Introduction
Ovarian cancer is considered to be the most lethal
gynecologic tumor. In China, ~52,100 new cases of ovarian cancer
were diagnosed and ~22,500 mortalities occurred due to ovarian
cancer in 2015 (1). Compared with
China, the incidence and mortality rates in Europe and North
America are higher (2). Epithelial
ovarian cancer is divided into the following five major
pathological subtypes: High-grade serous (70%), low-grade serous
(<5%), endometrioid (10%), clear cell (10%) and mucinous (3%)
(3,4). High-grade serous ovarian carcinoma
(HGSOC) accounts for 70–80% of ovarian cancer-associated
mortalities (5). Therefore,
gaining an improved understanding of the molecular mechanisms
underlying HGSOC is critical.
Forkhead box D1 (FOXD1), also known as FREAC-4,
belongs to the forkhead box family and functions as a transcription
factor (6). According to previous
reports (7,8), it serves an important role in the
formation of the retina and nephrons during embryogenesis. In
addition, downregulation of FOXD1 decreases the reprogramming
efficiency and inhibits the expression of reprogramming-associated
genes in mouse embryonic fibroblasts (9). FOXD1 also serves a role in the
development of different cancers. In lung, breast and brain
cancers, FOXD1 functions as an oncogene and promotes cell
proliferation (10–12). In hepatocellular carcinoma,
microarray analysis has demonstrated that FOXD1 is one of ten
transcription factors involved in tumorigenesis (13). By contrast, FOXD1 may function as a
tumor suppressor in ovarian cancer. Jiang et al (14) analyzed three GSE cohorts (GSE14001,
GSE15578 and GSE12172) and demonstrated that FOXD1 is downregulated
in ovarian cancer. The same study identified FOXD1 as one of the
top 20 differentially expressed genes between abnormal and normal
ovarian epithelial tissues (14).
p21 (also known as WAF1/CIP1 and cyclin-dependent
kinase inhibitor 1A) is a general G1 phase cell cycle inhibitor,
which is regulated by p53-dependent or p53-independent signaling
pathways (15). The function of
p21 as an inhibitor of the cell cycle via p53 was a landmark
discovery in molecular biology in the early 1990s (16,17).
Although p21 is widely known to be a crucial effector of p53 and an
inhibitor of the cell cycle, it is now evident that p21 is
regulated by a number of additional signaling pathways (18,19).
p21 mutations are rare in human cancer (20,21).
Increased expression of p21 has been associated with a favorable
outcome in many cancer types (22–25).
MicroRNAs (miRNAs/miRs) are endogenous short (~22
nucleotide) single-stranded RNAs that regulate the translation of
target genes by promoting mRNA decay. miRNAs recognize target mRNAs
by base-pairing with their complementary seed sequences (typically
nucleotides 2–7) in the 3′-untranslated region (UTR) (26). miRNAs function as tumor suppressors
or oncogenes during carcinogenesis and tumor development.
Specifically, the miR-30 and miR-200 families are known to function
as tumor suppressors in a number of cancer types by inhibiting
tumor cell proliferation and metastasis (27–29);
however, several studies have demonstrated that these miR families
may serve an oncogenic role in ovarian cancer (30–36).
In the present study, the expression of FOXD1 was
analyzed in patients with HGSOC, and its role in the proliferation
of ovarian cancer cells was investigated using in vitro and
in vivo studies. The results indicated that FOXD1 was
downregulated in HGSOC, and it suppressed ovarian cancer cell
proliferation via targeting p21. In addition, the expression of
miR-30a-5p and miR-200a-5p was elevated in patients with HGSOC, and
FOXD1 was demonstrated to be a direct target of these miRNA
sequences.
Materials and methods
Patients and tissue samples
A total of 140 HGSOC tissue specimens were collected
for the purposes of this study. The tissue microarray (TMA)
included 120 HGSOC samples and these tissues were collected from
female patients admitted to Qilu Hospital (Jinan, China) from May
2006 to July 2013. The HGSOC samples (n=20) used for western blot
and reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analyses were collected from patients admitted to Qilu
hospital from September 2014 to July 2015. All HGSOC specimens were
collected from patients with primary ovarian cancer that had not
received chemotherapy prior to surgery. The fresh-frozen normal
fallopian tube (FT, n=11) tissues were obtained from female
patients with benign gynecological tumors that had received a
hysterectomy with bilateral salpingo-oophorectomy at Qilu hospital
from September 2014 to July 2015. The tumor diagnoses were verified
by two gynecological pathologists. Ethical approval was obtained
from Ethics Committee of Shandong University Qilu Hospital (Jinan,
China). All patients provided written informed consent.
Cell lines and culture conditions
293T cells, and the human ovarian cancer cell lines,
A2780 and HO8910, were purchased from the China Center for Type
Culture Collection (Wuhan, China). Human ovarian cancer cell lines,
SKOV3, OVCAR3, CAOV3 and the p53-null H1299 human lung cancer cell
line, were purchased from the American Type Culture Collection
(Manassas, VA, USA). A2780, HO8910 and H1299 cells were cultured in
RPMI-1640 supplemented with 10% fetal bovine serum (FBS; both
purchased from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA); OVCAR3 cells were maintained in RPMI-1640 supplemented with
20% FBS and 0.01 mg/ml bovine insulin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany); SKOV3 cells were cultured in McCoy’s 5A medium
(Thermo Fisher Scientific, Inc.) supplemented with 10% FBS; CAOV3
cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. In
addition, 100 U/ml penicillin and 100 μg/ml streptomycin
(Sigma-Aldrich; Merck KGaA) were added to the complete culture
medium for all cell lines. All cells were cultured at 37°C with 5%
CO2 in a humidified incubator.
RNA extraction and RT-qPCR analysis
Total RNA was extracted from fresh-frozen HGSOC and
FT tissue samples, as well as OVCAR3 cells transfected with
miR-30a-5p/miR-200a-5p mimics or inhibitors, using TRIzol reagent
according to manufacturer’s instructions (Invitrogen; Thermo Fisher
Scientific, Inc.). Total RNA (1,000 ng) was reverse transcribed to
cDNA using the PrimeScript RT reagent kit or the Mir-X miRNA
First-Strand Synthesis kit (both purchased from Takara
Biotechnology, Co., Ltd., Dalian, China) according to
manufacturer’s instructions. qPCR analysis was performed using the
SYBR Premix Ex Taq (Takara Biotechnology, Co., Ltd.). The qPCR
reaction was performed using the StepOnePlus™ Real-Time PCR System
(Thermo Fisher Scientific, Inc.). The thermal cycling parameters
were as follows: 30 sec at 95°C followed by 43 cycles at 95°C for 5
sec and 60°C for 30 sec. The primer sequences employed were as
follows: FOXD1, forward, 5′-GATCTGTGAGTTCATCAGCGGC-3′ and reverse,
5′-TGACGAAGCAGTCGTTGAGCGA-3′; β-actin, forward,
5′-CATGTACGTTGCTATCCAGGC-3′ and reverse,
5′-CTCCTTAATGTCACGCACGAT-3′; miR-30a-5p, forward,
5′-TGTAAACATCCTCGACTGGAAG-3′ and reverse, mRQ 3′ Primer (provided
in Mir-X miRNA First-Strand Synthesis kit; Takara Biotechnology,
Co., Ltd.); miR-200a-5p, forward, 5′-CATCTTACCGGACAGTGCTGGA-3′ and
reverse, mRQ 3′ Primer; U6, forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, mRQ 3′ Primer. The
expression of β-actin and U6 was used as the internal control. Data
analysis was performed using the 2−ΔΔCq method (37).
Western blotting
Protein from fresh-frozen HGSOC and FT tissue
samples was isolated using TRIzol reagent according to
manufacturer’s instructions (Invitrogen; Thermo Fisher Scientific,
Inc.). The HGSOC and FT tissue samples were divided into smaller
sections (~0.1 g) and lysed with TRIzol for 10 min at room
temperature. The lysate was centrifuged at 12,000 × g for 10 min at
4°C and the phenol-ethanol supernatant was incubated with 100%
isopropanol for 10 min at room temperature. The mixture was then
centrifuged again at 12,000 × g for 10 min at 4°C to pellet the
protein. The protein pellet was washed twice with 0.3 M guanidine
hydrochloride and once with 100% ethanol. Then the protein pellet
was solubilized in 1% SDS. For isolating protein from cells, the
cells were first washed three times with cold PBS and lysed using
radioimmunoprecipitation assay buffer on ice for 30 min. The
protein concentration was determined using a BCA assay (Merck
KGaA). Protein samples (40 μg) were separated by 12%
SDS-PAGE and electro-transferred onto polyvinylidene fluoride
membranes using the semi-dry transfer method. The membranes were
blocked for 1 h with 5% non-fat milk at room temperature, and then
were incubated at 4°C overnight with the following primary
antibodies: Anti-FOXD1 (dilution, 1:500; cat. no. ab179940; Abcam,
Cambridge, USA), anti-p21 (dilution, 1:1,000; cat. no. 2947S; CST
Biological Reagents Co., Ltd., Shanghai, China), anti-β-actin
(dilution, 1:5,000; cat. no. ab8226; Abcam). The membranes were
subsequently washed with Tris-buffered saline with 0.1% Tween-20,
and then incubated with the secondary horseradish peroxidase
(HRP)-conjugated anti-mouse IgG antibody (dilution, 1:8,000; cat.
no. 074-1806) or anti-rabbit IgG antibody (dilution, 1:6,000; cat
no. 5220-0336) (both from Kirkegaard & Perry Laboratories,
Inc., Gaithersburg, MD, USA) for 2 h at room temperature. The
signals were detected using an enhanced chemiluminescence system
(GE Healthcare Life Sciences, Little Chalfont, UK). β-actin was
used as an endogenous control.
Plasmid construction, lentivirus
production and infection
For plasmid construction, the coding DNA sequence of
FOXD1 was cloned into the pLenti-C-Myc-DDK-IRES-Puro tagged vector
(PCMV; OriGene Technologies, Inc., Rockville, MD, USA). Short
hairpin RNA (shRNA) targeting FOXD1 was purchased from
Sigma-Aldrich; Merck KGaA, and cloned into the pLKO.1 vector
(Addgene, Inc., Cambridge, MA, USA). A scrambled shRNA control
(5′-CCTAAGGTTAAGTCGCCCTCGCTC GAGCGAGGGCGACTTAACCTTAGG-3′) was
cloned into the pLKO.1 vector and used as a negative control. The
lentiviral packaging and envelope plasmids used were pMD2.G and
psPAX2 (Addgene, Inc.). For lentivirus production, 293T cells were
seeded at a density of 4×106 in 10 cm culture dishes and
cultured for 16 h. The PCMV vector and lentiviral packaging and
envelope plasmids were co-transfected into 293T cells using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.).
For stable transfection, cells (A2780, HO8910, SKOV3, CAOV3, OVCAR3
and H1299) were infected with lentivirus at a multiplicity of
infection of 50 for 24 h, and the cells were selected in medium
containing puromycin (2 μg/ml) for 2 weeks.
Transfection of miRNA mimics and
inhibitors
OVCAR3 cells were seeded in 6 cm dishes at a density
of 5×105/dish at 24–36 h prior to transfection.
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) was
employed to transfect the cells according to manufacturer’s
instructions. The concentration of miRNA mimics and inhibitors used
were as follows: 80 nM miR-30a-5p/miR-200a-5p mimics; 80 nM mimics
control; 100 nM miR-30a-5p/miR-200a-5p inhibitors; and 100 nM
inhibitors control. The medium was refreshed at 6 h following
transfection. The sequences of miRNA mimics and inhibitors was as
follows: miR-30a-5p mimics, 5′-UGUAAACAUCCUCGACUGGAAG-3′;
miR-30a-5p inhibitors, 5′-CUUCCAGUCGAGGAUGUUUACA; miR-200a-5p
mimics, 5′-CAUCUUACCGGACAGUGCUGGA-3′; miR-200a-5p inhibitors,
5′-UCCAGCACUGUCCGGUAAGAUG-3′; miRNA mimics negative control,
5′-UUCUCCGAACGUGUCACGUTT-3′; miRNA inhibitors negative control,
5′-CAGUACUUUUGUGUAGUACAA-3′. All the miRNA mimics and inhibitors
were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou,
China).
Small interfering (si)RNA
transfection
A2780 cells (2.5×105) were plated in 6 cm
dishes and transfected with p21 siRNA sequences using Lipofectamine
3000 according to the manufacturer’s protocol (Invitrogen; Thermo
Fisher Scientific, Inc.). At 48 h following transfection, the cells
were harvested for MTT and clonogenic assay analysis. The sequences
of the p21 siRNAs were the same as described previously (38,39).
The sequences of the p21 siRNA sequences were as follows: sip21-1,
5′-AGCGAUGGAACUUCGACUUTT-3′; sip21-2,
5′-AAUGGCGGGCUGCAUCCAGGATT-3′; siCtrl, 5′-UUCUCCGAACGUGUCACGUTT-3′
(all purchased from Shanghai GenePharma Co., Ltd., Shanghai,
China).
Clonogenic assay
Cells (n=800–1,000/well) stably expressing FOXD1 or
transiently transfected with p21 siRNA and FOXD1 were plated in
6-well plates and cultured for 2–3 weeks. The colonies were fixed
with 100% methanol for 15 min at room temperature and then stained
with 1% crystal violet for 10 min at room temperature. The number
of colonies was assessed using ImageQuant TL software (version 8.1;
GE Healthcare Life Sciences). The data are presented as the mean ±
standard error of three independent experiments.
Cell proliferation assay
Cell proliferation was assessed using an MTT assay.
A2780 cells (800 cells/well) were first seeded in triplicate wells
of a 96-well plate. MTT reagent (5 mg/ml; cat no. M2128;
Sigma-Aldrich; Merck KGaA) was subsequently added to the wells and
incubated for 4 h. DMSO was subsequently added to dissolve the
formazan crystals and the absorbance at 490 nm was then read.
Dual-luciferase reporter assay
In order to investigate whether FOXD1 may be a
direct target of miR-30a-5p and miR-200-5p, the 3′-UTR of the FOXD1
mRNA sequence containing the putative miR-30a-5p and miR-200a-5p
binding sites were cloned into the pmirGLO Dual-Luciferase miRNA
Target Expression Vector (Promega Corporation, Madison, WI, USA).
The FOXD1 mutant sequences were constructed by Sangon Biotech, Co.,
Ltd. (Shanghai, China) and cloned into pmirGLO vector. 293T cells
were co-transfected with miR-30a-5p or miR-200a-5p mimics and the
pmirGLO vector containing either the wild-type or mutant FOXD1
sequence using Lipofectamine 3000 according to the manufacturer’s
protocol (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h
following transfection, luciferase activity was measured using the
Dual-Glo Luciferase Assay System (Promega Corporation).
To investigate FOXD1 binding sites in the p21
promoter, three predicted wild-type binding sites of FOXD1 were
separately cloned into the pGL4.26[luc2/minP/Hygro] vector (Promega
Corporation), and the mutant binding sites were constructed by
Sangon Biotech, Co., Ltd. and cloned into the pGL4.26 vector. The
pGL4.74[hRluc/TK] vector (Promega Corporation) was used as an
endogenous control. 293T cells (4×104/well) were seeded
in a 96-well plate and transfected with vectors using Lipofectamine
3000 according to the manufacturer’s protocol (Invitrogen; Thermo
Fisher Scientific, Inc.). Cells were transfected with vectors at
the following concentration ratio: PCMV (75 ng): pGL4.26 (25 ng):
pGL4.74 (2.5 ng). At 48 h following transfection, luciferase
activity was measured.
Immunohistochemical staining
The TMA of HGSOC samples were incubated at 65°C for
30 min and deparaffinized in xylene immediately. The TMA was
subsequently rehydrated in a graded ethanol series. Antigen
retrieval was performed in citric acid buffer (pH 6.0) by heat
treatment (98°C) for 15 min. This assay was performed using the
SP-9000 IHC reagent kit (cat. no. SP-9000; OriGene Technologies,
Inc.). The samples were blocked in 1.5% goat serum from the SP-9000
IHC reagent kit at 37°C for 20 min, and the TMA was then incubated
with primary antibodies against FOXD1 (dilution, 1:1,000; cat. no.
ab179940; Abcam) at 4°C for 14 h. The TMA was subsequently washed
and incubated with a HRP-conjugated secondary antibody from the
SP-9000 IHC reagent kit (OriGene Technologies, Inc.) for 30 min at
37°C. Staining was detected using a 3,3′-diaminobenzidine detection
system. The Eclipse Ni-E microscope (Nikon Corporation, Tokyo,
Japan) was used to visualize the stained tissue sections. As
transcription factors modulate gene expression by binding to the
promoter of target genes in the nucleus, the expression of FOXD1 in
the nucleus was determined in the present study (40–42).
The final score of each sample was decided by two pathologists and
was dependent on the extent and intensity of staining. The
intensity of staining was evaluated and scored as follows: 0,
negative; 1, weak; 2, moderate; or 3, strong. The proportion of
positively stained cells was divided into four levels and scored as
follows: 1, 0–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. Staining
scores for FOXD1 were calculated by multiplying the score assigned
to the intensity of staining (0–3) by the score assigned to the
percentage of positively stained cells (1–4).
According to the staining scores, the samples were divided into the
low expression group (score <6) and high expression group (score
≥6). For the immunohistochemical staining of xenograft tumor
samples from mice, the tumor tissues were fixed in 10% formalin for
2 days at room temperature and then embedded in paraffin. The
samples were then sectioned (4-μm thickness), and then
incubated at 65°C for 30 min before they were deparaffinized and
rehydrated in a graded ethanol series. For antigen retrieval, the
tissue sections were immersed in the citric acid buffer (pH 6.0)
and incubated at 98°C for 15 min. The samples were blocked in 1.5%
goat serum (SP-9000 IHC reagent kit; OriGene Technologies, Inc.) at
37°C for 20 min. The tissue sections were then incubated with
primary antibodies against FOXD1 (dilution, 1:1,000; cat. no.
ab179940; Abcam) and p21 (dilution, 1:1,000; cat. no. 2947S; CST
Biological Reagents Co., Ltd.) at 4°C for 14 h. The sections were
washed and incubated with a HRP-conjugated secondary antibody from
the SP-9000 IHC reagent kit (OriGene Technologies, Inc.) for 30 min
at 37°C. Staining was detected using a 3,3′-diaminobenzidine
detection system.
Cell cycle synchronization and
analysis
Cells (A2780, HO8910, SKOV3, OVCAR3, CAOV3 and
H1299) stably over-expressing FOXD1 were seeded in 6 cm dishes at a
density of 1.2–2.0×105 cells/well and cultured until
they were 30–40% confluent. The cells were then synchronized using
a double thymidine block as described previously (43) in order to synchronize cells in G1
phase. The cells were then released from this phase at different
intervals (A2780, 3 h; HO8910, 6 h; SKOV3, 12 h; OVCAR3, 6 h;
CAOV3, 12 h; H1299, 10 h) by replacing the medium with complete
culture medium without thymidine. The cells were harvested for cell
cycle analysis using the BD FACSCalibur Flow Cytometer (BD
Biosciences, Franklin Lakes, NJ, USA) and the results were analyzed
using ModFit LT software (version 2.0; Verity Software House,
Topsham, ME, USA).
Chromatin immunoprecipitation (ChIP)
assay
A ChIP assay was performed to enrich the specific
regions of chromatin immunoprecipitated by a specific antibody via
using the ChIP-IT Express Chromatin Immunoprecipitation kit (Active
Motif, Carlsbad, CA, USA) according to the manufacturer’s protocol.
A2780 cells (1×107) stably overexpressing FOXD1 were
first cross-linked and sheared into ~500-bp DNA fragments by
sonication. The DNA fragments were then selectively
immunoprecipitated from the cell debris by incubating with a
DYKDDDDK Tag antibody (dilution, 1:50; cat. no. 14793S), or a
rabbit IgG antibody (dilution, 1:50; cat. no. 2729) (both from CST
Biological Reagents Co., Ltd.), which was used as negative control,
for 16 h at 4°C. The purified DNA was amplified by PCR using the
Phusion High-Fidelity PCR Master Mix with HF Buffer (cat no. M0531;
New England Biolabs, Inc., Ipswich, MA, USA) and the following
primers: Site B, forward, 5′-GTCGTGGTGGTGGTGA-3′ and reverse,
5′-CTGCTTTCAGGCATTTC-3′; site C, forward,
5′-ATGTCATCCTCCTGATCTTT-3′ and reverse, 5′-AGTCCCTCGCCTGCGTTGGT-3′.
The thermal cycling parameters were as follows: 30 sec at 98°C
followed by 40 cycles at 98°C for 10 sec, 60°C for 30 sec, 72°C for
15 sec, and then a final step at 72°C for 6 min. The PCR products
were separated by 2% agarose gel electrophoresis and stained with
ethidium bromide.
In vivo studies
All experiments involving animals were approved by
the Ethics Committee on Animal Experiments of Shandong University.
A total of 20 BALB/c nu/nu female mice (age, 6–7 weeks; weight,
20.3±1.5 g) were maintained in a pathogen-free facility
(temperature, 25°C; humidity, 50–60%; 12 h light/day cycles) and
had free access to water and food. The mice were equally divided
into A2780 and OVCAR3 groups, and each group was further subdivided
into a control and FOXD1 overexpression group. For the in
vivo xenograft experiments, A2780 and OVCAR3 cells transfected
with PCMV or PCMV-FOXD1 vectors (6×106 cells) were
injected subcutaneously into the right flank region of the mice.
Following 4 weeks (A2780 cells) or 5 weeks (OVCAR3 cells), the mice
were sacrificed to determine tumor weight and volume. Tumor volume
was calculated using the following formula: Volume = [(long
diameter) × (short diameter)2] ×1/2. Immunohistochemical
staining was employed for the detection of FOXD1 and p21 using the
aforementioned methods.
Bioinformatics analyses
The GSE9891 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE9891)
contains gene expression data for 285 ovarian carcinoma samples
(44). Following the removal of 7
samples without corresponding clinical survival information, and
two samples derived from patients that succumbed due to unrelated
causes, the remaining 276 samples were included in the current
study. The inclusion criteria for HGSOC were as follows: Serous
histological subtype and a pathological grade of 3. A total of 152
patients were identified and divided into high and low FOXD1
expression groups. The significance cut-off value was calculated
using the X-Tile (version 3.6.1) bioinformatics tool (45). To identify predicted miRNA
sequences that may regulate FOXD1, TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org) were employed (46,47).
A total of 142 candidate miRNAs binding sites were identified by
TargetScan (context, ++; score >90; version 7.1) and 146
candidate miRNA binding sites were identified by miRanda (version:
August 2010 Release; mirSVR score ≤−0.1). To identify target genes
that may be regulated by FOXD1, MatInspector (version 8.4;
http://www.genomatix.de) and JASPAR (version
2016; http://jaspar.genereg.net) were employed
(48,49). A region 2 kb upstream of target
gene promoters was screened for FOXD1-binding sites.
Statistical analysis
Data are expressed as the mean ± standard deviation
of three independent experiments. Overall survival and
progression-free survival analysis was performed using Kaplan-Meier
plots and a log-rank test. A Student’s t-test was selected to
analyze significant differences between two groups. Analysis of
variance and the Dunnett’s post hoc test was used to compare the
means among >2 groups. The χ2 test was used to
compare clinical characteristics. P<0.05 was considered to
indicate a statistically significant difference.
Results
FOXD1 is downregulated in HGSOC and
elevated FOXD1 predicts good prognosis
To determine the protein and mRNA expression levels
of FOXD1 in HGSOC tissue samples and normal FT samples, western
blot and RT-qPCR analyses were performed, respectively. At the
protein level, the expression of FOXD1 was higher in FT (n=7) when
compared with HGSOC (n=8) samples (Fig. 1A). At the mRNA level, the
expression of FOXD1 was significantly higher in FT (n=11) compared
with HGSOC (n=20) samples (Fig.
1B). To further confirm the role of FOXD1 in HGSOC,
immunohistochemical staining was performed to measure its
expression in the HGSOC TMA (n=120). As expected, the FOXD1
transcription factor was primarily located in the nucleus (Fig. 1C). The stained HGSOC tissue samples
were scored and divided into high (n=58) and low (n=62) FOXD1
expression groups. The overall survival rate of the high expression
group was significantly higher when compared with the low
expression group (P=0.018; Fig.
1D). The association between FOXD1 expression and
clinicopathological parameters was also analyzed; however, no
statistically significant associations were identified (Table I). To investigate these
observations further, the GSE9891 dataset was used to evaluate the
prognostic role of FOXD1 at the mRNA level (44). The expression of FOXD1 in HGSOC
samples alone was first determined. As demonstrated in Fig. 1E, the overall survival (P=0.030)
and progression-free survival (P=0.003) of the low FOXD1 expression
group was significantly reduced when compared with the high FOXD1
expression group. Samples of 276 in the GSE9891 cohort were then
divided into high and low FOXD1 expression groups. As shown in
Fig. 1F, the overall survival
(P=0.010) and PFS (P=0.011) of the low FOXD1 expression group was
significantly reduced when compared with the high FOXD1 expression
group. These results provide evidence to suggest that reduced FOXD1
expression may predict poor prognosis in HGSOC patients.
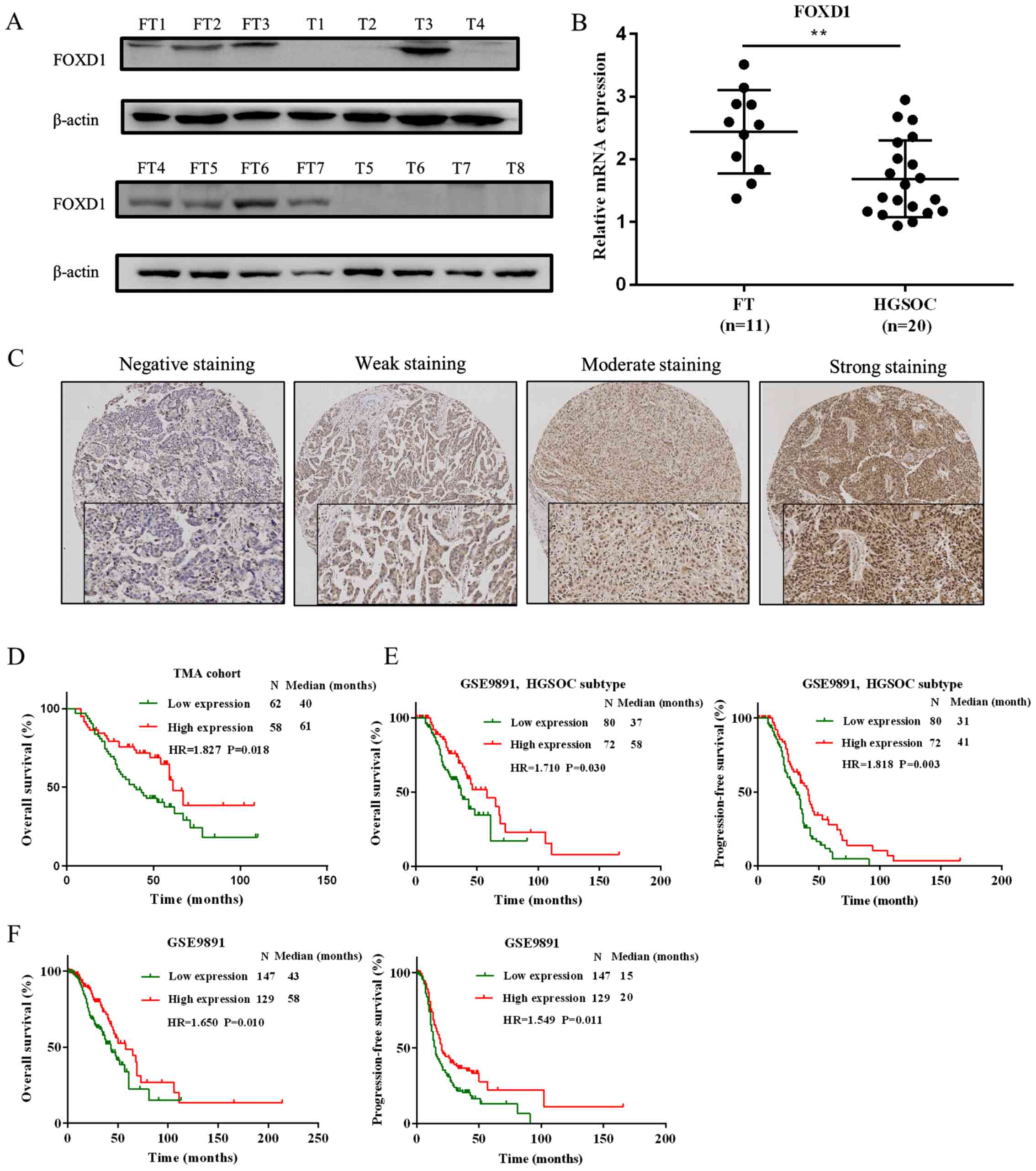 | Figure 1Expression of FOXD1 in HGSOC. (A) The
protein expression levels of FOXD1 in HGSOC samples (n=8) and
normal FT samples (n=7). (B) The mRNA levels of FOXD1 in HGSOC
samples (n=20) and normal FT samples (n=11). (C) Representative
immunohistochemical staining images of negative, weak, moderate and
strong FOXD1 expression in the TMA cohort (magnification, ×100).
(D) Kaplan-Meier curves demonstrating overall survival rates of
patients with low and high FOXD1 expression in the TMA cohort. (E)
Overall survival and progression-free survival of patients with
HGSOC from the GSE9891 cohort, divided into high and low FOXD1
expression groups. (F) Overall survival and progression-free
survival of 276 ovarian cancer patients from the GSE9891 cohort,
divided into high and low FOXD1 expression groups.
**P<0.01, as indicated. FOXD1, forkhead box D1;
HGSOC, high-grade serous ovarian carcinoma; FT, fallopian tube;
TMA, tissue microarray; T, HGSOC tumor samples. |
 | Table IAssociation between FOXD1 expression
and the clinicopathological features of patients with high-grade
serous ovarian carcinoma. |
Table I
Association between FOXD1 expression
and the clinicopathological features of patients with high-grade
serous ovarian carcinoma.
| Clinicopathological
feature | FOXD1 expression
|
|---|
| Low | High | P-value |
|---|
| Age (years) | | | |
| <55 | 29 | 33 | 0.724 |
| ≥55 | 29 | 29 | |
| FIGO stage | | | |
| I-II | 14 | 13 | 0.983 |
| III-IV | 48 | 45 | |
| CA125 (U/ml) | | | |
| <600 | 22 | 18 | 0.605 |
| ≥600 | 40 | 40 | |
| Platinum
status | | | |
| Sensitive | 27 | 25 | 0.099 |
| Resistant | 2 | 7 | |
| Residual
lesions | | | |
| <1 | 39 | 37 | 0.903 |
| ≥1 | 21 | 19 | |
FOXD1 influences the proliferation of
ovarian cancer and H1299 cells
To investigate the effect of FOXD1 expression on
ovarian cancer cells, five ovarian cancer cell lines, including
A2780, HO8910, SKOV3, OVCAR3, CAOV3 and one lung cancer cell line,
H1299, were employed and transfected with a FOXD1 overexpression
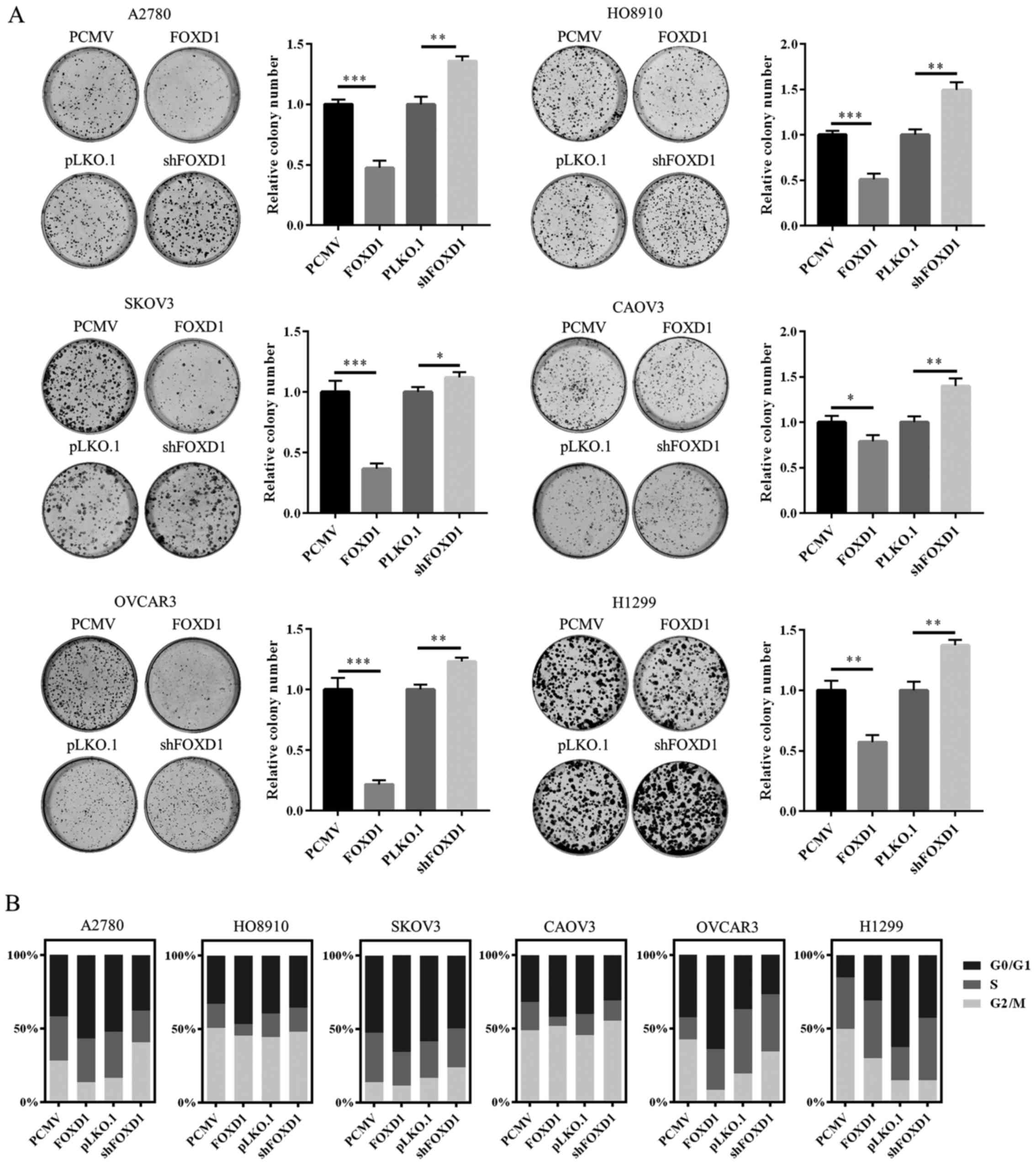
vector or an shRNA targeting FOXD1. As demonstrated in Fig. 2A, overexpression of FOXD1
significantly inhibited the colony-forming efficiency of all six
cell lines when compared with empty vector controls. By contrast,
downregulation of FOXD1 significantly increased their clonogenicity
compared with an empty vector control (Fig. 2A). The cell cycle distribution
among these transfected cell lines was then measured. For cell
cycle analysis, the cells were synchronized in G1 phase using the
double thymidine block method. Then the cells were released by
replacing the medium with thymidine-free complete culture medium.
As shown in Fig. 2B, upregulated
FOXD1 expression induced cell cycle arrest at the G0/G1 phase and
downregulation of FOXD1 reversed this effect. The results indicate
that FOXD1 may suppress the proliferation of ovarian cancer cells
by inducing cell cycle arrest in G1 phrase in vitro.
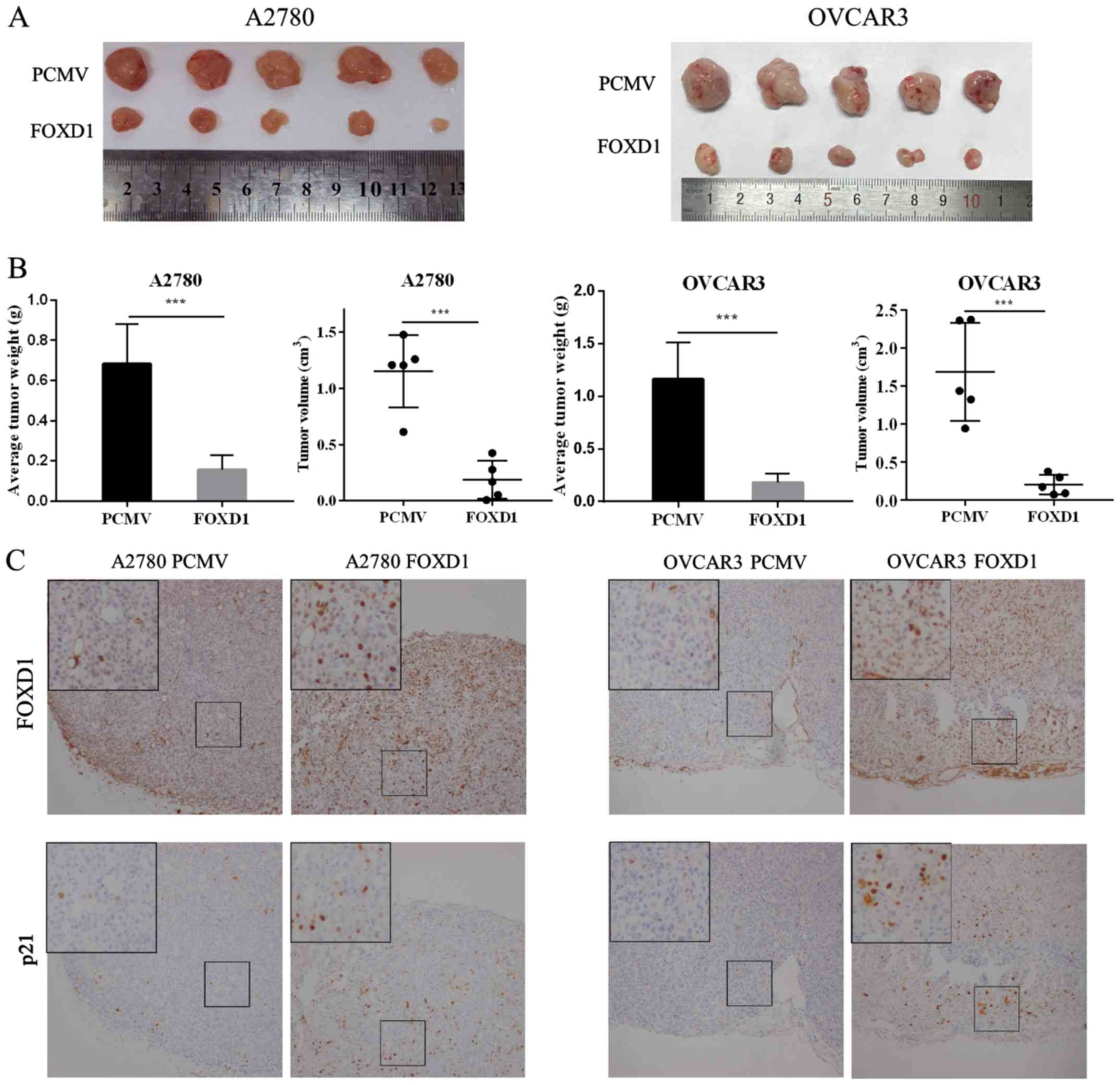
FOXD1 inhibits tumor growth in vivo
The Cancer Genome Atlas Research Network reported
that HGSOC is characterized by TP53 mutations in ~96% of tumors
(50). To further validate the
role of FOXD1 in vivo, OVCAR3 (p53 mutant) and A2780 (p53
wild-type) cells with or without FOXD1 overexpression, were
subcutaneously inoculated into nude mice (n=5 per group). Following
4–5 weeks, the mice were sacrificed, and the tumor weight and
volume were measured. As demonstrated in Fig. 3A and B, the weight and volume of
tumors from mice in the FOXD1 overexpression group was
significantly lower when compared with the control group for both
cell lines. Immunohistochemical staining of the xenograft tumor
tissues was then performed to examine the expression of FOXD1 and
p21. As shown in Fig. 3C, the
expression of FOXD1 in nucleus was positively associated with p21
expression in the nucleus. The results indicate that FOXD1 may
inhibit the proliferation of ovarian cancer cells in
vivo.
miR-30a-5p and miR-200a-5p are direct
regulators of FOXD1 expression
From the results presented thus far, the authors
hypothesized that FOXD1 may function as a tumor suppressor in
ovarian cancer. To investigate the molecular mechanisms underlying
the downregulation of FOXD1 expression in ovarian cancer, miRNA
target prediction tools (miRanda and TargetScan) were used to
identify putative miRNA sequences that may regulate FOXD1. Among
all candidate miRNAs identified, miR-30a-5p and miR-200a-5p were
selected for further investigation. According to previous studies,
miR-30a-5p and miR-200a-5p have been observed to be upregulated in
ovarian cancer (30–36). As demonstrated in Fig. 4A and B, the expression levels of
miR-30a-5p and miR-200a-5p were determined in HGSOC and normal FT
tissue samples. HGSOC samples were observed to exhibit
significantly higher levels of miR-30a-5p and miR-200a-5p
expression when compared with normal FT samples (Fig. 4A and B). A dual-luciferase reporter
assay was subsequently employed to verify the binding sites for
miR-30a-5p and miR-200a-5p in the FOXD1 3′-UTR region. Wild-type or
mutant FOXD1 3′-UTR sequences were cloned into the luciferase
vector, and then co-transfected with miR-30a-5p or miR-200a-5p
mimics into 293T cells. Upregulation of miR-30a-5p or miR-200a-5p
significantly decreased the luciferase activity of cells
transfected with wild-type but not the mutant FOXD1 vector
(Fig. 4C and D). The role of
miR-30a-5p and miR-200a-5p in ovarian cancer cells was then
investigated. OVCAR3 cells were transfected with miR-30a-5p and
miR-200a-5p mimics or inhibitors. As shown in Fig. 4E and F, the expression of FOXD1 at
mRNA and protein levels demonstrated a positive association with
p21 expression. These results indicated that miR-30a-5p and
miR-200a-5p may directly downregulate FOXD1 expression.
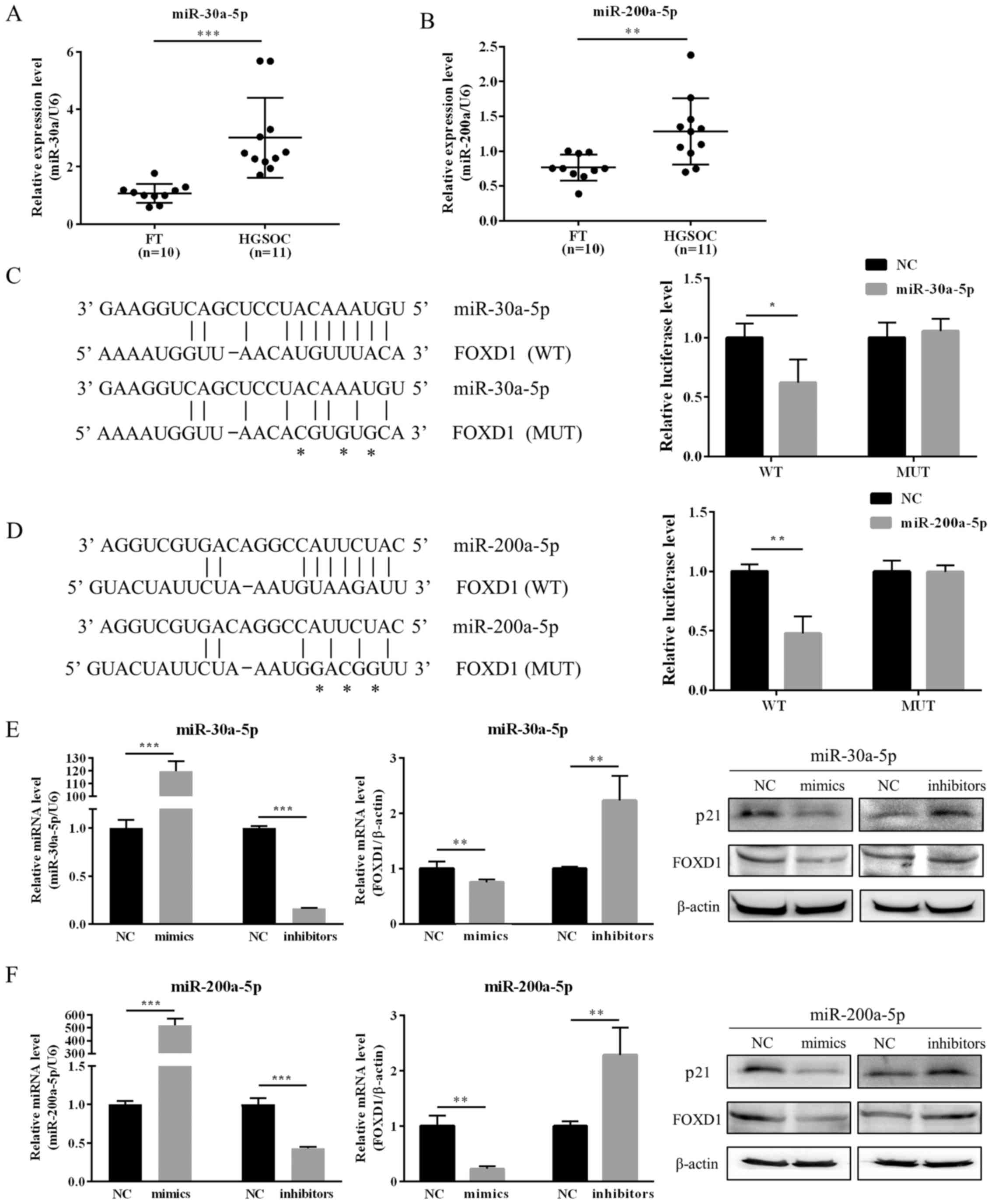 | Figure 4Expression of miR-30a-5p and
miR-200a-5p is upregulated in HGSOC, and these miRNAs directly
target FOXD1 via predicted binding sites. RT-qPCR analysis
demonstrated that (A) miR-30a-5p and (B) miR-200a-5p were
upregulated in HGSOC (n=11) compared with normal FT samples (n=10).
Schematics indicating the predicted binding sites of (C) miR-30a-5p
and (D) miR-200a-5p in the 3′-UTR of FOXD1 mRNA. Asterisks indicate
the mutated binding sites. Reporter vectors with wild-type or
mutant FOXD1 3′-UTRs were co-transfected with
miR-30a-5p/miR-200a-5p mimics in 293T cells and the relative
luciferase activities are shown. The transfection efficiency was
examined by RT-qPCR. Western blotting and RT-qPCR were employed to
evaluate the expression of p21 and FOXD1 in OVCAR3 cells
transfected with (E) miR-30a-5p mimics and inhibitors and (F)
miR-200a-5p mimics and inhibitors. The results are expressed as the
mean ± standard deviation. *P<0.05,
**P<0.01 and ***P<0.001, as indicated.
miR, microRNA, FOXD1, forkhead box D1; HGSOC, high-grade serous
ovarian carcinoma; RT-qPCR, reverse transcription, quantitative
polymerase chain reaction; FT, fallopian tube; UTR, untranslated
region; WT, wild-type; MUT, mutant; NC, negative control. |
FOXD1 directly promotes p21 expression in
vitro and in vivo
A region 2 kb upstream of the p21 promoter was
screened for FOXD1-binding sites using online promoter prediction
tools such as MatInspector and JASPAR. FOXD1 was identified as a
putative regulator of p21 by JASPAR. A total of three candidate
transcription factor binding sites (TFBSs) were also identified
(Fig. 5A). A dual-luciferase
reporter assay was employed to verify these TFBSs. As shown in
Fig. 5B, two out of the three
TFBSs in the FOXD1 overexpression group (sites B and C)
demonstrated a significant increase in luciferase activity compared
with the empty vector controls. When sites B and C were mutated, no
significant difference in luciferase activity was observed between
the FOXD1 over-expression group and control group (Fig. 5C). A ChIP assay and PCR were
subsequently performed to further verify the TFBSs. As shown in
Fig. 5D, enrichment of the p21
promoter fragments were confirmed by PCR using primers flanking
FOXD1 binding sites (sites B and C). The expression of p21 in five
ovarian cancer cell lines and one lung carcinoma cell line
following overexpression or silencing of FOXD1 expression was then
evaluated. As demonstrated in Fig.
5E, FOXD1 overexpression was associated with an increase in p21
protein expression, while a reduction in FOXD1 was associated with
a decrease in p21 expression. These results, together with those
obtained from the immunohistochemical analysis of p21 expression in
ovarian tumor xenografts, suggest that FOXD1 may bind to the
promoter of p21 directly to regulate its expression in vitro
and in vivo.
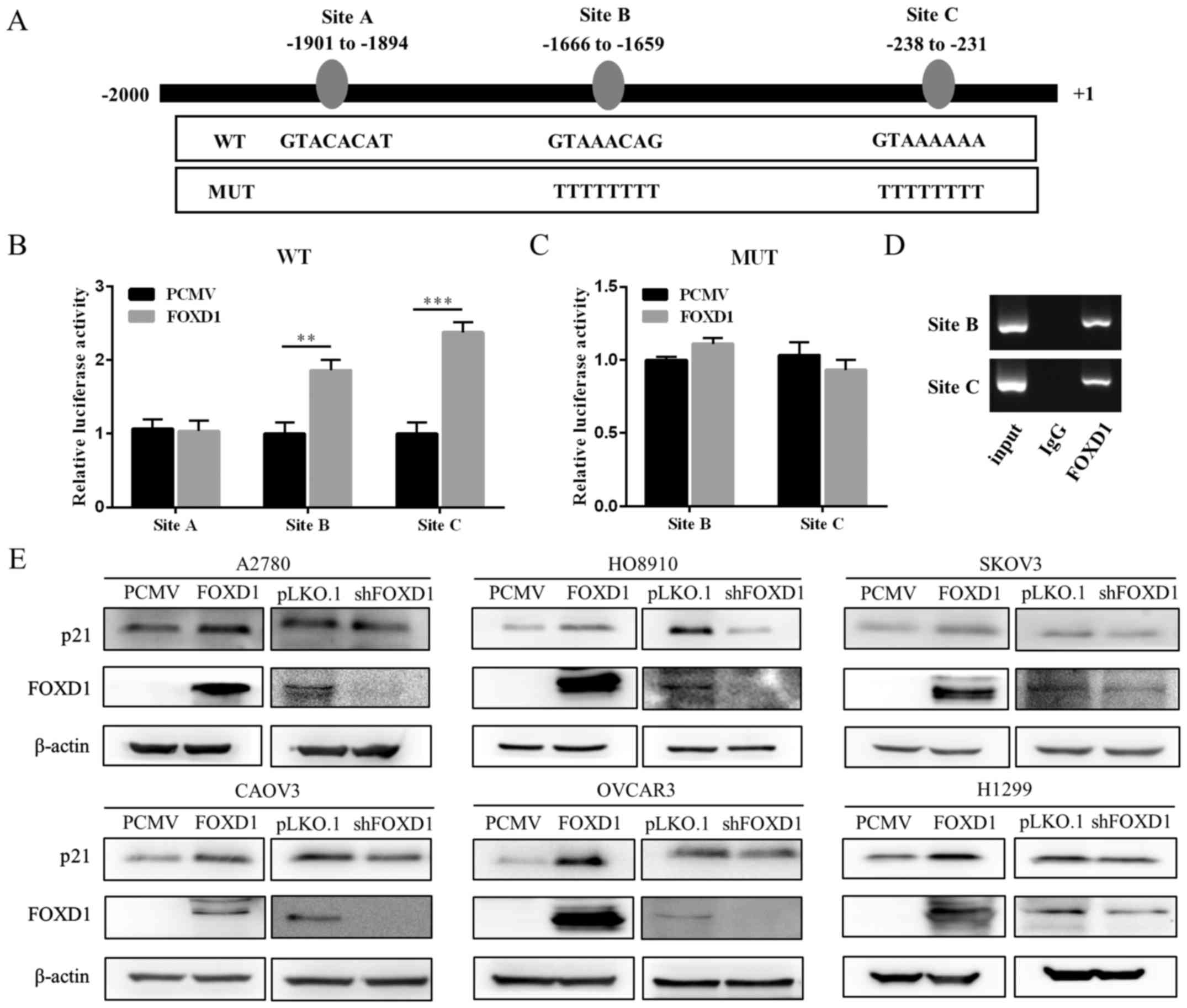 | Figure 5FOXD1 binds to the p21 promoter and
induces its expression. (A) Three putative TFBSs of FOXD1 in the
p21 promoter and the corresponding mutant sequence. (B) 293T cells
were co-transfected with luciferase reporter vectors, each
containing one of the TFBSs, and a FOXD1 overexpression vector or
empty control vector. Luciferase activity was measured at 48 h
following transfection. (C) 293T cells were co-transfected with
luciferase reporters containing mutant TFBSs and a FOXD1
overexpression vector or empty vector control. Luciferase activity
was determined at 48 h following transfection. (D) A2780 cells
stably expressing FOXD1 were used to perform chromatin
immunoprecipitation assay analysis. The immunoprecipitated
protein-chromatin structures were then subjected to PCR analysis.
(E) Western blot analysis demonstrated that manipulation of FOXD1
expression modulated p21 expression in six cancer cell lines. The
results are expressed as the mean ± standard deviation.
**P<0.01 and ***P<0.001, as indicated.
FOXD1, forkhead box D1; TFBSs, transcription factor binding sites;
PCMV, pLenti-C-Myc-DDK-IRES-Puro tagged vector; WT, wild-type; MUT,
mutant; IgG, immunoglobulin G; shFOXD1, short hairpin RNA targeting
FOXD1. |
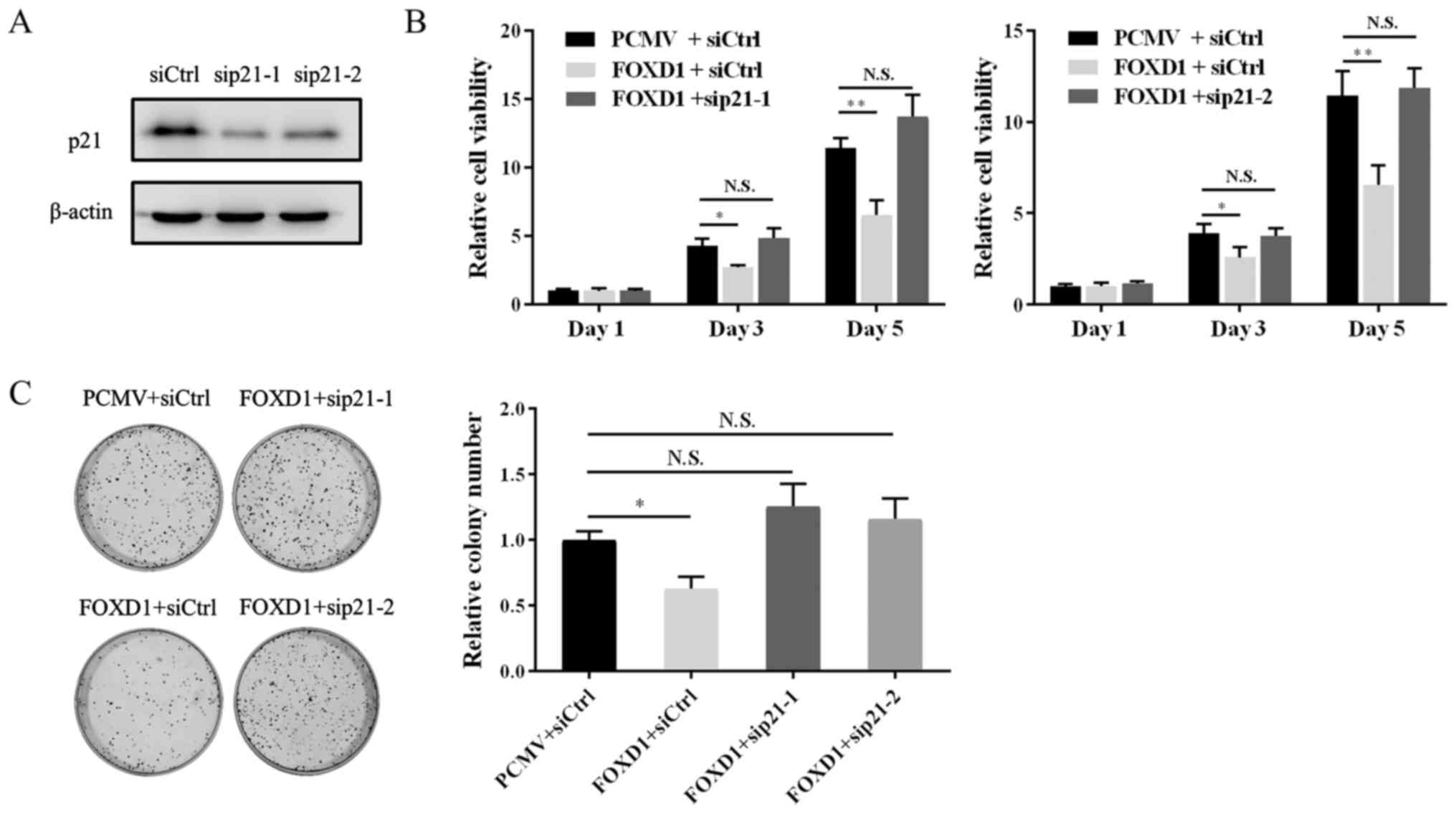
Knockdown of p21 in FOXD1 overexpressing
cells restores cell viability
To further investigate the role of FOXD1 in ovarian
cancer cell proliferation via increasing p21 transcription, a
rescue assay was performed by co-transfecting FOXD1 and
p21-specific siRNA sequences in A2780 cells. The knockdown
efficiency of the two p21-siRNAs is demonstrated in Fig. 6A. As indicated in Fig. 6B and C, the viability and
clonogenicity of cells transfected with p21 siRNA and FOXD2
overexpression vectors was restored when compared with FOXD1
overexpressing cells transfected with siRNA controls. The results
suggest that FOXD1 inhibits the proliferation of ovarian cancer
cells via the direct targeting of p21.
Discussion
FOXD1 is known to function as an important
transcription factor, which is involved in a number of biological
processes (51). It has been
demonstrated to function as an oncogene in lung, breast, brain and
liver cancers (10–12). By contrast, microarray analyses
have indicated that FOXD1 is downregulated in ovarian cancer
patients (14). Consistently, the
results of the current study demonstrated that FOXD1 expression is
decreased at the mRNA and protein levels in HGSOC patients. In
addition, a previous study observed downregulation of FOXD1 in
chemo-resistant ovarian cancer patients (52). Previous reports have also
demonstrated that overexpression of p21 in SKOV3 and OVCAR3 cells
lead to increased cisplatin sensitivity (53). The results of the present study
indicated that FOXD1 directly promotes p21 expression, which may
explain the association between FOXD1 and chemosensitivity in
ovarian cancer. However further validation is required to establish
this relationship.
miR-30a-5p serves an important role in cancer
development and primarily influences the proliferation, metastasis,
and autophagy of cancer cells (29). In the majority of cancer types, it
functions as a tumor suppressor (29); however, in ovarian serous
adenocarcinoma patients, miR-30a-5p was observed to be upregulated
in urine samples, tissue samples and cell lines (30). Consistent with these observations,
the results of the current study indicated that miR-30a-5p is
elevated in HGSOC patient tissue samples. A previous report
demonstrated that breast cancer cells co-transfected with
miR-30a-5p and a luciferase reporter plasmid containing the 3′-UTR
of FOXD1 mRNA, exhibited decreased luciferase activity (54). The current study identified the
miR-30a-5p binding site in the 3′-UTR of FOXD1 mRNA. Previous
studies have indicated that miR-30a-5p is upregulated in
drug-resistant ovarian cancer cell lines (31,32).
As FOXD1 has been observed to be downregulated in chemo-resistant
ovarian cancer patients (52), the
association between miR-30a-5p, FOXD1 and p21 may explain the drug
resistance in ovarian cancer and provide a basis for further
investigation. According to previous reports, miR-200a-5p is
upregulated in HGSOC tissues (35). In addition, the expression of serum
miR-200a-5p levels was significantly higher in patients with the
serous subtype when compared with those presenting with clear cell,
endometrioid or undifferentiated subtypes ovarian cancer subtypes
(34). The results of the current
study verified that miR-200a-5p is upregulated in HGSOC tissues.
The elevated expression of these two miRNAs may explain why FOXD1
is downregulated in HGSOC.
p21 was the first p53-effector gene discovered;
however, it is also known to be induced by p53-independent
signaling pathways (16–19). The results of the present study
demonstrated that FOXD1 inhibited the proliferation of five ovarian
cancer cell lines and binds to the promoter of p21 in 293T cells.
The p53 status of these five ovarian cancer cell lines are as
follows: A2780 and HO8910, wild-type; OVCAR3 and CAOV3, mutant;
SKOV3, controversial (null or mutant) (55–57).
Therefore, FOXD1 induces p21 expression in ovarian cancer cells
irrespective of the p53 status. To confirm that the inhibitory
effects of FOXD1 overexpression on cancer cell proliferation is
p53-independent and involves p21, the p53-null H1299 human lung
cancer cell line was used. The results indicated that the
inhibitory effect of FOXD1 on the growth of H1299 cells was similar
to that of ovarian cancer cells, and that the regulation of p21 by
FOXD1 does not involve p53. p21 is a multi-functional genome
guardian depending on its subcellular localization (19). The nuclear accumulation of p21
induces cell cycle arrest and growth inhibition, while cytoplasmic
accumulation promotes cell growth and survival (58,59).
Nuclear p21 therefore functions as a tumor suppressor. The results
of the current study demonstrated that p21 expression was increased
with forced FOXD1 expression, and that this interaction likely
occurred in the nucleus. Therefore, these results suggest that the
nuclear accumulation of p21 may function as a tumor suppressor in
ovarian cancer.
In conclusion, the results of the present study
demonstrate that FOXD1 is downregulated in HGSOC, and the decreased
level of FOXD1 predicts poor prognosis. In addition, FOXD1 was
observed to inhibit the proliferation of ovarian cancer cells in
vivo and in vitro, and was demonstrated to bind to the
promoter of p21 directly. Furthermore, the expression of miR-30a-5p
and miR-200a-5p was observed to be upregulated in HGSOC. These
miRNA sequences were demonstrated to bind to the 3′-UTR of FOXD1
mRNA. Therefore, these results suggest that FOXD1 functions as a
tumor suppressor in HGSOC and may serve as a biomarker or
therapeutic target for HGSOC.
Acknowledgments
The authors would like to thank the staff at the
Department of Pathology of Qilu Hospital (Shandong, China) for
their technical support.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81572554) and the National
Clinical Research Center for Gynecological Oncology (grant no.
2015BAI13B05).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author’s contributions
YW was a major contributor in designing the
research, performing statistical analysis and writing the
manuscript. CQ collected the tumor samples and interpreted data
from immunohistochemistry analyses. NL and ZL provided technical
support. CJ produced the lentiviruses and performed cell infection
studies. CS performed experiments involving miRNA sequences. HB and
SD preformed the chromatin immunoprecipitation experiments. HY
performed the immunohistochemistry experiments. BK conceived and
supervised the study. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from Ethics Committee
of Shandong University Qilu Hospital and Ethics Committee on Animal
Experiments of Shandong University Qilu Hospital (Shandong,
China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar
|
|
2
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017. View Article : Google Scholar
|
|
3
|
McCluggage WG: Morphological subtypes of
ovarian carcinoma: A review with emphasis on new developments and
pathogenesis. Pathology. 43:420–432. 2011. View Article : Google Scholar
|
|
4
|
Prat J: Ovarian carcinomas: Five distinct
diseases with different origins, genetic alterations, and
clinicopathological features. Virchows Arch. 460:237–249. 2012.
View Article : Google Scholar
|
|
5
|
Bowtell DD, Böhm S, Ahmed AA, Aspuria PJ,
Bast RC Jr, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et
al: Rethinking ovarian cancer II: Reducing mortality from
high-grade serous ovarian cancer. Nat Rev Cancer. 15:668–679. 2015.
View Article : Google Scholar
|
|
6
|
Carlsson P and Mahlapuu M: Forkhead
transcription factors: Key players in development and metabolism.
Dev Biol. 250:1–23. 2002. View Article : Google Scholar
|
|
7
|
Carreres MI, Escalante A, Murillo B,
Chauvin G, Gaspar P, Vegar C and Herrera E: Transcription factor
Foxd1 is required for the specification of the temporal retina in
mammals. J Neurosci. 31:5673–5681. 2011. View Article : Google Scholar
|
|
8
|
Fetting JL, Guay JA, Karolak MJ, Iozzo RV,
Adams DC, Maridas DE, Brown AC and Oxburgh L: FOXD1 promotes
nephron progenitor differentiation by repressing decorin in the
embryonic kidney. Development. 141:17–27. 2014. View Article : Google Scholar
|
|
9
|
Koga M, Matsuda M, Kawamura T, Sogo T,
Shigeno A, Nishida E and Ebisuya M: Foxd1 is a mediator and
indicator of the cell reprogramming process. Nat Commun.
5:31972014. View Article : Google Scholar
|
|
10
|
Zhao YF, Zhao JY, Yue H, Hu KS, Shen H,
Guo ZG and Su XJ: FOXD1 promotes breast cancer proliferation and
chemotherapeutic drug resistance by targeting p27. Biochem Biophys
Res Commun. 456:232–237. 2015. View Article : Google Scholar
|
|
11
|
Nakayama S, Soejima K, Yasuda H, Yoda S,
Satomi R, Ikemura S, Terai H, Sato T, Yamaguchi N, Hamamoto J, et
al: FOXD1 expression is associated with poor prognosis in non-small
cell lung cancer. Anticancer Res. 35:261–268. 2015.
|
|
12
|
Gao YF, Zhu T, Mao XY, Mao CX, Li L, Yin
JY, Zhou HH and Liu ZQ: Silencing of Forkhead box D1 inhibits
proliferation and migration in glioma cells. Oncol Rep.
37:1196–1202. 2017. View Article : Google Scholar
|
|
13
|
Chen J, Qian Z, Li F, Li J and Lu Y:
Integrative analysis of microarray data to reveal regulation
patterns in the pathogenesis of hepatocellular carcinoma. Gut
Liver. 11:112–120. 2017. View
Article : Google Scholar
|
|
14
|
Jiang X, Zhu T, Yang J, Li S, Ye S, Liao
S, Meng L, Lu Y and Ma D: Identification of novel epithelial
ovarian cancer biomarkers by cross-laboratory microarray analysis.
J Huazhong Univ Sci Technolog Med Sci. 30:354–359. 2010. View Article : Google Scholar
|
|
15
|
Liu S, Bishop WR and Liu M: Differential
effects of cell cycle regulatory protein p21(WAF1/Cip1) on
apoptosis and sensitivity to cancer chemotherapy. Drug Resist
Updat. 6:183–195. 2003. View Article : Google Scholar
|
|
16
|
el-Deiry WS, Tokino T, Velculescu VE, Levy
DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW and
Vogelstein B: WAF1, a potential mediator of p53 tumor suppression.
Cell. 75:817–825. 1993. View Article : Google Scholar
|
|
17
|
el-Deiry WS, Harper JW, O’Connor PM,
Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill
DE, Wang Y, et al: WAF1/CIP1 is induced in p53-mediated G1 arrest
and apoptosis. Cancer Res. 54:1169–1174. 1994.
|
|
18
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View
Article : Google Scholar
|
|
19
|
Georgakilas AG, Martin OA and Bonner WM:
p21: A two-faced genome guardian. Trends Mol Med. 23:310–319. 2017.
View Article : Google Scholar
|
|
20
|
Shiohara M, el-Deiry WS, Wada M, Nakamaki
T, Takeuchi S, Yang R, Chen DL, Vogelstein B and Koeffler HP:
Absence of WAF1 mutations in a variety of human malignancies.
Blood. 84:3781–3784. 1994.
|
|
21
|
McKenzie KE, Siva A, Maier S, Runnebaum
IB, Seshadri R and Sukumar S: Altered WAF1 genes do not play a role
in abnormal cell cycle regulation in breast cancers lacking p53
mutations. Clin Cancer Res. 3:1669–1673. 1997.
|
|
22
|
Bukholm IK and Nesland JM: Protein
expression of p53, p21 (WAF1/CIP1), bcl-2, Bax, cyclin D1 and pRb
in human colon carcinomas. Virchows Arch. 436:224–228. 2000.
View Article : Google Scholar
|
|
23
|
Caffo O, Doglioni C, Veronese S, Bonzanini
M, Marchetti A, Buttitta F, Fina P, Leek R, Morelli L, Palma PD, et
al: Prognostic value of p21(WAF1) and p53 expression in breast
carcinoma: An immunohistochemical study in 261 patients with
long-term follow-up. Clin Cancer Res. 2:1591–1599. 1996.
|
|
24
|
Ogawa M, Onoda N, Maeda K, Kato Y, Nakata
B, Kang SM, Sowa M and Hirakawa K: A combination analysis of p53
and p21 in gastric carcinoma as a strong indicator for prognosis.
Int J Mol Med. 7:479–483. 2001.
|
|
25
|
Lu X, Toki T, Konishi I, Nikaido T and
Fujii S: Expression of p21WAF1/CIP1 in adenocarcinoma of the
uterine cervix: A possible immunohistochemical marker of a
favorable prognosis. Cancer. 82:2409–2417. 1998. View Article : Google Scholar
|
|
26
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar
|
|
27
|
Yang SJ, Yang SY, Wang DD, Chen X, Shen
HY, Zhang XH, Zhong SL, Tang JH and Zhao JH: The miR-30 family:
Versatile players in breast cancer. Tumour Biol.
39:1010428317692204. 2017. View Article : Google Scholar
|
|
28
|
Feng X, Wang Z, Fillmore R and Xi Y:
MiR-200, a new star miRNA in human cancer. Cancer Lett.
344:166–173. 2014. View Article : Google Scholar
|
|
29
|
Yang X, Chen Y and Chen L: The versatile
role of microRNA-30a in human cancer. Cell Physiol Biochem.
41:1616–1632. 2017. View Article : Google Scholar
|
|
30
|
Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen
Y, Wang J, Liu Y, Chen P, Wu X, et al: Urinary microRNA-30a-5p is a
potential biomarker for ovarian serous adenocarcinoma. Oncol Rep.
33:2915–2923. 2015. View Article : Google Scholar
|
|
31
|
Chen N, Chon HS, Xiong Y, Marchion DC,
Judson PL, Hakam A, Gonzalez-Bosquet J, Permuth-Wey J, Wenham RM,
Apte SM, et al: Human cancer cell line microRNAs associated with in
vitro sensitivity to paclitaxel. Oncol Rep. 31:376–383. 2014.
View Article : Google Scholar
|
|
32
|
Liu J, Wu X, Liu H, Liang Y, Gao X, Cai Z,
Wang W and Zhang H: Expression of microRNA-30a-5p in drug-resistant
and drug-sensitive ovarian cancer cell lines. Oncol Lett.
12:2065–2070. 2016. View Article : Google Scholar
|
|
33
|
Wyman SK, Parkin RK, Mitchell PS, Fritz
BR, O’Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS and
Tewari M: Repertoire of microRNAs in epithelial ovarian cancer as
determined by next generation sequencing of small RNA cDNA
libraries. PLoS One. 4:e53112009. View Article : Google Scholar
|
|
34
|
Zuberi M, Mir R, Das J, Ahmad I, Javid J,
Yadav P, Masroor M, Ahmad S, Ray PC and Saxena A: Expression of
serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in
epithelial ovarian cancer and their association with
clinicopathological features. Clin Transl Oncol. 17:779–787. 2015.
View Article : Google Scholar
|
|
35
|
Vilming Elgaaen B, Olstad OK, Haug KB,
Brusletto B, Sandvik L, Staff AC, Gautvik KM and Davidson B: Global
miRNA expression analysis of serous and clear cell ovarian
carcinomas identifies differentially expressed miRNAs including
miR-200c-3p as a prognostic marker. BMC Cancer. 14:802014.
View Article : Google Scholar
|
|
36
|
Katz B, Tropé CG, Reich R and Davidson B:
MicroRNAs in ovarian cancer. Hum Pathol. 46:1245–1256. 2015.
View Article : Google Scholar
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
38
|
Wu X, Yang N, Zhou WH, Xu J, Chen JJ,
Zheng FM, Long ZJ, Yue CF, Ai KX, Liu LL, et al: Up-regulation of
p21 inhibits TRAIL-mediated extrinsic apoptosis, contributing
resistance to SAHA in acute myeloid leukemia cells. Cell Physiol
Biochem. 34:506–518. 2014. View Article : Google Scholar
|
|
39
|
Rao A, Coan A, Welsh JE, Barclay WW,
Koumenis C and Cramer SD: Vitamin D receptor and p21/WAF1 are
targets of genistein and 1,25-dihydroxyvitamin D3 in human prostate
cancer cells. Cancer Res. 64:2143–2147. 2004. View Article : Google Scholar
|
|
40
|
Kobayashi A, Mugford JW, Krautzberger AM,
Naiman N, Liao J and McMahon AP: Identification of a multipotent
self-renewing stromal progenitor population during mammalian kidney
organogenesis. Stem Cell Reports. 3:650–662. 2014. View Article : Google Scholar
|
|
41
|
Filtz TM, Vogel WK and Leid M: Regulation
of transcription factor activity by interconnected
post-translational modifications. Trends Pharmacol Sci. 35:76–85.
2014. View Article : Google Scholar
|
|
42
|
Gan L, Zheng W, Chabot JG, Unterman TG and
Quirion R: Nuclear/cytoplasmic shuttling of the transcription
factor FoxO1 is regulated by neurotrophic factors. J Neurochem.
93:1209–1219. 2005. View Article : Google Scholar
|
|
43
|
Harper JV: Synchronization of cell
populations in G1/S and G2/M phases of the cell cycle. Methods Mol
Biol. 296:157–166. 2005.
|
|
44
|
Tothill RW, Tinker AV, George J, Brown R,
Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro
B, et al: Novel molecular subtypes of serous and endometrioid
ovarian cancer linked to clinical outcome. Clin Cancer Res.
14:5198–5208. 2008. View Article : Google Scholar
|
|
45
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar
|
|
46
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:42015. View Article : Google Scholar
|
|
47
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The http://microRNA.orgurisimplemicroRNA.org resource:
Targets and expression. Nucleic Acids Res. 36:D149–D153. 2008.
View Article : Google Scholar
|
|
48
|
Cartharius K, Frech K, Grote K, Klocke B,
Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M and Werner T:
MatInspector and beyond: Promoter analysis based on transcription
factor binding sites. Bioinformatics. 21:2933–2942. 2005.
View Article : Google Scholar
|
|
49
|
Mathelier A, Fornes O, Arenillas DJ, Chen
CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al:
JASPAR 2016: A major expansion and update of the open-access
database of transcription factor binding profiles. Nucleic Acids
Res. 44:D110–D115. 2016. View Article : Google Scholar
|
|
50
|
Bell D, Berchuck A, Birrer M, Chien J,
Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P, et al Cancer
Genome Atlas Research Network: Integrated genomic analyses of
ovarian carcinoma. Nature. 474:609–615. 2011. View Article : Google Scholar
|
|
51
|
Pierrou S, Hellqvist M, Samuelsson L,
Enerbäck S and Carlsson P: Cloning and characterization of seven
human forkhead proteins: Binding site specificity and DNA bending.
EMBO J. 13:5002–5012. 1994.
|
|
52
|
Ju W, Yoo BC, Kim IJ, Kim JW, Kim SC and
Lee HP: Identification of genes with differential expression in
chemoresistant epithelial ovarian cancer using high-density
oligonucleotide microarrays. Oncol Res. 18:47–56. 2009. View Article : Google Scholar
|
|
53
|
Lincet H, Poulain L, Remy JS, Deslandes E,
Duigou F, Gauduchon P and Staedel C: The p21(cip1/waf1)
cyclin-dependent kinase inhibitor enhances the cytotoxic effect of
cisplatin in human ovarian carcinoma cells. Cancer Lett. 161:17–26.
2000. View Article : Google Scholar
|
|
54
|
Ouzounova M, Vuong T, Ancey PB, Ferrand M,
Durand G, Le-Calvez Kelm F, Croce C, Matar C, Herceg Z and
Hernandez-Vargas H: MicroRNA miR-30 family regulates non-attachment
growth of breast cancer cells. BMC Genomics. 14:1392013. View Article : Google Scholar
|
|
55
|
Forbes S, Clements J, Dawson E, Bamford S,
Webb T, Dogan A, Flanagan A, Teague J, Wooster R, Futreal PA, et
al: COSMIC 2005. Br J Cancer. 94:318–322. 2006. View Article : Google Scholar
|
|
56
|
Soussi T: Handbook of p53 Mutation in Cell
Lines. Version 1. 2007
|
|
57
|
Su WJ, Fang JS, Cheng F, Liu C, Zhou F and
Zhang J: RNF2/Ring1b negatively regulates p53 expression in
selective cancer cell types to promote tumor development. Proc Natl
Acad Sci USA. 110:1720–1725. 2013. View Article : Google Scholar
|
|
58
|
Piccolo MT and Crispi S: The dual role
played by p21 may influence the apoptotic or anti-apoptotic fate in
cancer. J Cancer Res Updates. 1:189–202. 2012.
|
|
59
|
Gawriluk TR, Simkin J, Thompson KL, Biswas
SK, Clare-Salzler Z, Kimani JM, Kiama SG, Smith JJ, Ezenwa VO and
Seifert AW: Comparative analysis of ear-hole closure identifies
epimorphic regeneration as a discrete trait in mammals. Nat Commun.
7:111642016. View Article : Google Scholar
|