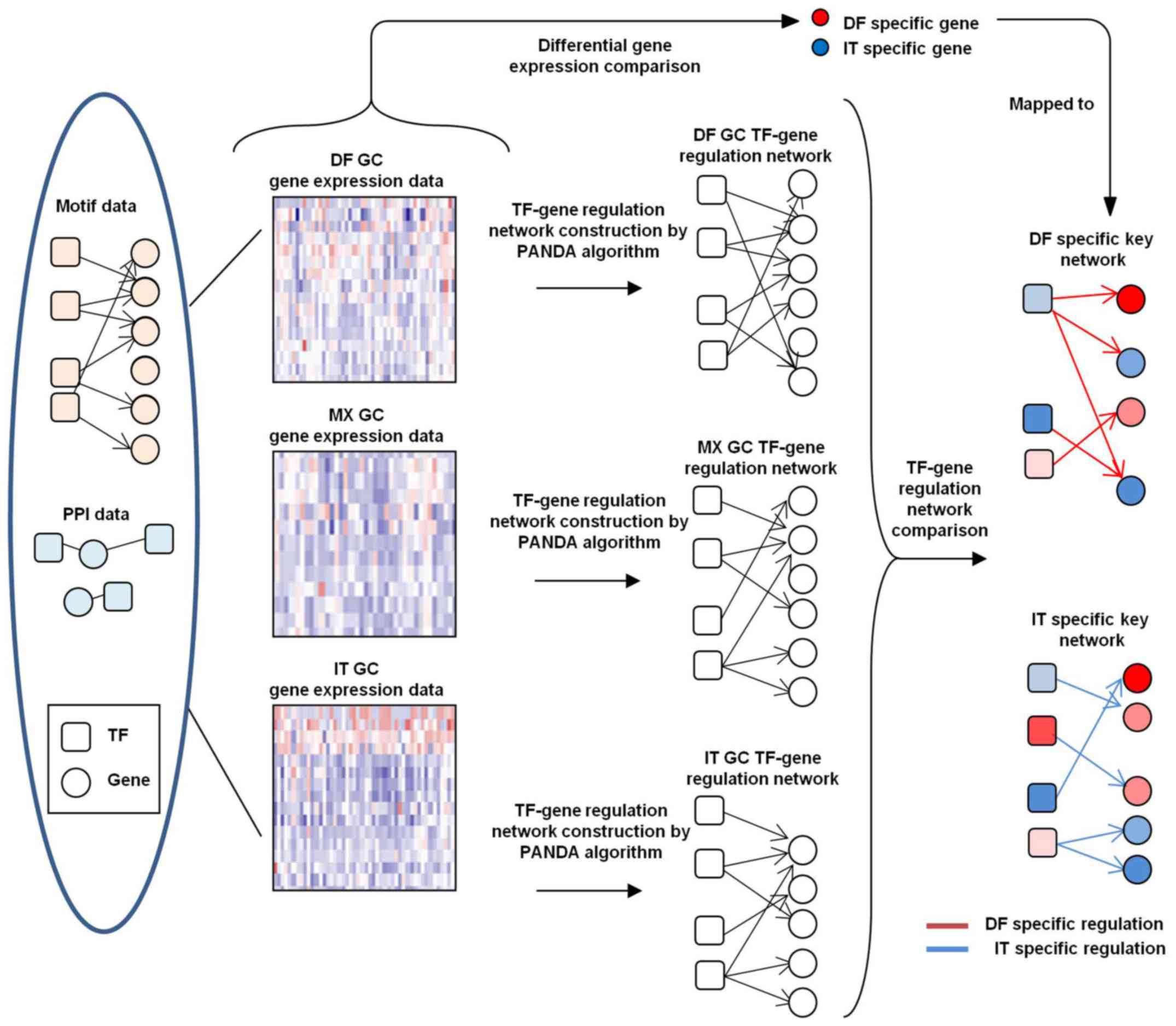
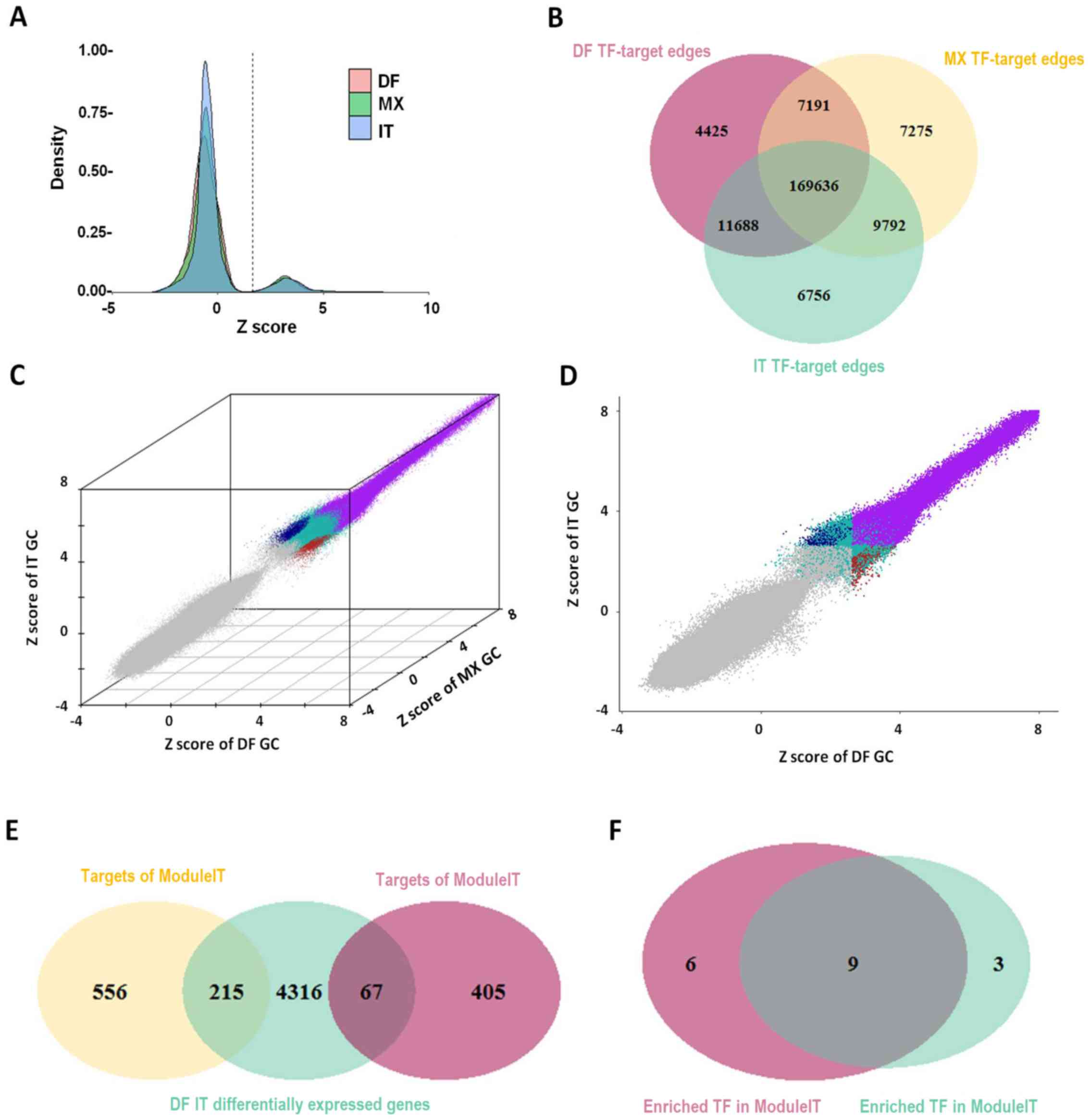
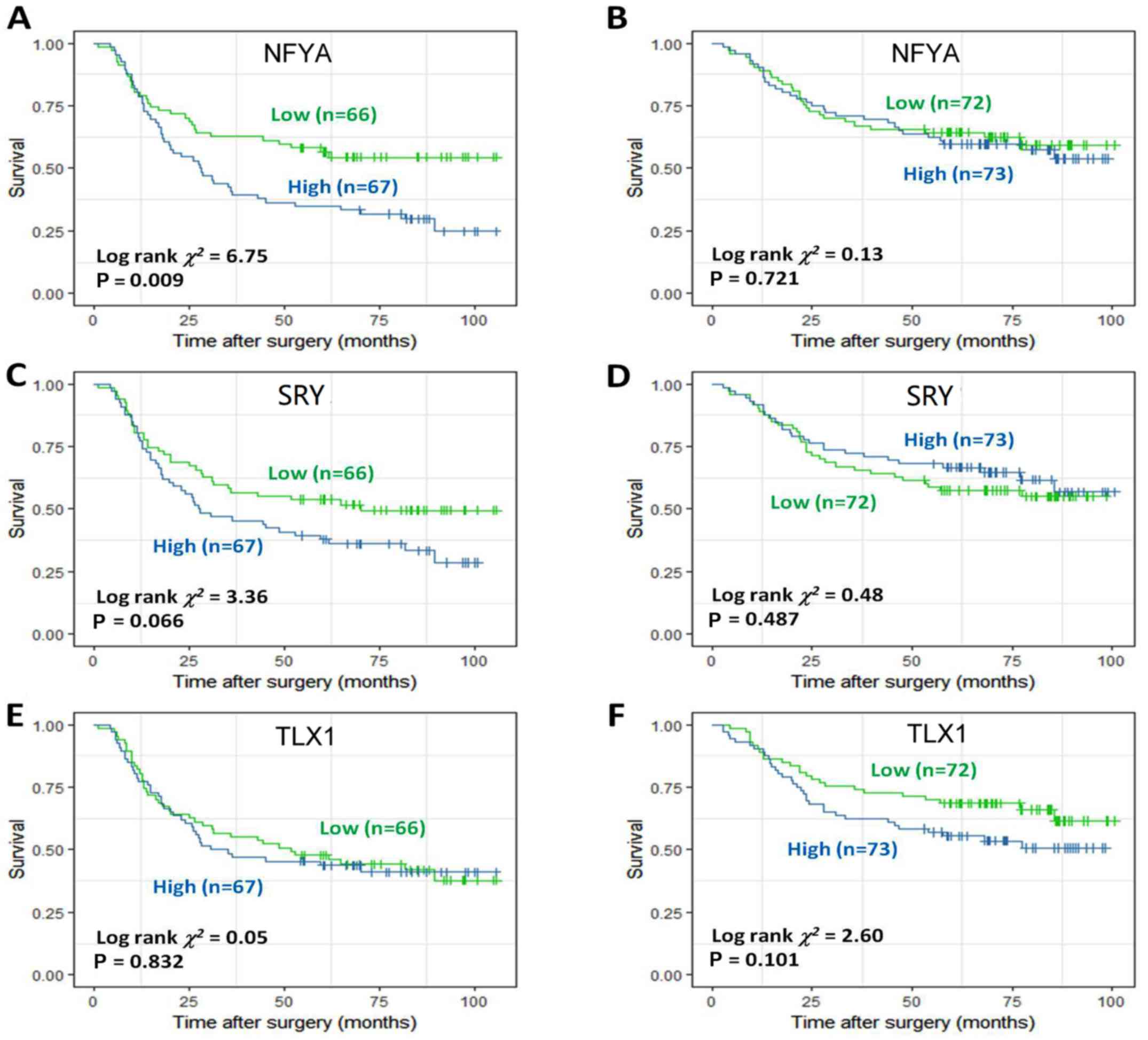
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A; Global cancer statistics: 2012.CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J; Cancer statistics in China:
2015.CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar
|
|
3
|
Wong SS, Kim KM, Ting JC, Yu K, Fu J, Liu
S, Cristescu R, Nebozhyn M, Gong L, Yue YG, et al: Genomic
landscape and genetic heterogeneity in gastric adenocarcinoma
revealed by whole-genome sequencing. Nat Commun. 5:54772014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lauren T: The two histological main types
of gastric carcinoma, an attempt at a histoclinical classification.
Acta Pathol Microbiol Scand. 64:191965. View Article : Google Scholar
|
|
5
|
Hartgrink HH, Jansen EP, van Grieken NC
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He Z and Li B: Recent progress in genetic
and epigenetic profile of diffuse gastric cancer. Cancer Transl
Med. 1:80–93. 2015. View Article : Google Scholar
|
|
7
|
Shah MA, Khanin R, Tang L, Janjigian YY,
Klimstra DS, Gerdes H and Kelsen DP: Molecular classification of
gastric cancer: A new paradigm. Clin Cancer Res. 17:2693–2701.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fléjou JF: WHO Classification of digestive
tumors: The fourth edition. Ann Pathol. 31(Suppl 5): pp. S27–S31.
2011, (In French). View Article : Google Scholar
|
|
9
|
Palli D, Bianchi S, Cipriani F, Duca P,
Amorosi A, Avellini C, Russo A, Saragoni A, Todde P and Valdes E:
Reproducibility of histologic classification of gastric cancer. Br
J Cancer. 63:765–768. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jinawath N, Furukawa Y, Hasegawa S, Li M,
Tsunoda T, Satoh S, Yamaguchi T, Imamura H, Inoue M, Shiozaki H, et
al: Comparison of gene-expression profiles between diffuse- and
intestinal-type gastric cancers using a genome-wide cDNA
microarray. Oncogene. 23:6830–6844. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee YS, Cho YS, Lee GK, Lee S, Kim YW, Jho
S, Kim HM, Hong SH, Hwang JA, Kim SY, et al: Genomic profile
analysis of diffuse-type gastric cancers. Genome Biol. 15:R552014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tanabe S, Aoyagi K, Yokozaki H and Sasaki
H: Gene expression signatures for identifying diffuse-type gastric
cancer associated with epithelial-mesenchymal transition. Int J
Oncol. 44:1955–1970. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Förster S, Gretschel S, Jöns T, Yashiro M
and Kemmner W: THBS4, a novel stromal molecule of diffuse-type
gastric adenocarcinomas, identified by transcriptome-wide
expression profiling. Mod Pathol. 24:1390–1403. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim B, Bang S, Lee S, Kim S, Jung Y, Lee
C, Choi K, Lee SG, Lee K, Lee Y, et al: Expression profiling and
subtype-specific expression of stomach cancer. Cancer Res.
63:8248–8255. 2003.PubMed/NCBI
|
|
15
|
Yoon C, Cho SJ, Aksoy BA, Park DJ, Schultz
N, Ryeom S and Yoon SS: Chemotherapy resistance in diffuse type
gastric adenocarcinoma is mediated by RhoA activation in cancer
stem-like cells. Clin Cancer Res. 22:971–983. 2016. View Article : Google Scholar :
|
|
16
|
Min L, Zhao Y, Zhu S, Qiu X, Cheng R, Xing
J, Shao L, Guo S and Zhang S: Integrated analysis identifies
molecular signatures and specific prognostic factors for different
gastric cancer subtypes. Transl Oncol. 10:99–107. 2017. View Article : Google Scholar :
|
|
17
|
Ritchie MD, Holzinger ER, Li R,
Pendergrass SA and Kim D: Methods of integrating data to uncover
genotype-phenotype interactions. Nat Rev Genet. 16:85–97. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vaquerizas JM, Kummerfeld SK, Teichmann SA
and Luscombe NM: A census of human transcription factors: Function,
expression and evolution. Nat Rev Genet. 10:252–263. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Buckingham M and Rigby PW: Gene regulatory
networks and transcriptional mechanisms that control myogenesis.
Dev Cell. 28:225–238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Banks CA, Lee ZT, Boanca G,
Lakshminarasimhan M, Groppe BD, Wen Z, Hattem GL, Seidel CW,
Florens L and Washburn MP: Controlling for gene expression changes
in transcription factor protein networks. Mol Cell Proteomics.
13:1510–1522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Glass K, Huttenhower C, Quackenbush J and
Yuan GC: Passing messages between biological networks to refine
predicted interactions. PLoS One. 8:pp. e648322013, View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lao T, Glass K, Qiu W, Polverino F, Gupta
K, Morrow J, Mancini JD, Vuong L, Perrella MA, Hersh CP, et al:
Haploinsufficiency of Hedgehog interacting protein causes increased
emphysema induced by cigarette smoke through network rewiring.
Genome Med. 7:122015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Glass K, Quackenbush J, Spentzos D,
Haibe-Kains B and Yuan GC: A network model for angiogenesis in
ovarian cancer. BMC Bioinformatics. 16:1152015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Min L, Zhang C, Qu L, Huang J, Jiang L,
Liu J, Pinello L, Yuan GC and Shou C: Gene regulatory pattern
analysis reveals essential role of core transcriptional factors’
activation in triple- negative breast cancer. Oncotarget.
8:21938–21953. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cristescu R, Lee J, Nebozhyn M, Kim K-M,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Workman C, Jensen LJ, Jarmer H, Berka R,
Gautier L, et al: A new non-linear normalization method for
reducing variability in DNA microarray experiments. Genome Biol:
Aug. 30:pp. 2002Epub ahead of print.
|
|
28
|
Li C and Wong WH: Model-based analysis of
oligonucleotide arrays: Expression index computation and outlier
detection. Proc Natl Acad Sci USA. 98:31–36. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu C, Macleod I and Su AI: BioGPS and
MyGene.info: Organizing online, gene-centric information. Nucleic
Acids Res. 41:D561–D565. 2013. View Article : Google Scholar :
|
|
30
|
Mathelier A, Fornes O, Arenillas DJ, Chen
CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, et al:
JASPAR 2016: A major expansion and update of the open-access
database of transcription factor binding profiles. Nucleic Acids
Res. 44:D110–D115. 2016. View Article : Google Scholar :
|
|
31
|
Zhao Y, Min L, Xu C, Shao L, Guo S, Cheng
R, Xing J, Zhu S and Zhang S: Construction of disease-specific
transcriptional regulatory networks identifies co-activation of
four gene in esophageal squamous cell carcinoma. Oncol Rep.
38:411–417. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ravasi T, Suzuki H, Cannistraci CV,
Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S,
Kanamori-Katayama M, Bertin N, et al: An atlas of combinatorial
transcriptional regulation in mouse and man. Cell. 140:744–752.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ernst J, Beg QK, Kay KA, Balázsi G, Oltvai
ZN and Bar-Joseph Z: A semi-supervised method for predicting
transcription factor- gene interactions in Escherichia coli. PLoS
Comput Biol. 4:pp. e10000442008, View Article : Google Scholar
|
|
34
|
Lemmens K, Dhollander T, De Bie T,
Monsieurs P, Engelen K, Smets B, Winderickx J, De Moor B and
Marchal K: Inferring transcriptional modules from ChIP-chip, motif
and microarray data. Genome Biol. 7:R372006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Faith JJ, Hayete B, Thaden JT, Mogno I,
Wierzbowski J, Cottarel G, Kasif S, Collins JJ and Gardner TS:
Large-scale mapping and validation of Escherichia coli
transcriptional regulation from a compendium of expression
profiles. PLoS Biol. 5:e82007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Altay G and Emmert-Streib F: Structural
influence of gene networks on their inference: Analysis of C3NET.
Biol Direct. 6:312011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Benatti P, Chiaramonte ML, Lorenzo M,
Hartley JA, Hochhauser D, Gnesutta N, Mantovani R, Imbriano C and
Dolfini D: NF-Y activates genes of metabolic pathways altered in
cancer cells. Oncotarget. 7:1633–1650. 2016. View Article : Google Scholar :
|
|
38
|
Murakami S, Ninomiya W, Sakamoto E,
Shibata T, Akiyama H and Tashiro F: SRY and OCT4 are required for
the acquisition of cancer stem cell-like properties and are
potential differentiation therapy targets. Stem Cells.
33:2652–2663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Asuthkar S, Gogineni VR, Rao JS and
Velpula KK: Nuclear translocation of Hand-1 acts as a molecular
switch to regulate vascular radiosensitivity in medulloblastoma
tumors: The protein uPAR is a cytoplasmic sequestration factor for
Hand-1. Mol Cancer Ther. 13:1309–1322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang G, Cheng Z, Liu F, Zhang H, Li J and
Li F: CREB is a key negative regulator of carbonic anhydrase IX
(CA9) in gastric cancer. Cell Signal. 27:1369–1379. 2015.
View Article : Google Scholar
|
|
41
|
Dadi S, Le Noir S, Payet-Bornet D,
Lhermitte L, Zacarias- Cabeza J, Bergeron J, Villarèse P, Vachez E,
Dik WA, Millien C, et al: TLX homeodomain oncogenes mediate T cell
maturation arrest in T-ALL via interaction with ETS1 and
suppression of TCRα gene expression. Cancer Cell. 21:563–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zheng L, Pu J, Jiang G, Weng M, He J, Mei
H, Hou X and Tong Q: Abnormal expression of early growth response 1
in gastric cancer: Association with tumor invasion, metastasis and
heparanase transcription. Pathol Int. 60:268–277. 2010. View Article : Google Scholar : PubMed/NCBI
|