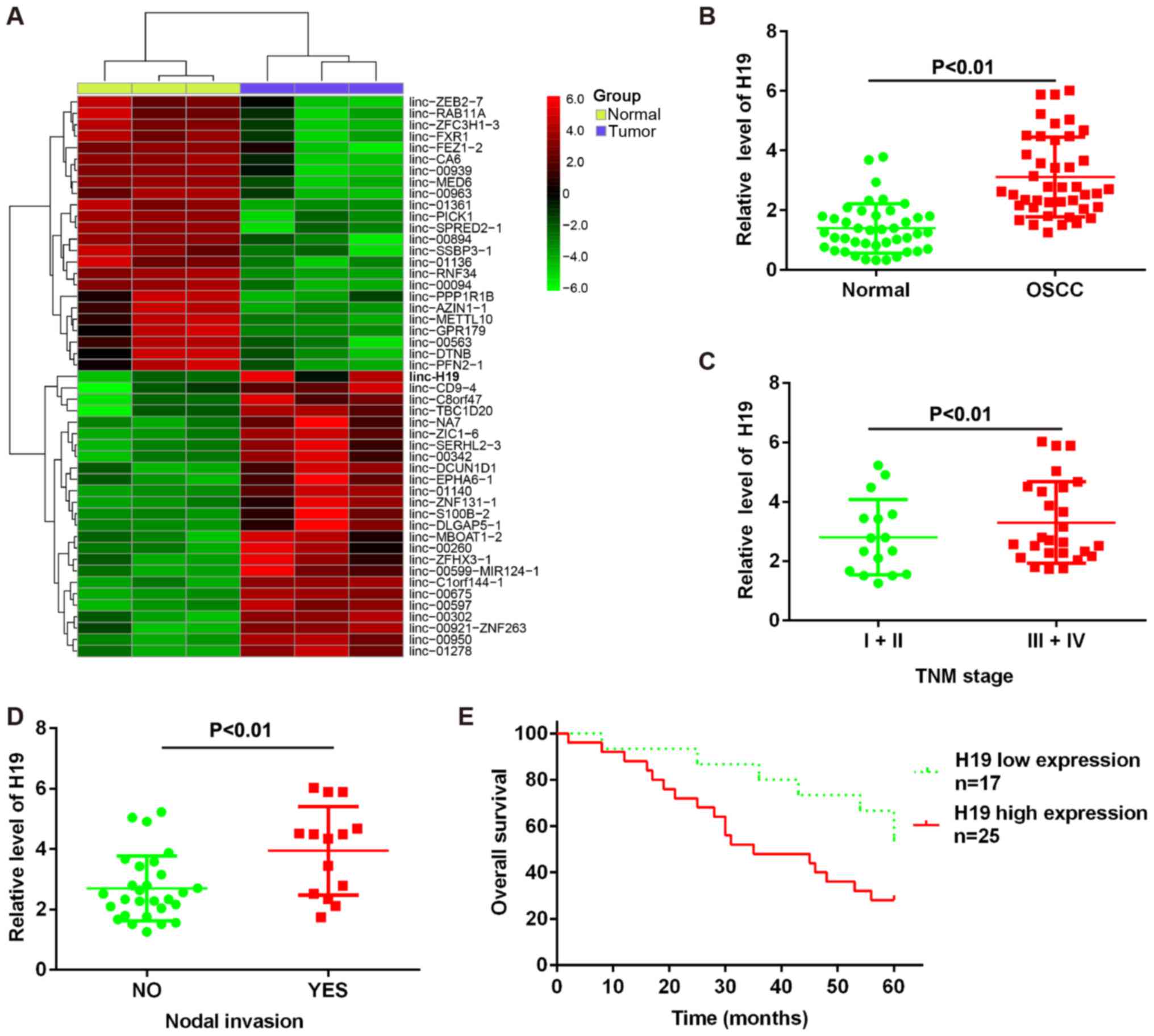
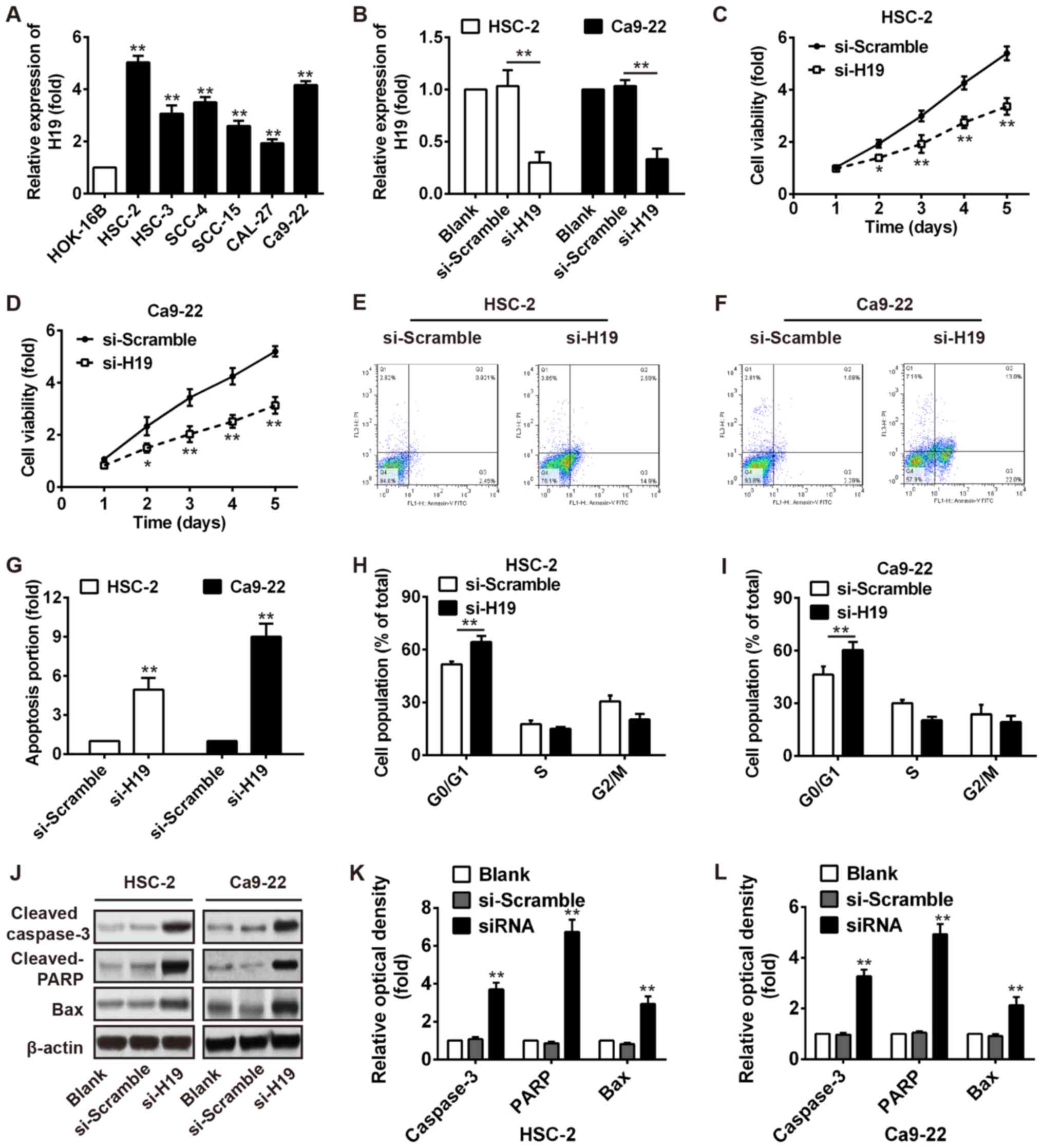
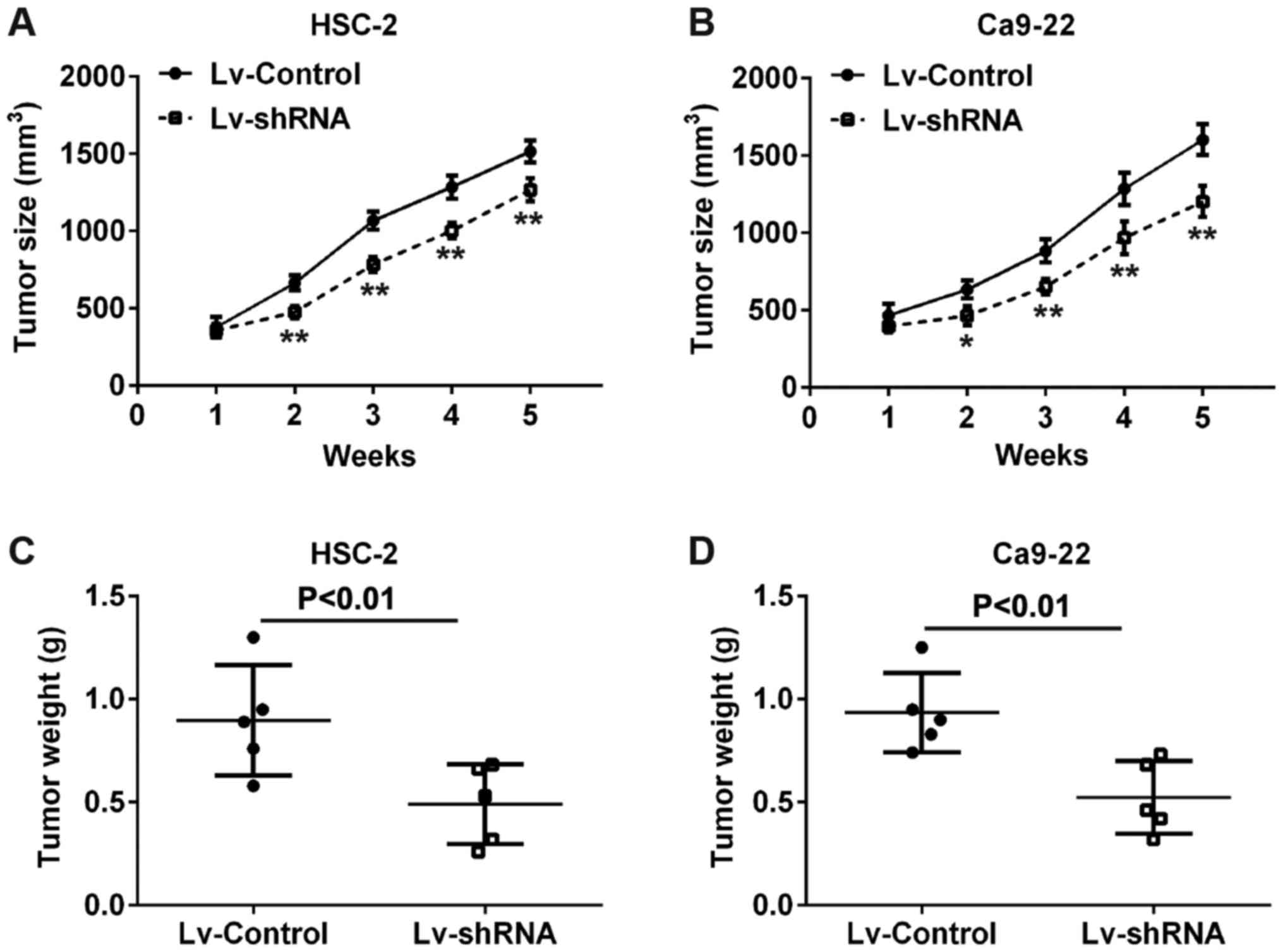
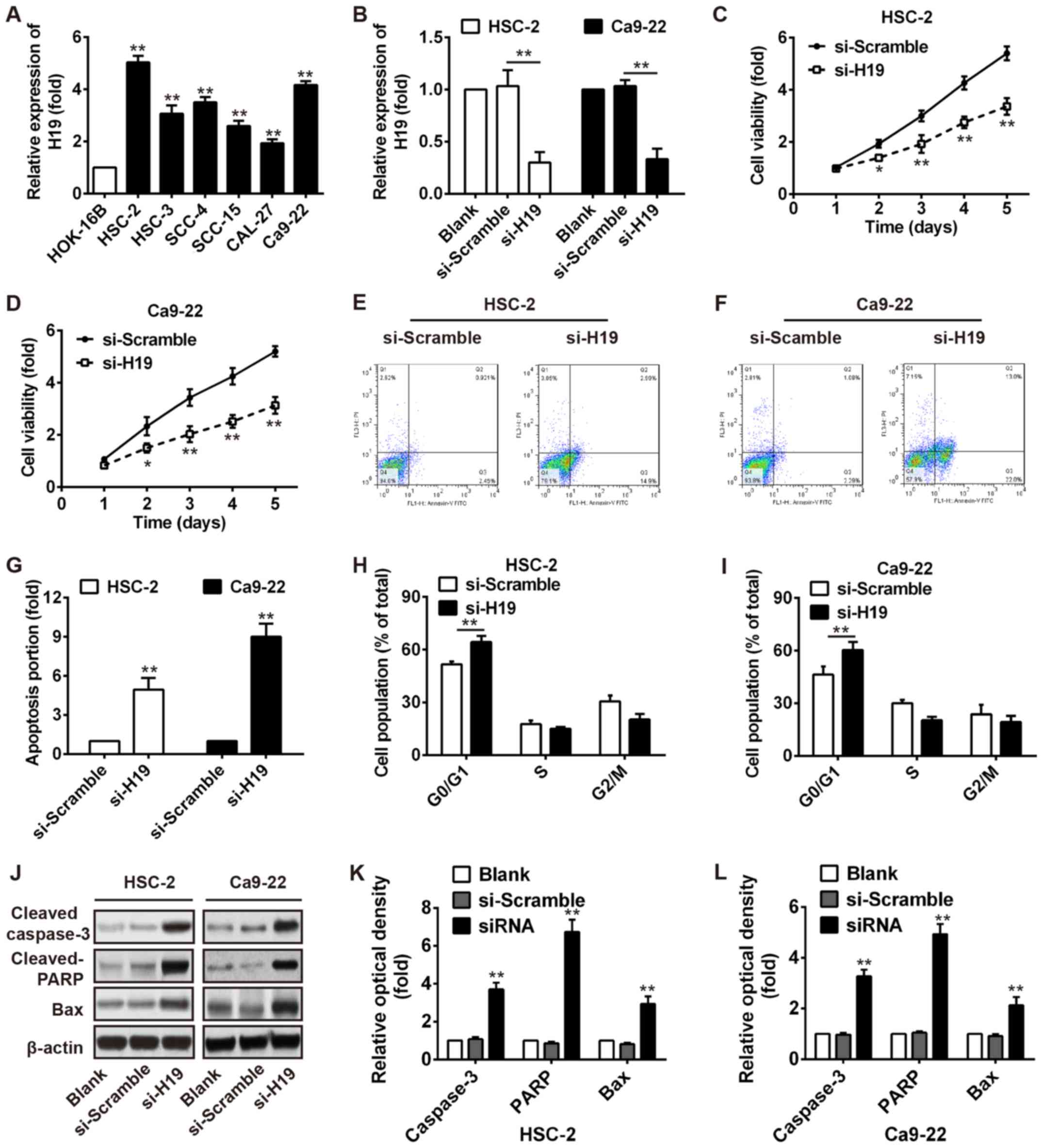
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chi AC, Day TA and Neville BW: Oral cavity
and oropharyngeal squamous cell carcinoma - an update. CA Cancer J
Clin. 65:401–421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kang YY, Sun FL, Zhang Y and Wang Z: SIRT1
acts as a potential tumor suppressor in oral squamous cell
carcinoma. J Chin Med Assoc. S1726–4901. (17): 30270–8.
2017.PubMed/NCBI
|
|
4
|
Sharma A, Boaz K and Natarajan S:
Understanding patterns of invasion: A novel approach to assessment
of podoplanin expression in prediction of lymph node metastasis in
OSCC. Histopathology. 2017. View Article : Google Scholar
|
|
5
|
Rao SJ, Rao JBM and Rao PJ:
Immunohistochemical analysis of stromal fibrocytes and
myofibroblasts to envision the invasion and lymph node metastasis
in oral squamous cell carcinoma. J Oral Maxillofac Pathol.
21:218–223. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al
RIKEN Genome Exploration Research Group and Genome Science Group
(Genome Network Project Core Group): The transcriptional landscape
of the mammalian genome. Science. 309:1559–1563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krzyzanowski PM, Muro EM and
Andrade‑Navarro MA: Computational approaches to discovering
noncoding RNA. Wiley Interdiscip Rev RNA. 3:567–579. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Zhao Z, Feng W, Ye Z, Dai W, Zhang
C, Peng J and Wu K: Long non-coding RNA TUG1 promotes colorectal
cancer metastasis via EMT pathway. Oncotarget. 7:51713–51719.
2016.PubMed/NCBI
|
|
11
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ariel I, Ayesh S, Perlman EJ, Pizov G,
Tanos V, Schneider T, Erdmann VA, Podeh D, Komitowski D, Quasem AS,
et al: The product of the imprinted H19 gene is an oncofetal RNA.
Mol Pathol. 50:34–44. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Berteaux N, Lottin S, Monté D, Pinte S,
Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T and
Adriaenssens E: H19 mRNA-like noncoding RNA promotes breast cancer
cell proliferation through positive control by E2F1. J Biol Chem.
280:29625–29636. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu‑lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan
Z and Ai K: H19 promotes pancreatic cancer metastasis by
derepressing let-7′s suppression on its target HMGA2-mediated EMT.
Tumour Biol. 35:9163–9169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al: The lncRNA H1
promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo QY, Wang H and Wang Y: lncRNA H1
polymorphisms associated with the risk of OSCC in Chinese
population. Eur Rev Med Pharmacol Sci. 21:3770–3774.
2017.PubMed/NCBI
|
|
19
|
Zhang DM, Lin ZY, Yang ZH, Wang YY, Wan D,
Zhong JL, Zhuang PL, Huang ZQ, Zhou B and Chen WL: IncRNA H1
promotes tongue squamous cell carcinoma progression through
β-catenin/GSK3β/EMT signaling via association with EZH2. Am J
Transl Res. 9:3474–3486. 2017.
|
|
20
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zeng G, Xun W, Wei K, Yang Y and Shen H:
MicroRNA‑27a-3p regulates epithelial to mesenchymal transition via
targeting YAP1 in oral squamous cell carcinoma cells. Oncol Rep.
36:1475–1482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu B, Lei D, Wang L, Yang X, Jia S, Yang
Z, Shan C, Yang X, Zhang C and Lu B: miRNA‑101 inhibits oral
squamous‑cell carcinoma growth and metastasis by targeting zinc
finger E‑box binding homeobox 1. Am J Cancer Res. 6:1396–1407.
2016.
|
|
23
|
Xu R, Zeng G, Gao J, Ren Y, Zhang Z, Zhang
Q, Zhao J, Tao H and Li D: miR-138 suppresses the proliferation of
oral squamous cell carcinoma cells by targeting Yes-associated
protein 1. Oncol Rep. 34:2171–2178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Toruner GA, Ulger C, Alkan M, Galante AT,
Rinaggio J, Wilk R, Tian B, Soteropoulos P, Hameed MR, Schwalb MN,
et al: Association between gene expression profile and tumor
invasion in oral squamous cell carcinoma. Cancer Genet Cytogenet.
154:27–35. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Kuo HF, Liu PL, Chong IW, Liu YP, Chen YH,
Ku PM, Li CY, Chen HH, Chiang HC, Wang CL, et al: Pigment
epithelium-derived factor mediates autophagy and apoptosis in
myocardial hypoxia/reoxygenation injury. PLoS One. 11:e01560592016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li YC, Bu LL, Mao L, Ma SR, Liu JF, Yu GT,
Deng WW, Zhang WF and Sun ZJ: SATB1 promotes tumor metastasis and
invasiveness in oral squamous cell carcinoma. Oral Dis. 23:247–254.
2017. View Article : Google Scholar
|
|
28
|
Qu J, Li M, Zhong W and Hu C: Competing
endogenous RNA in cancer: A new pattern of gene expression
regulation. Int J Clin Exp Med. 8:17110–17116. 2015.
|
|
29
|
Wang H, Shen Q, Zhang X, Yang C, Cui S,
Sun Y, Wang L, Fan X and Xu S: The long non-coding RNA XIST
controls non-small cell lung cancer proliferation and invasion by
modulating miR-186-5p. Cell Physiol Biochem. 41:2221–2229. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sauvageau M and Sauvageau G: Polycomb
group proteins: Multi-faceted regulators of somatic stem cells and
cancer. Cell Stem Cell. 7:299–313. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Su KJ, Lin CW, Chen MK, Yang SF and Yu YL:
Effects of EZH2 promoter polymorphisms and methylation status on
oral squamous cell carcinoma susceptibility and pathology. Am J
Cancer Res. 5:3475–3484. 2015.
|
|
32
|
Kidani K, Osaki M, Tamura T, Yamaga K,
Shomori K, Ryoke K and Ito H: High expression of EZH2 is associated
with tumor proliferation and prognosis in human oral squamous cell
carcinomas. Oral Oncol. 45:39–46. 2009. View Article : Google Scholar
|
|
33
|
Zhu Z, Tang J, Wang J, Duan G, Zhou L and
Zhou X: miR‑138 acts as a tumor suppressor by targeting EZH2 and
enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS
One. 11:e01500262016. View Article : Google Scholar
|
|
34
|
Liang J, Zhang Y, Jiang G, Liu Z, Xiang W,
Chen X, Chen Z and Zhao J: miR‑138 induces renal carcinoma cell
senescence by targeting EZH2 and is downregulated in human clear
cell renal cell carcinoma. Oncol Res. 21:83–91. 2013. View Article : Google Scholar
|
|
35
|
Liu Q, Huang J, Zhou N, Zhang Z, Zhang A,
Lu Z, Wu F and Mo YY: lncRNA loc285194 is a p53-regulated tumor
suppressor. Nucleic Acids Res. 41:4976–4987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gibb EA, Vucic EA, Enfield KS, Stewart GL,
Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S,
Brown CJ, et al: Human cancer long non-coding RNA transcriptomes.
PLoS One. 6:e259152011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meng J, Li P, Zhang Q, Yang Z and Fu S: A
four‑long non‑coding RNA signature in predicting breast cancer
survival. J Exp Clin Cancer Res. 33:842014. View Article : Google Scholar
|
|
39
|
Zhou M, Zhao H, Wang Z, Cheng L, Yang L,
Shi H, Yang H and Sun J: Identification and validation of potential
prognostic lncRNA biomarkers for predicting survival in patients
with multiple myeloma. J Exp Clin Cancer Res. 34:1022015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang YT, Wang YF, Lai JY, Shen SY, Wang F,
Kong J, Zhang W and Yang HY: Long non-coding RNA UCA1 contributes
to the progression of oral squamous cell carcinoma by regulating
the WNT/β-catenin signaling pathway. Cancer Sci. 107:1581–1589.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren
Y, Wu Y, Mei M, Zhang L and Wang X: Long non coding RNA MALAT1
promotes tumor growth and metastasis by inducing
epithelial-Mesenchymal transition in oral squamous cell carcinoma.
Sci Rep. 5:159722015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tan D, Wu Y, Hu L, He P, Xiong G, Bai Y
and Yang K: Long noncoding RNA H1 is up-regulated in esophageal
squamous cell carcinoma and promotes cell proliferation and
metastasis. Dis Esophagus. 30:1–9. 2017.
|
|
44
|
Xu Y, Wang Z, Jiang X and Cui Y:
Overexpression of long noncoding RNA H1 indicates a poor prognosis
for cholangiocarcinoma and promotes cell migration and invasion by
affecting epithelial-mesenchymal transition. Biomed Pharmacother.
92:17–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non‑coding RNA H1 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E‑cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sui CJ, Zhou YM, Shen WF, Dai BH, Lu JJ,
Zhang MF and Yang JM: Long noncoding RNA GIHCG promotes
hepatocellular carcinoma progression through epigenetically
regulating miR-200b/a/429. J Mol Med (Berl). 94:1281–1296. 2016.
View Article : Google Scholar
|
|
49
|
Zhang S, Zhang G and Liu J: Long noncoding
RNA VT1 promotes cervical cancer progression through epigenetically
silencing miR-200b. APMIS. 124:649–658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu L, Yang J, Zhu X, Li D, Lv Z and Zhang
X: Long noncoding RNA H1 competitively binds miR-17-5p to regulate
YES1 expression in thyroid cancer. FEBS J. 283:2326–2339. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang H, Zhang H, Zhao M, Lv Z, Zhang X,
Qin X, Wang H, Wang S, Su J, Lv X, et al: miR-138 inhibits tumor
growth through repression of EZH2 in non‑small cell lung cancer.
Cell Physiol Biochem. 31:56–65. 2013. View Article : Google Scholar
|
|
52
|
Li X, Lin Y, Yang X, Wu X and He X: Long
noncoding RNA H1 regulates EZH2 expression by interacting with
miR‑630 and promotes cell invasion in nasopharyngeal carcinoma.
Biochem Biophys Res Commun. 473:913–919. 2016. View Article : Google Scholar : PubMed/NCBI
|