the HSC-2
cells (Fig. 5D), whereas the
miR-138 level was downregulated after H19 was overexpressed in the
CAL-27 cells (Fig. 5E).
Furthermore, we detected miR-138 expression in 42 pairs of OSCC
tissues and normal tissues. Consistent with the results obtained
with the OSCC cell lines, the expression of miR-138 was
downregulated in the OSCC tissues (Fig. 5F), and an inverse correlation was
observed between the expression of miR-138 and the expression of
H19 (Fig. 5G). All these data
suggest that H19 negatively regulates the expression of miR-138 in
OSCC.
Suppression of miR-138 attenuates the
si-H19-induced inhibitory effects on OSCC cells by targeting
EZH2
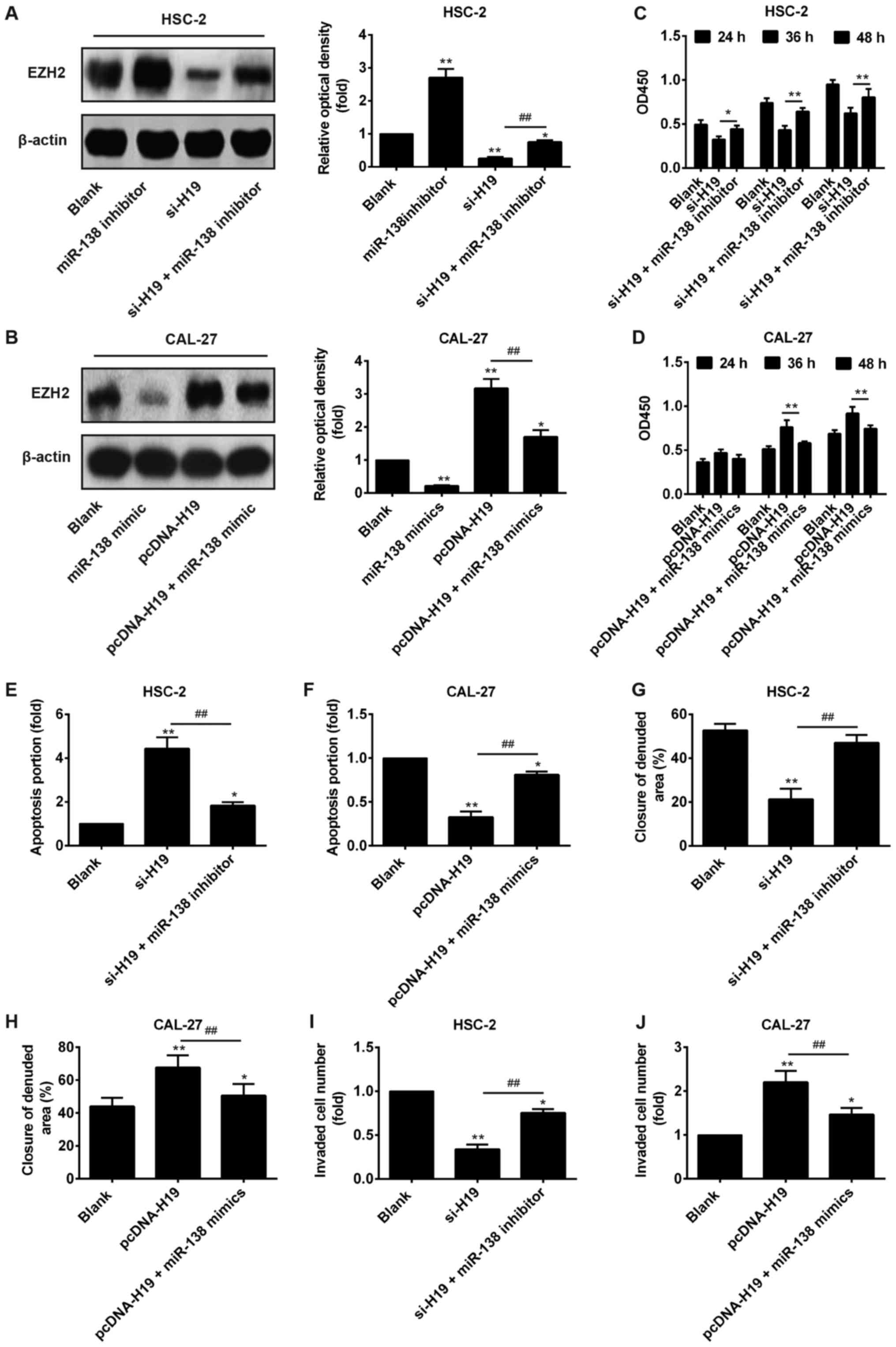
Recent findings have demonstrated that EZH2 acts as
an oncogene and correlates with the malignant potential and a poor
prognosis in a wide range of cancer types, including OSCC (30–32).
Importantly, EZH2 has been identified as a target of miR‑138 in
several cancer cells (33–35). Therefore, we sought to determine
whether H19 functions as a ceRNA for miR-138 to regulate the
expression of EZH2 in OSCC. Western blot analysis revealed that the
knockdown of H19 in the HSC‑2 cells significantly decreased the
expression levels of EZH2, while transfection with miR‑138
inhibitor restored EZH2 expression in the OSCC cells in which H19
was knocked down (Fig. 6A). By
contrast, the overexpression of H19 in the CAL-27 cells increased
the expression levels of EZH2, while transfection with miR‑138
mimic inhibited the promotion of EZH2 expression in the cells
overexpressing H19 (Fig. 6B).
These results suggest that H19 regulates the expression of the
oncogene EZH2 by competing with miR-138 in OSCC cells.
In order to analyze the importance of miR‑138 in
H19-mediated OSCC proliferation, apoptosis and invasion, we knocked
down the expression of miR-138 in the HSC-2 cells transfected with
si-H19 and overexpressed its expression in the CAL-27 cells
transfected with pcDNA-H19, separately. The results revealed that
miR-138 knockdown blocked the inhibitory effects of H19 on cell
proliferation, apoptosis and invasion (Fig. 6C–J), suggesting that the effects of
H19 on OSCC growth and invasion are partially mediated by miR-138.
Taken together, these data indicated that the effects of H19 on the
growth and metastasis of OSCC are partially mediated by regulating
the expression of miR-138.
Discussion
In the present study, we found that lncRNA H1 was
upregulated in OSCC tissues and cell lines and that a high H19
expression was associated with a poor clinical outcome. Moreover,
the knockdown of H19 inhibited OSCC cell proliferation, migration
and invasion, induced cell apoptosis and decreased the tumor growth
in vivo. Mechanistically, we demonstrated that H19 affects
the the biological characteristics of OSCC cells by positively
modulating EZH2 expression through competition for miR-138.
Collectively, our results demonstrated the roles and functional
mechanisms of H19 in OSCC and provide novel insight into potential
therapeutic targets for OSCC.
Recent experimental studies have demonstrated that
lncRNAs play various roles in tumorigenesis, including OSCC
(36–40). For example, HOX transcript
antisense RNA (HOTAIR) has been reported to be upregulated in OSCC
and its expression has been shown to be associated with the
metastasis and poor prognosis of OSCC (41). Metastasis associated lung
adenocarcinoma transcript 1 (MALAT1) is another reported lncRNA,
which contributes to EMT-mediated metastasis in OSCC by modulating
the activation of β-catenin and NF-κB pathways (42). However, the roles of lncRNAs in
OSCC remain largely unknown. In this study, we analyzed and
validated a list of significantly dysregulated lncRNAs in OSCC
tissues by retrieving the microarray data in the GEO dataset
(accession no. GSE3524). In this study, we found that lncRNA H1 was
one of the most significantly differentially expressed lncRNA.
Moreover, a high level of H19 positively correlated with clinical
stages and was identified as a prognostic parameter for patient
survival. These data indicate that H19 may serve as a biomarker for
the diagnosis and prognosis of OSCC.
A large body of evidence has indicated that H19 is
involved in cancer invasion and metastasis. In esophageal squamous
cell carcinoma (ESCC), H19 has been shown to be upregu-lated and to
promote cell proliferation and metastasis (43). Xu et al found that H19
functioned as a marker of poor prognosis in cholangiocarcinoma
(CCA) and H19 enhanced cell migration and invasion by affecting EMT
(44). H19 can also activate
Wnt/β-catenin signaling to affect cell proliferation and metastasis
in bladder cancer (45). In this
study, we proved that the knockdown of H19 inhibited OSCC cell
proliferation, migration and invasion, induced cell apoptosis,
arrested the cells in the G0/G1 phase and decreased the tumor
volume in vivo. Accordingly, apoptosis was increased,
conferred by the upregulation of cleaved caspase-3, cleaved PARP
and Bax. Moreover, we found that the ectopic expression of H19
decreased the expression of E‑cadherin and ZEB1, and increased the
expression of vimentin and N-cadherin in OSCC cells, which
suggested that H19 may promote OSCC cell invasion by inducing EMT.
Therefore, these data suggest that H19 may serve as an oncogene
that promotes OSCC malignant progression.
Recently, increasing evidence has indicated that
lncRNAs function as ceRNAs to silence target mRNAs by sponging
target miRNAs (46,47). Sui et al found that lncRNA
GIHCG promoted hepatocellular carcinoma progression by
epige-netically regulating miR-200b/a/429 (48). Another study demonstrated that
lncRNA PVT1 promoted cervical cancer progression through the
silencing of miR-200b (49).
Similarly, lncRNA H1 competitively binds miR-17-5p to regulate YES1
expression in thyroid cancer (50). In this study, we demonstrated that
H19 directly targeted miR-138 by bioinformatics analysis and
luciferase reporter assays. We also confirmed that miR‑138 was
significantly decreased in OSCC tissues and inversely correlated
with the expression level of H19. However, the ceRNA mechanisms for
H19 deregulation in OSCC have not been thoroughly elucidated.
Previous studies have indicated that miR-138 plays
critical roles in various types of cancer by targeting EZH2. For
example, miR-138 acts as a tumor suppressor miRNA in human clear
cell renal cell carcinoma (ccRCC), induces SN-12 cell senescence by
downregulating EZH2 expression (34). The study by Zhang et al
demonstrated that miR-138 inhibited tumor growth through the
repression of EZH2 in non‑small cell lung cancer (51). A recent study identified EZH2 as a
target of miR-138 in osteosarcoma cells (33). In addition, Li et al found
that lncRNA H1 regulated EZH2 expression by interacting with
miR-630 and promoted cell invasion in nasopharyngeal carcinoma
(52). Therefore, it was
hypothesized that H19 may also serve as a ceRNA to regulate EZH2
expression by sponging miR‑138. Consistent with the findings of
previous studies, we confirmed that H19 regulated the expression of
EZH2, and that miR‑138 attenuated the effects of H19 on the
expression of EZH2 in OSCC cells. Notably, all the effects of H19
on the biological characteristics of the OSCC cells were blocked by
miR-138. Taken together, these data strongly suggest that lncRNA H1
functions as a ceRNA for miR-138 in OSCC.
In this study, the detection of H19 level in OSCC
cell lines indicated that H19 had the highest level in HSC-2 cells
and the lowest level in CAL-27 cells. Therefore, the HSC-2 cells
were selected for the loss-of-function experiments and the CAL-27
cells for gain-of-function experiments. The results revealed that
the knockdown of H19 inhibited OSCC cell proliferation and
invasion, and induced cell apoptosis, whereas the overexpression of
H19 had an opposite result. In addition, the expression of H19 in 6
OSCC cell lines was markedly upregulated compared with that in the
HOK-16B cells. However, we did not find OSCC tumor cells in which
the expression of H19 was similar to that in the HOK-16B cells. In
the future, we aim to find a cell line with which to explore
whether this approach would yield the same results in normal
H-19-expressing cells in which H19 is knocked down or
overexpressed.
In conclusion, in this study, demonstrate that H19
promotes EZH2 expression by competitively binding miR‑138,
contributing to the induction of the EMT process in OSCC. We also
identified the H19/miR‑138/EZH2 axis as a novel signaling network
in OSCC, which may provide a novel therapeutic strategy for the
targeted treatment of OSCC.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma - an update. CA Cancer J
Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kang YY, Sun FL, Zhang Y and Wang Z: SIRT1
acts as a potential tumor suppressor in oral squamous cell
carcinoma. J Chin Med Assoc. S1726–4901. (17): 30270–8.
2017.PubMed/NCBI
|
|
4
|
Sharma A, Boaz K and Natarajan S:
Understanding patterns of invasion: A novel approach to assessment
of podoplanin expression in prediction of lymph node metastasis in
OSCC. Histopathology. 2017. View Article : Google Scholar
|
|
5
|
Rao SJ, Rao JBM and Rao PJ:
Immunohistochemical analysis of stromal fibrocytes and
myofibroblasts to envision the invasion and lymph node metastasis
in oral squamous cell carcinoma. J Oral Maxillofac Pathol.
21:218–223. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al
RIKEN Genome Exploration Research Group and Genome Science Group
(Genome Network Project Core Group): The transcriptional landscape
of the mammalian genome. Science. 309:1559–1563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krzyzanowski PM, Muro EM and
Andrade‑Navarro MA: Computational approaches to discovering
noncoding RNA. Wiley Interdiscip Rev RNA. 3:567–579. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Zhao Z, Feng W, Ye Z, Dai W, Zhang
C, Peng J and Wu K: Long non-coding RNA TUG1 promotes colorectal
cancer metastasis via EMT pathway. Oncotarget. 7:51713–51719.
2016.PubMed/NCBI
|
|
11
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ariel I, Ayesh S, Perlman EJ, Pizov G,
Tanos V, Schneider T, Erdmann VA, Podeh D, Komitowski D, Quasem AS,
et al: The product of the imprinted H19 gene is an oncofetal RNA.
Mol Pathol. 50:34–44. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu‑lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan
Z and Ai K: H19 promotes pancreatic cancer metastasis by
derepressing let-7′s suppression on its target HMGA2-mediated EMT.
Tumour Biol. 35:9163–9169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al: The lncRNA H1
promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo QY, Wang H and Wang Y: lncRNA H1
polymorphisms associated with the risk of OSCC in Chinese
population. Eur Rev Med Pharmacol Sci. 21:3770–3774.
2017.PubMed/NCBI
|
|
19
|
Zhang DM, Lin ZY, Yang ZH, Wang YY, Wan D,
Zhong JL, Zhuang PL, Huang ZQ, Zhou B and Chen WL: IncRNA H1
promotes tongue squamous cell carcinoma progression through
β-catenin/GSK3β/EMT signaling via association with EZH2. Am J
Transl Res. 9:3474–3486. 2017.
|
|
20
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zeng G, Xun W, Wei K, Yang Y and Shen H:
MicroRNA‑27a-3p regulates epithelial to mesenchymal transition via
targeting YAP1 in oral squamous cell carcinoma cells. Oncol Rep.
36:1475–1482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu B, Lei D, Wang L, Yang X, Jia S, Yang
Z, Shan C, Yang X, Zhang C and Lu B: miRNA‑101 inhibits oral
squamous‑cell carcinoma growth and metastasis by targeting zinc
finger E‑box binding homeobox 1. Am J Cancer Res. 6:1396–1407.
2016.
|
|
23
|
Xu R, Zeng G, Gao J, Ren Y, Zhang Z, Zhang
Q, Zhao J, Tao H and Li D: miR-138 suppresses the proliferation of
oral squamous cell carcinoma cells by targeting Yes-associated
protein 1. Oncol Rep. 34:2171–2178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Toruner GA, Ulger C, Alkan M, Galante AT,
Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN,
et al: Association between gene expression profile and tumor
invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet.
154:27–35. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Kuo HF, Liu PL, Chong IW, Liu YP, Chen YH,
Ku PM, Li CY, Chen HH, Chiang HC, Wang CL, et al: Pigment
epithelium-derived factor mediates autophagy and apoptosis in
myocardial hypoxia/reoxygenation injury. PLoS One. 11:e01560592016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li YC, Bu LL, Mao L, Ma SR, Liu JF, Yu GT,
Deng WW, Zhang WF and Sun ZJ: SATB1 promotes tumor metastasis and
invasiveness in oral squamous cell carcinoma. Oral Dis. 23:247–254.
2017. View Article : Google Scholar
|
|
28
|
Qu J, Li M, Zhong W and Hu C: Competing
endogenous RNA in cancer: A new pattern of gene expression
regulation. Int J Clin Exp Med. 8:17110–17116. 2015.
|
|
29
|
Wang H, Shen Q, Zhang X, Yang C, Cui S,
Sun Y, Wang L, Fan X and Xu S: The long non-coding RNA XIST
controls non-small cell lung cancer proliferation and invasion by
modulating miR-186-5p. Cell Physiol Biochem. 41:2221–2229. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sauvageau M and Sauvageau G: Polycomb
group proteins: Multi-faceted regulators of somatic stem cells and
cancer. Cell Stem Cell. 7:299–313. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Su KJ, Lin CW, Chen MK, Yang SF and Yu YL:
Effects of EZH2 promoter polymorphisms and methylation status on
oral squamous cell carcinoma susceptibility and pathology. Am J
Cancer Res. 5:3475–3484. 2015.
|
|
32
|
Kidani K, Osaki M, Tamura T, Yamaga K,
Shomori K, Ryoke K and Ito H: High expression of EZH2 is associated
with tumor proliferation and prognosis in human oral squamous cell
carcinomas. Oral Oncol. 45:39–46. 2009. View Article : Google Scholar
|
|
33
|
Zhu Z, Tang J, Wang J, Duan G, Zhou L and
Zhou X: miR‑138 acts as a tumor suppressor by targeting EZH2 and
enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS
One. 11:e01500262016. View Article : Google Scholar
|
|
34
|
Liang J, Zhang Y, Jiang G, Liu Z, Xiang W,
Chen X, Chen Z and Zhao J: miR‑138 induces renal carcinoma cell
senescence by targeting EZH2 and is downregulated in human clear
cell renal cell carcinoma. Oncol Res. 21:83–91. 2013. View Article : Google Scholar
|
|
35
|
Liu Q, Huang J, Zhou N, Zhang Z, Zhang A,
Lu Z, Wu F and Mo YY: lncRNA loc285194 is a p53-regulated tumor
suppressor. Nucleic Acids Res. 41:4976–4987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gibb EA, Vucic EA, Enfield KS, Stewart GL,
Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S,
Brown CJ, et al: Human cancer long non-coding RNA transcriptomes.
PLoS One. 6:e259152011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meng J, Li P, Zhang Q, Yang Z and Fu S: A
four‑long non‑coding RNA signature in predicting breast cancer
survival. J Exp Clin Cancer Res. 33:842014. View Article : Google Scholar
|
|
39
|
Zhou M, Zhao H, Wang Z, Cheng L, Yang L,
Shi H, Yang H and Sun J: Identification and validation of potential
prognostic lncRNA biomarkers for predicting survival in patients
with multiple myeloma. J Exp Clin Cancer Res. 34:1022015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang YT, Wang YF, Lai JY, Shen SY, Wang F,
Kong J, Zhang W and Yang HY: Long non-coding RNA UCA1 contributes
to the progression of oral squamous cell carcinoma by regulating
the WNT/β-catenin signaling pathway. Cancer Sci. 107:1581–1589.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren
Y, Wu Y, Mei M, Zhang L and Wang X: Long non coding RNA MALAT1
promotes tumor growth and metastasis by inducing
epithelial-Mesenchymal transition in oral squamous cell carcinoma.
Sci Rep. 5:159722015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tan D, Wu Y, Hu L, He P, Xiong G, Bai Y
and Yang K: Long noncoding RNA H1 is up-regulated in esophageal
squamous cell carcinoma and promotes cell proliferation and
metastasis. Dis Esophagus. 30:1–9. 2017.
|
|
44
|
Xu Y, Wang Z, Jiang X and Cui Y:
Overexpression of long noncoding RNA H1 indicates a poor prognosis
for cholangiocarcinoma and promotes cell migration and invasion by
affecting epithelial-mesenchymal transition. Biomed Pharmacother.
92:17–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non‑coding RNA H1 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E‑cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sui CJ, Zhou YM, Shen WF, Dai BH, Lu JJ,
Zhang MF and Yang JM: Long noncoding RNA GIHCG promotes
hepatocellular carcinoma progression through epigenetically
regulating miR-200b/a/429. J Mol Med (Berl). 94:1281–1296. 2016.
View Article : Google Scholar
|
|
49
|
Zhang S, Zhang G and Liu J: Long noncoding
RNA VT1 promotes cervical cancer progression through epigenetically
silencing miR-200b. APMIS. 124:649–658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu L, Yang J, Zhu X, Li D, Lv Z and Zhang
X: Long noncoding RNA H1 competitively binds miR-17-5p to regulate
YES1 expression in thyroid cancer. FEBS J. 283:2326–2339. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang H, Zhang H, Zhao M, Lv Z, Zhang X,
Qin X, Wang H, Wang S, Su J, Lv X, et al: miR-138 inhibits tumor
growth through repression of EZH2 in non‑small cell lung cancer.
Cell Physiol Biochem. 31:56–65. 2013. View Article : Google Scholar
|
|
52
|
Li X, Lin Y, Yang X, Wu X and He X: Long
noncoding RNA H1 regulates EZH2 expression by interacting with
miR‑630 and promotes cell invasion in nasopharyngeal carcinoma.
Biochem Biophys Res Commun. 473:913–919. 2016. View Article : Google Scholar : PubMed/NCBI
|