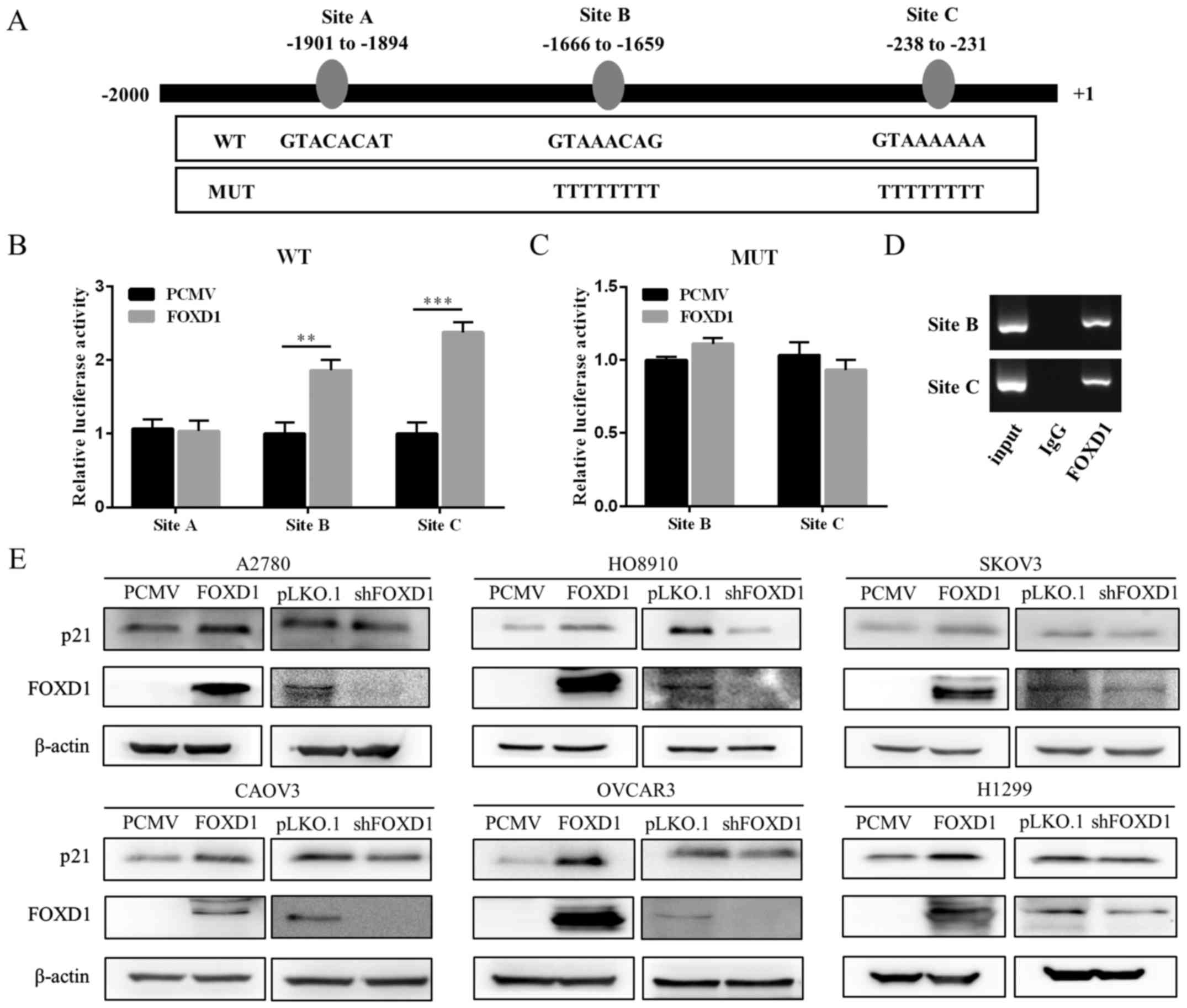
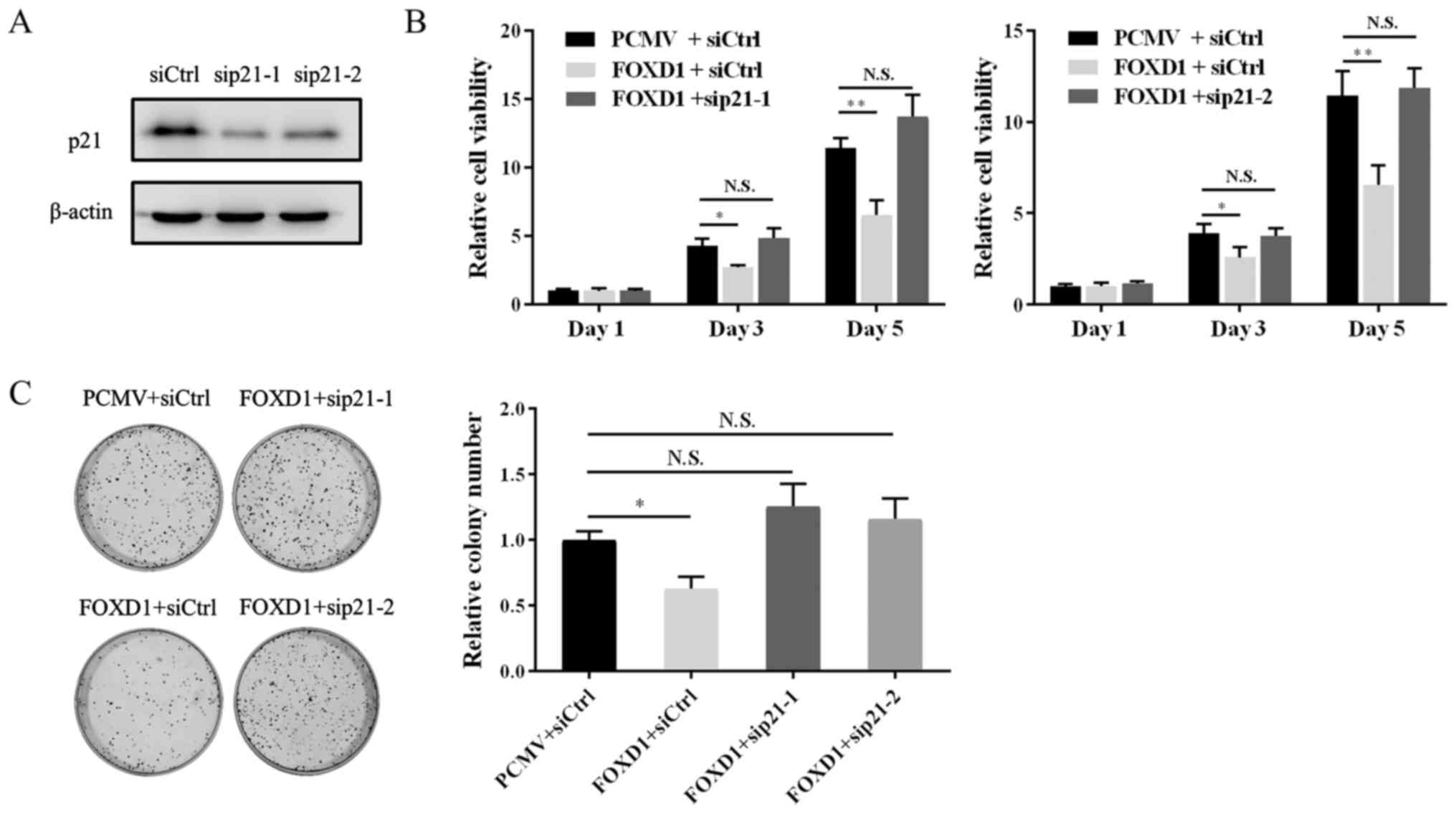
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar
|
|
2
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017. View Article : Google Scholar
|
|
3
|
McCluggage WG: Morphological subtypes of
ovarian carcinoma: A review with emphasis on new developments and
pathogenesis. Pathology. 43:420–432. 2011. View Article : Google Scholar
|
|
4
|
Prat J: Ovarian carcinomas: Five distinct
diseases with different origins, genetic alterations, and
clinicopathological features. Virchows Arch. 460:237–249. 2012.
View Article : Google Scholar
|
|
5
|
Bowtell DD, Böhm S, Ahmed AA, Aspuria PJ,
Bast RC Jr, Beral V, Berek JS, Birrer MJ, Blagden S, Bookman MA, et
al: Rethinking ovarian cancer II: Reducing mortality from
high-grade serous ovarian cancer. Nat Rev Cancer. 15:668–679. 2015.
View Article : Google Scholar
|
|
6
|
Carlsson P and Mahlapuu M: Forkhead
transcription factors: Key players in development and metabolism.
Dev Biol. 250:1–23. 2002. View Article : Google Scholar
|
|
7
|
Carreres MI, Escalante A, Murillo B,
Chauvin G, Gaspar P, Vegar C and Herrera E: Transcription factor
Foxd1 is required for the specification of the temporal retina in
mammals. J Neurosci. 31:5673–5681. 2011. View Article : Google Scholar
|
|
8
|
Fetting JL, Guay JA, Karolak MJ, Iozzo RV,
Adams DC, Maridas DE, Brown AC and Oxburgh L: FOXD1 promotes
nephron progenitor differentiation by repressing decorin in the
embryonic kidney. Development. 141:17–27. 2014. View Article : Google Scholar
|
|
9
|
Koga M, Matsuda M, Kawamura T, Sogo T,
Shigeno A, Nishida E and Ebisuya M: Foxd1 is a mediator and
indicator of the cell reprogramming process. Nat Commun.
5:31972014. View Article : Google Scholar
|
|
10
|
Zhao YF, Zhao JY, Yue H, Hu KS, Shen H,
Guo ZG and Su XJ: FOXD1 promotes breast cancer proliferation and
chemotherapeutic drug resistance by targeting p27. Biochem Biophys
Res Commun. 456:232–237. 2015. View Article : Google Scholar
|
|
11
|
Nakayama S, Soejima K, Yasuda H, Yoda S,
Satomi R, Ikemura S, Terai H, Sato T, Yamaguchi N, Hamamoto J, et
al: FOXD1 expression is associated with poor prognosis in non-small
cell lung cancer. Anticancer Res. 35:261–268. 2015.
|
|
12
|
Gao YF, Zhu T, Mao XY, Mao CX, Li L, Yin
JY, Zhou HH and Liu ZQ: Silencing of Forkhead box D1 inhibits
proliferation and migration in glioma cells. Oncol Rep.
37:1196–1202. 2017. View Article : Google Scholar
|
|
13
|
Chen J, Qian Z, Li F, Li J and Lu Y:
Integrative analysis of microarray data to reveal regulation
patterns in the pathogenesis of hepatocellular carcinoma. Gut
Liver. 11:112–120. 2017. View
Article : Google Scholar
|
|
14
|
Jiang X, Zhu T, Yang J, Li S, Ye S, Liao
S, Meng L, Lu Y and Ma D: Identification of novel epithelial
ovarian cancer biomarkers by cross-laboratory microarray analysis.
J Huazhong Univ Sci Technolog Med Sci. 30:354–359. 2010. View Article : Google Scholar
|
|
15
|
Liu S, Bishop WR and Liu M: Differential
effects of cell cycle regulatory protein p21(WAF1/Cip1) on
apoptosis and sensitivity to cancer chemotherapy. Drug Resist
Updat. 6:183–195. 2003. View Article : Google Scholar
|
|
16
|
el-Deiry WS, Tokino T, Velculescu VE, Levy
DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW and
Vogelstein B: WAF1, a potential mediator of p53 tumor suppression.
Cell. 75:817–825. 1993. View Article : Google Scholar
|
|
17
|
el-Deiry WS, Harper JW, O’Connor PM,
Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill
DE, Wang Y, et al: WAF1/CIP1 is induced in p53-mediated G1 arrest
and apoptosis. Cancer Res. 54:1169–1174. 1994.
|
|
18
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View
Article : Google Scholar
|
|
19
|
Georgakilas AG, Martin OA and Bonner WM:
p21: A two-faced genome guardian. Trends Mol Med. 23:310–319. 2017.
View Article : Google Scholar
|
|
20
|
Shiohara M, el-Deiry WS, Wada M, Nakamaki
T, Takeuchi S, Yang R, Chen DL, Vogelstein B and Koeffler HP:
Absence of WAF1 mutations in a variety of human malignancies.
Blood. 84:3781–3784. 1994.
|
|
21
|
McKenzie KE, Siva A, Maier S, Runnebaum
IB, Seshadri R and Sukumar S: Altered WAF1 genes do not play a role
in abnormal cell cycle regulation in breast cancers lacking p53
mutations. Clin Cancer Res. 3:1669–1673. 1997.
|
|
22
|
Bukholm IK and Nesland JM: Protein
expression of p53, p21 (WAF1/CIP1), bcl-2, Bax, cyclin D1 and pRb
in human colon carcinomas. Virchows Arch. 436:224–228. 2000.
View Article : Google Scholar
|
|
23
|
Caffo O, Doglioni C, Veronese S, Bonzanini
M, Marchetti A, Buttitta F, Fina P, Leek R, Morelli L, Palma PD, et
al: Prognostic value of p21(WAF1) and p53 expression in breast
carcinoma: An immunohistochemical study in 261 patients with
long-term follow-up. Clin Cancer Res. 2:1591–1599. 1996.
|
|
24
|
Ogawa M, Onoda N, Maeda K, Kato Y, Nakata
B, Kang SM, Sowa M and Hirakawa K: A combination analysis of p53
and p21 in gastric carcinoma as a strong indicator for prognosis.
Int J Mol Med. 7:479–483. 2001.
|
|
25
|
Lu X, Toki T, Konishi I, Nikaido T and
Fujii S: Expression of p21WAF1/CIP1 in adenocarcinoma of the
uterine cervix: A possible immunohistochemical marker of a
favorable prognosis. Cancer. 82:2409–2417. 1998. View Article : Google Scholar
|
|
26
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar
|
|
27
|
Yang SJ, Yang SY, Wang DD, Chen X, Shen
HY, Zhang XH, Zhong SL, Tang JH and Zhao JH: The miR-30 family:
Versatile players in breast cancer. Tumour Biol.
39:1010428317692204. 2017. View Article : Google Scholar
|
|
28
|
Feng X, Wang Z, Fillmore R and Xi Y:
MiR-200, a new star miRNA in human cancer. Cancer Lett.
344:166–173. 2014. View Article : Google Scholar
|
|
29
|
Yang X, Chen Y and Chen L: The versatile
role of microRNA-30a in human cancer. Cell Physiol Biochem.
41:1616–1632. 2017. View Article : Google Scholar
|
|
30
|
Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen
Y, Wang J, Liu Y, Chen P, Wu X, et al: Urinary microRNA-30a-5p is a
potential biomarker for ovarian serous adenocarcinoma. Oncol Rep.
33:2915–2923. 2015. View Article : Google Scholar
|
|
31
|
Chen N, Chon HS, Xiong Y, Marchion DC,
Judson PL, Hakam A, Gonzalez-Bosquet J, Permuth-Wey J, Wenham RM,
Apte SM, et al: Human cancer cell line microRNAs associated with in
vitro sensitivity to paclitaxel. Oncol Rep. 31:376–383. 2014.
View Article : Google Scholar
|
|
32
|
Liu J, Wu X, Liu H, Liang Y, Gao X, Cai Z,
Wang W and Zhang H: Expression of microRNA-30a-5p in drug-resistant
and drug-sensitive ovarian cancer cell lines. Oncol Lett.
12:2065–2070. 2016. View Article : Google Scholar
|
|
33
|
Wyman SK, Parkin RK, Mitchell PS, Fritz
BR, O’Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS and
Tewari M: Repertoire of microRNAs in epithelial ovarian cancer as
determined by next generation sequencing of small RNA cDNA
libraries. PLoS One. 4:e53112009. View Article : Google Scholar
|
|
34
|
Zuberi M, Mir R, Das J, Ahmad I, Javid J,
Yadav P, Masroor M, Ahmad S, Ray PC and Saxena A: Expression of
serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in
epithelial ovarian cancer and their association with
clinicopathological features. Clin Transl Oncol. 17:779–787. 2015.
View Article : Google Scholar
|
|
35
|
Vilming Elgaaen B, Olstad OK, Haug KB,
Brusletto B, Sandvik L, Staff AC, Gautvik KM and Davidson B: Global
miRNA expression analysis of serous and clear cell ovarian
carcinomas identifies differentially expressed miRNAs including
miR-200c-3p as a prognostic marker. BMC Cancer. 14:802014.
View Article : Google Scholar
|
|
36
|
Katz B, Tropé CG, Reich R and Davidson B:
MicroRNAs in ovarian cancer. Hum Pathol. 46:1245–1256. 2015.
View Article : Google Scholar
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
38
|
Wu X, Yang N, Zhou WH, Xu J, Chen JJ,
Zheng FM, Long ZJ, Yue CF, Ai KX, Liu LL, et al: Up-regulation of
p21 inhibits TRAIL-mediated extrinsic apoptosis, contributing
resistance to SAHA in acute myeloid leukemia cells. Cell Physiol
Biochem. 34:506–518. 2014. View Article : Google Scholar
|
|
39
|
Rao A, Coan A, Welsh JE, Barclay WW,
Koumenis C and Cramer SD: Vitamin D receptor and p21/WAF1 are
targets of genistein and 1,25-dihydroxyvitamin D3 in human prostate
cancer cells. Cancer Res. 64:2143–2147. 2004. View Article : Google Scholar
|
|
40
|
Kobayashi A, Mugford JW, Krautzberger AM,
Naiman N, Liao J and McMahon AP: Identification of a multipotent
self-renewing stromal progenitor population during mammalian kidney
organogenesis. Stem Cell Reports. 3:650–662. 2014. View Article : Google Scholar
|
|
41
|
Filtz TM, Vogel WK and Leid M: Regulation
of transcription factor activity by interconnected
post-translational modifications. Trends Pharmacol Sci. 35:76–85.
2014. View Article : Google Scholar
|
|
42
|
Gan L, Zheng W, Chabot JG, Unterman TG and
Quirion R: Nuclear/cytoplasmic shuttling of the transcription
factor FoxO1 is regulated by neurotrophic factors. J Neurochem.
93:1209–1219. 2005. View Article : Google Scholar
|
|
43
|
Harper JV: Synchronization of cell
populations in G1/S and G2/M phases of the cell cycle. Methods Mol
Biol. 296:157–166. 2005.
|
|
44
|
Tothill RW, Tinker AV, George J, Brown R,
Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro
B, et al: Novel molecular subtypes of serous and endometrioid
ovarian cancer linked to clinical outcome. Clin Cancer Res.
14:5198–5208. 2008. View Article : Google Scholar
|
|
45
|
Camp RL, Dolled-Filhart M and Rimm DL:
X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004. View Article : Google Scholar
|
|
46
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:42015. View Article : Google Scholar
|
|
47
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The http://microRNA.orgurisimplemicroRNA.org resource:
Targets and expression. Nucleic Acids Res. 36:D149–D153. 2008.
View Article : Google Scholar
|
|
48
|
Cartharius K, Frech K, Grote K, Klocke B,
Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M and Werner T:
MatInspector and beyond: Promoter analysis based on transcription
factor binding sites. Bioinformatics. 21:2933–2942. 2005.
View Article : Google Scholar
|
|
49
|
Mathelier A, Fornes O, Arenillas DJ, Chen
CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al:
JASPAR 2016: A major expansion and update of the open-access
database of transcription factor binding profiles. Nucleic Acids
Res. 44:D110–D115. 2016. View Article : Google Scholar
|
|
50
|
Bell D, Berchuck A, Birrer M, Chien J,
Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P, et al Cancer
Genome Atlas Research Network: Integrated genomic analyses of
ovarian carcinoma. Nature. 474:609–615. 2011. View Article : Google Scholar
|
|
51
|
Pierrou S, Hellqvist M, Samuelsson L,
Enerbäck S and Carlsson P: Cloning and characterization of seven
human forkhead proteins: Binding site specificity and DNA bending.
EMBO J. 13:5002–5012. 1994.
|
|
52
|
Ju W, Yoo BC, Kim IJ, Kim JW, Kim SC and
Lee HP: Identification of genes with differential expression in
chemoresistant epithelial ovarian cancer using high-density
oligonucleotide microarrays. Oncol Res. 18:47–56. 2009. View Article : Google Scholar
|
|
53
|
Lincet H, Poulain L, Remy JS, Deslandes E,
Duigou F, Gauduchon P and Staedel C: The p21(cip1/waf1)
cyclin-dependent kinase inhibitor enhances the cytotoxic effect of
cisplatin in human ovarian carcinoma cells. Cancer Lett. 161:17–26.
2000. View Article : Google Scholar
|
|
54
|
Ouzounova M, Vuong T, Ancey PB, Ferrand M,
Durand G, Le-Calvez Kelm F, Croce C, Matar C, Herceg Z and
Hernandez-Vargas H: MicroRNA miR-30 family regulates non-attachment
growth of breast cancer cells. BMC Genomics. 14:1392013. View Article : Google Scholar
|
|
55
|
Forbes S, Clements J, Dawson E, Bamford S,
Webb T, Dogan A, Flanagan A, Teague J, Wooster R, Futreal PA, et
al: COSMIC 2005. Br J Cancer. 94:318–322. 2006. View Article : Google Scholar
|
|
56
|
Soussi T: Handbook of p53 Mutation in Cell
Lines. Version 1. 2007
|
|
57
|
Su WJ, Fang JS, Cheng F, Liu C, Zhou F and
Zhang J: RNF2/Ring1b negatively regulates p53 expression in
selective cancer cell types to promote tumor development. Proc Natl
Acad Sci USA. 110:1720–1725. 2013. View Article : Google Scholar
|
|
58
|
Piccolo MT and Crispi S: The dual role
played by p21 may influence the apoptotic or anti-apoptotic fate in
cancer. J Cancer Res Updates. 1:189–202. 2012.
|
|
59
|
Gawriluk TR, Simkin J, Thompson KL, Biswas
SK, Clare-Salzler Z, Kimani JM, Kiama SG, Smith JJ, Ezenwa VO and
Seifert AW: Comparative analysis of ear-hole closure identifies
epimorphic regeneration as a discrete trait in mammals. Nat Commun.
7:111642016. View Article : Google Scholar
|