Introduction
Lung cancer is the leading cause of cancer-related
mortality worldwide, with non-small cell lung cancer (NSCLC)
accounting for almost 80% of all lung cancer cases (1,2). In
China, the overall 5-year survival rate for NSCLC patients is
approximately 15% and the rate of recurrence remains high (3–7).
Despite advances in early detection and diagnosis, chemical and
immunotherapy, precision radiotherapy, and expert surgical
intervention, eradicating cancer in patients remains a major
challenge. Although our understanding of cancer cell biology has
made significant progress, a definite cure for most types of cancer
does not exist at present (8–11).
Therefore, there is urgent need to discover novel biomarkers for
diagnosis and treatment.
MicroRNAs (miRNAs or miRs) are small non-coding
single-stranded RNAs of approximately 20-23 nt in length that
regulate gene expression by binding to the 3′-untranslated regions
(3′-UTR) of mRNAs (12-14). The expression levels of miRNAs have
profound effects on cancer progression and human carcinogenesis
(15). The expression of
miR-296-5p has been shown to be significantly downregulated in lung
cancer (3,16,17),
breast cancer (18), diabetes
(19-21) and other types of cancer and
diseases (17,22,23).
Therefore, exploring the function of miR-296-5p and the role of its
possible target genes is essential to the understanding of the
molecular mechanism of this miRNA in NSCLC. Sodium-glucose
co-transporter-2 [SGLT2, also known as solute carrier family
5 member 2 (SLC5A2)], a sodium-dependent glucose
transporter, is a common therapeutic target in the treatment of
diabetes (24). In addition,
SGLT2 promotes the development of pancreatic and prostate
adenocarcinomas (25) and
increases lung cancer metastasis (26). Therefore, we hypothesized that
miR-296-5p may play a pivotal role in lung cancer tumorigenesis by
targeting SGLT2.
In this study, we aimed to investigate this
hypothesis. We demonstrate that miR-296-5p is downregulated in
NSCLC patient samples and NSCLC cell lines. Moreover, we
demonstrate that miR-296-5p directly targets SGLT2. These
results may provide a potential molecular therapeutic target for
the occurrence and development of NSCLC.
Materials and methods
Tissue samples
All NSCLC samples and non-tumor samples (also termed
paracancerous tissue, i.e., tissue adjacent to cancerous tissue)
were obtained from the Department of Oncology, Shanghai Chest
Hospital (Shanghai, China) from October, 2014 to August, 2016 and
their use was approved by the Ethics Committee of Shanghai Hospital
and consent was obtained from all the patients. All the details of
the tissue samples used in this study are listed in Table I. We obtained the corresponding
results by analyzing the specific information of the patient tissue
samples, such as sex, stage and size. Tumor size is the primary
determinant of prognosis and metastasis. A previous study
demonstrated that a small tumor size (≤3 cm) is associated with a
lower probability of metastasis (27). Moreover, in a previous study, we
demonstrated that miR-18a-targeted interferon regulatory factor 2
(IRF2) expression was downregulated in tumor tissues, and likely
related to tumor size (28). Thus,
in this study, we also used this method to assay the function of
miR-296 in lung cancer.
 | Table IThe clinicopathological
characteristics of the 46 patients with non-small cell lung
cancer. |
Table I
The clinicopathological
characteristics of the 46 patients with non-small cell lung
cancer.
| No. | Sex | Age | Specimen type | Histological
type | Lymphatic
invasion | TNM |
|---|
| 1 | M | 47 | Pneumonectomy | Adenocarcinoma | | T1bN0M0 |
| 2 | M | 78 | Lobectomy | Adenocarcinoma | | T2aN0M0 |
| 3 | F | 67 | Pneumonectomy | Adenocarcinoma | | T2N1M0 |
| 4 | M | 54 | Lobectomy | Adenocarcinoma | | T4N0M0 |
| 5 | M | 67 | Lobectomy | Adenocarcinoma | Absent | T2aN0M0 |
| 6 | F | 53 | Lobectomy | Squamous cell
carcinoma | Absent | T3N0M0 |
| 7 | M | 67 | Lobectomy | Adenocarcinoma | | T2aN0M0 |
| 8 | F | 62 | Lobectomy | Adenocarcinoma | Absent | T2aN0M0 |
| 9 | F | 75 | Lobectomy | Adenocarcinoma | Absent | T2aN0M0 |
| 10 | F | 58 | Lobectomy | Adenocarcinoma | | T2aN1M0 |
| 11 | F | 65 | Lobectomy | Adenocarcinoma | Present | T2aN0M0 |
| 12 | M | 72 | Lobectomy | Adenocarcinoma | | T2aN0M0 |
| 13 | M | 64 | Lobectomy | Adenocarcinoma | Absent | T1aN0M0 |
| 14 | F | 62 | Lobectomy | Adenocarcinoma | Present | T2aN0M0 |
| 15 | M | 65 | Lobectomy | Adenocarcinoma | | T4N2M0 |
| 16 | M | 58 | Lobectomy | Squamous
cellcarcinoma | Present | T3N2M0 |
| 17 | M | 50 | Lobectomy | Adenocarcinoma | | T2aN0M0 |
| 18 | M | 60 | Lobectomy | Adenocarcinoma | Absent | T1bN0M0 |
| 19 | M | 67 | Lobectomy | Adenocarcinoma | Absent | T2aN0M0 |
| 20 | M | 71 | Lobectomy | Adenocarcinoma | Present | T1bN0M0 |
| 21 | F | 64 | Lobectomy | Adenocarcinoma | Absent | T2aN0M0 |
| 22 | M | 62 | Lobectomy | Adenocarcinoma | Absent | T2aN0M0 |
| 23 | M | 58 | Lobectomy | Adenocarcinoma | Present | T2aN0M0 |
| 24 | M | 70 | Lobectomy | Adenocarcinoma | Present | T3N0M0 |
| 25 | M | 59 | Lobectomy | Adenocarcinoma | Present | T2aN0M1a |
| 26 | F | 72 | Lobectomy | Adenocarcinoma | Absent | T2aN0M0 |
| 27 | M | 51 | Lobectomy | Adenocarcinoma | Absent | T1bN0M0 |
| 28 | M | 51 | Lobectomy | Adenocarcinoma | Absent | T1aN0M0 |
| 29 | F | 55 | Lobectomy | Adenocarcinoma | Absent | T1bN0M0 |
| 30 | F | 53 | Lobectomy | Squamous cell
carcinoma | | T3N0M0 |
| 31 | M | 54 | Lobectomy | Squamous cell
carcinoma | | T4N2M0 |
| 32 | M | 67 | Lobectomy | Adenocarcinoma | Absent | T2aN0M0 |
| 33 | M | 49 | Lobectomy | Adenocarcinoma | Present | T2bN2M0 |
| 34 | M | 65 | Lobectomy | Squamous cell
carcinoma | Absent | T1bN0M0 |
| 35 | M | 65 | Lobectomy | Squamous cell
carcinoma | | T2aN2M0 |
| 36 | M | 67 | Lobectomy | Squamous cell
carcinoma | Absent | T2aN1M0 |
| 37 | M | 51 | Lobectomy | Adenocarcinoma | Absent | T1bN0M0 |
| 38 | F | 71 | Lobectomy | Adenocarcinoma | Absent | T3N0M0 |
| 39 | F | 58 | Lobectomy | Adenocarcinoma | Present | T2aN1M0 |
| 40 | F | 67 | Lobectomy | Adenocarcinoma | Present | T2N1M0 |
| 41 | M | 68 | Lobectomy | Adenocarcinoma | Absent | T3N0M0 |
| 42 | M | 70 | Lobectomy | Adenocarcinoma | Absent | T3N0M0 |
| 43 | M | 71 | Lobectomy | Adenocarcinoma | Absent | T1bN0M0 |
| 44 | M | 67 | Lobectomy | Squamous cell
carcinoma | Present | T2aN1M0 |
| 45 | F | 77 | Lobectomy | Adenocarcinoma | Present | T4N3M0 |
| 46 | F | 55 | Lobectomy | Adenocarcinoma | Absent | T1bN0M0 |
Cell culture and cell transfection
The BEAS-2B, 16HBE and 293T cells were obtained from
the Cell Bank, China Academy of Sciences (Shanghai, China). The
A549, H1975, PC-9 and H1299 cells were purchased from the American
Type Culture Collection (ATCC, Manassas, VA, USA). The A549,
BEAS-2B, 293T and PC-9 cells were cultured in DMEM (Gibco). The
H1975 and H1299 cells were cultured in RPMI-1640 medium. The
culture conditions were set at 37°C in a 5% CO2
humidified environment. NSCLC cells were cultivated in 90% medium,
10% fetal bovine serum (FBS, HyClone Laboratories, Logan, UT, USA),
100 µg/ml penicillin, 100 µg/ml streptomycin (Gibco) and antibiotic
cocktail. The BEAS-2B cell line was originally isolated from the
normal bronchial epithelium.
The A549 and H1299 cells were transiently
transfected with 30 nM miR-296-5p mimic, a negative control mimic
(NC) and 100 nM SGLT2 siRNA (3 siSGLT2), or negative control siRNA
(siNC) (Guangzhou RiboBio Co., Ltd., Guangzhou, China) using
Invitrogen™ Lipofectamine 2000 (Thermo Fisher Scientific, Waltham,
MA, USA) kit according to the manufacturer’s instructions. The
sequences of the mimics and the siRNAs are presented in Table II. It is worth noting that we
selected siSGLT2-3, the most effective from the 3 siRNAs against
SGLT2, for use in the follow-up experiments. At 24 to 48 h
post-transfection, the cells were used for RT-qPCR, cell
proliferation analysis, colony formation analysis, cell cycle
analysis and western blot analysis.
 | Table IIThe sequences of the mimic and siRNAs
used in this study. |
Table II
The sequences of the mimic and siRNAs
used in this study.
| Mimic/siRNA | Sequence |
|---|
| NC mimic |
UUUGUACUACACAAAAGUACUG |
| miR-296-5p
mimic |
AGGGCCCCCCCUCAAUCCUGU |
| siNC |
TTCTCCGAACGTGTCACGT |
| siSGLT2-1 |
CGGGTCTCTTCGACAAATA |
| siSGLT2-2 |
GGCCCTGATTGACAATCCT |
| siSGLT2-3 |
CCTGCATCCTCATGGGTTA |
RNA isolation, reverse transcription and
RT-qPCR analysis
Total RNA was extracted from the cells and the
patient tissues using TRIzol reagent (Sangon Biotech, Shanghai,
China). The PrimeScript™ 1st Strand cDNA Synthesis kit (Takara,
Dalian, China) was used for reverse transcription. At the same
time, the PrimeScript®miRNA First-Strand cDNA Synthesis
SuperMixQuantiMir cDNA kit (TransgenBiotec, Beijing, China) was
used to synthesize a cDNA library of miRNAs. mRNA and miRNA
expression levels were quantified by RT-qPCR using SYBR-GreenⅡ
(Takara) and a CFX96™ Real-time System (Bio-Rad, Hercules, CA,
USA). The annealing temperature of the SGLT2 mRNA was 53°C,
for the duration of 1 min at 72°C, for 37 cycles, while the
annealing temperature of miR-296 was 56°C, for the duration of 1
min at 72°C, for 39 cycles. The data for relative quantification of
mRNAs and miRNAs were normal-ized to 18S RNA and U6 snRNA,
respectively. The expression was determined using the relative
quantification (2−∆∆Cq) method (29). The primer sequences are presented
in Table III.
 | Table IIIThe sequence of the primers used in
this study. |
Table III
The sequence of the primers used in
this study.
| Primer | Sequence |
|---|
| U6 snRNA (F) |
5′-CTCGCTTCGGCAGCACA-3′ |
| U6 snRNA (R) |
5′-AACGCTTCACGAATTTGCGT-3′ |
| 18S RNA (F) | 5′-
AGGAATTCCCAGTAAGTGCG-3′ |
| 18S RNA (R) | 5′-
GCCTCACTAAACCATCCAA-3′ |
| miR-296-5p
(qPCR) |
5′-AGGGCCCCCCCTCAATCCUGT-3 |
| SGLT2 qPCR (F) |
5′-GATTTACACGGTGACAGGAGGG-3′ |
| SGLT2 qPCR (R) |
5′-CGGAGCAGGTGGTAGGAGT-3′′ |
| SGLT2-3′-UTR
(F) |
5′-AGATCGCCGTGTAATTCTAGAGTCAACCTCAATGCCCTGCT-3′ |
| SGLT2-3′-UTR
(R) |
5′-TCCGGCTGCAGTGCCGAATTCAGGGGAAAGGCAGCTTTATTT-3′ |
| SGLT2-mut3′-UTR
(F) |
5′-TGCCTCGCGGCGACTGCATCTGATTGGCAGTCA-3′ |
| SGLT2-mut3′-UTR
(R) |
5′-CAGTCGCCGCGAGGCAGAGGAAGGCCGGGAGAA-3′ |
Cell proliferation assay
The cell proliferation assay was performed as
previously described (30). The
NSCLC cells were plated on a 96-well microplate. The density was
2×103 or 4×103 cells per well and the cells
were incubated at 37°C in 5% CO2. After 24, 48 and 72 h
of culture, 8 µl of CCK-8 (Dojindo, Tokyo, Japan) were added. The
cultures were then returned to the incubation conditions (37°C in
5% CO2) for 2 h. The light absorbance at 450 nm was
measured daily with a microplate reader, FLx8 (BioTek, Winooski,
VT, USA). Each point was measured from 3 replicate wells.
Colony formation assay
The colony formation assay was performed as
previously described (31). At
37°C in a 5% CO2 humidified environment, the cells were
plated in 6-well plates at 300 or 600 cells per well and incubated
for 2 weeks. Colonies were stained with crystal violet (Haoranbio,
Shanghai, China) (0.5% w/v) for 12 min at room temperature
following fixation with methanol, and then counted. Experiments
were performed in triplicate.
Cell cycle analysis
Cell cycle analysis was performed as previously
described (32). The cells
(106/ml) were seeded in 6-well plates and transfected
after 24–48 h in culture. The cells were then subjected to
propidium iodide (PI) (BD Pharmingen, San Diego, CA, USA) staining
at 4°C for 15 min. The results were determined with a MoFlo XDP
flow cytometer (Beckman Coulter, Inc., Brea, CA, USA). FlowJo
software (Tree Star Inc., Brea, CA, USA) was used for data
analysis. The experiments were performed in triplicate.
Dual luciferase reporter assay
The dual luciferase reporter assay was performed as
previously described (33). The
3′-UTR of the target gene (SGLT2) was amplified and inserted
downstream of the Firefly luciferase reporter gene in the pGL3
miReport vector (Promega, Madison, WI, USA), named
pGL3-SGLT2-3′-UTR. Mutated SGLT2 sequences were also constructed
(pGL3-SGLT2-3′-mUTR), and confirmed by sequencing (Sangon Biotech).
Moreover, the QuikChange Mutagenesis kit was used to conduct
site-directed mutagenesis of the 3′-UTR. To measure luciferase
activity, the 293T cells were cultured in 24-well plates. At 60-80%
confluence, the 293T cells were co-transfected with 400 ng of
luciferase vector pGL3-SGLT2-3′-UTR or pGL3-SGLT2-3′-mUTR and
miR-296-5p mimic or NC miRNA. We also used 100 nM with 20 ng
plasmid expressing the Renilla luciferase gene (pRL,
Promega) as a final concentration for transfection efficiency as a
control. Following incubation for 48 h, the luciferase activity was
determined by an Orion II Microplate Illuminometer
(Titertek-Berthold, South San Francisco, CA, USA). The primer
sequences are presented in Table
III.
Western blot analysis
Western blot analysis was performed as previously
described (28). Total protein
from the A549 and H1299 cells was extracted using RIPA lysis buffer
(CWBIO, Beijing, China) and a Protein BCA Assay kit (Bio-Rad) was
used to quantify the protein content. Protein samples were
separated by 10% SDS-PAGE and transferred to polyvinylidene
difluoride (PVDF) membranes (Millipore Corporation, Billerica, MA,
USA). After blocking in 5% powdered milk for at least 1 h at room
temperature, the membranes were incubated with rabbit anti-SGLT2
(ab37296; Abcam, Cambridge, MA, USA), and anti-β-actin antibodies
(CST 4970L; Cell Signaling Technology, Danvers, MA, USA) at 1:1,000
overnight at 4°C. The membranes were washed and incubated with a
horseradish peroxidase (HRP)-conjugated secondary antibody
(1:10,000; CST 7074S; Cell Signaling Technology) for 1 h at room
temperature. Subsequent visualization was detected using a
chemiluminescent HRP substrate (Millipore Corporation) and imaging
with an E-Gel Imager. Densitometry (ImageJ software, 1.51d 16;
June, 2016) was used to quantify the relative protein expression of
SGLT2, following normalization to β-actin.
Statistical analysis
The results are expressed as the group means ± SEM
and analyzed using GraphPad Prism 5 software (GraphPad Software,
Inc., La Jolla, CA, USA), using t-tests for two-group comparisons.
The expression of miR-296-5p in the different cell lines was
determined by one-way ANOVA followed by Tukey’s Honest Significant
difference post-hoc test. Moreover, miRNA target prediction
databases were used, including TargetScan (http://www.targetscan.org/vert_71/), RNA22 (https://omictools.com/rna22-tool) and miRDB
(http://www.mirdb.org/). Moreover, the data
analysis website ‘TCGA’ (http://www.kmplot.com), including 1,926 NSCLC patients
cases, was used for survival analysis with respect to SGLT2
expression. The negative correlation of miR-296-5p and SGLT2
was determined by Pearson’s correlation coefficient. Differences
were considered statistically significant at a value of
P<0.05.
Results
miR-296-5p is downregulated in NSCLC
tissues and cells
The investigation of 46 lung cancer patient datasets
(Table I) revealed that miR-296-5p
expression was downregulated compared with the corresponding
non-tumor lung tissues (Fig. 1A).
The analysis of several other datasets (we obtained the
corresponding results by analyzing the specific information of the
patient tissue samples, such as sex, stage and size), including
miRNA expression revealed that the downregulation of miR-296-5p was
associated with tumor size (Fig.
1D), but was not associated with pathological stage (Fig. 1B) or sex (Fig. 1C). Moreover, we probed for the
expression of miR-296-5p in NSCLC cell lines and found that
miR-296-5p was significantly downregulated in the A549, H1299
(P<0.001) and PC-9 cells (P<0.01), compared with the BEAS-2B
control normal lung cells (Fig.
1E). These findings indicate that the downregulation of
miR-296-5p is associated with NSCLC carcinogenesis.
miR-296-5p inhibits cell proliferation
and cell cycle progression
To determine whether miR-296-5p affects NSCLC cell
proliferation, we first transfected the A549 and H1299 cells with
the miR-296-5p mimic. The results of RT-qPCR analysis indicated
that miR-296-5p levels were significantly increased in the cells
transfected with the miR-296-5p mimic (Fig. 2A). Furthermore, we observed that
cellular proliferation gradually decreased with miR-296-5p
overexpression in the A549 and H1299 cells. The upregulation of
miR-296-5p led to a significant decrease in NSCLC cell growth
during a 24-72 h period, when compared with that of the negative
control (NC), as assessed by CCK-8 assay (Fig. 2B). Furthermore, the overexpression
of miR-296-5p significantly decreased colony-forming ability of the
NSCLC cells, as determined by cell colony assays (Fig. 2C). Additionally, G1 arrest was
mediated by the upregulation of miR-296-5p after 48 h (Fig. 2D). Overall, our findings
demonstrated that miR-296-5p exerts a potential inhibitory effect
on tumor formation.
miR-296-5p directly targets SGLT2
Having identified miR-296-5p as a regulator in NSCLC
tissues, we aimed to identify the miR-296-5p targets that mediate
the observed effects. We first identified predicted targets of
miR-296-5p using 3 different publicly available miRNA target
prediction databases (TargetScan, RNA22 and miRDB). Three genes,
namely bromo adjacent homology domain containing 1 (BAHD1),
signal transducer and activator of transcription 3 (STAT3)
and SGLT2 were among the predicted miR-296-5p targets
present in the 3 databases. SGLT2 was selected for further
analysis (Fig. 3A). To further
verify that SGLT2 is targeted by miR-296-5p, we cloned the
3′-UTR of SGLT2 into the pGL3 vector, downstream of the
luciferase open reading frame (ORF). In addition, to destroy the
miR-296-5p binding site, we conducted site-directed mutagenesis of
the 3′-UTR using the QuikChange Mutagenesis kit (Fig. 3B). Co-transfection of the 3′-UTR
vectors with the miR-296-5p mimic led to a decrease in luciferase
activity compared to the miR-control transfection of both the A549
and H1299 cells. By contrast, co-transfection of miR-296-5p mimic
with the mutated form of the 3′-UTR resulted in no significant
change in luciferase activity (Fig.
3C). Finally, we confirmed the silencing of SGLT2 by the
miR-296-5p mimic at the mRNA and protein levels. The mRNA levels of
SGLT2 decreased in the A549 cells following transfection
with the miR-296-5p mimic (Fig.
3D). Of note, the levels of SGLT2 increased in the H1299
cells, mainly owing to miRNAs being involved in the regulation of
the post-transcriptional of gene expression. The mechanisms of
action are complex. miRNAs function mainly in two ways, one is the
degradation of mRNA, and the other is to inhibit translation,
playing a role in protein levels. Western blot analysis clearly
indicated that the inhibitory effects of miR-296-5p are mediated,
at least in part, by the targeting of SGLT2, as the protein
expression of SGLT2 markedly decreased in the cells transfected
with the miR-296-5p mimic (Fig. 3E and
F). On the whole, these data suggest that miRNA can
specifically target the 3′-UTR of SGLT2 in the A549 and
H1299 cells.
SGLT2 is upregulated in NSCLC tissues and
cells
To determine whether the expression of SGLT2
is affected by miR-296-5p, the expression of SGLT2 was
detected in 46 tissues (Table I).
Among the pairs of tissues, SGLT2 expression was
upregu-lated relative to the matched para-cancerous tissues
(P<0.05; Fig. 4A), but was not
associated with pathological stage (Fig. 4B), sex (Fig. 4C), or tumor size (Fig. 4D). Therefore, based on the
Pearson’s correlation coefficient, miR-296-5p was found to
negatively correlate with SGLT2 (Fig. 4E).
Finally, we examined the effects of SGLT2
expression in lung cancer patients. We used the Kaplan-Meier
Plotter online database (www.kmplot.com/analysis) (Fig. 4F) to generate a Kaplan-Meier
survival curve of the patients with NSCLC with a low or high
expression of SGLT2. Among the 1,926 cases, patients with
NSCLC with a high expression of SGLT2 had lower survival
rates.
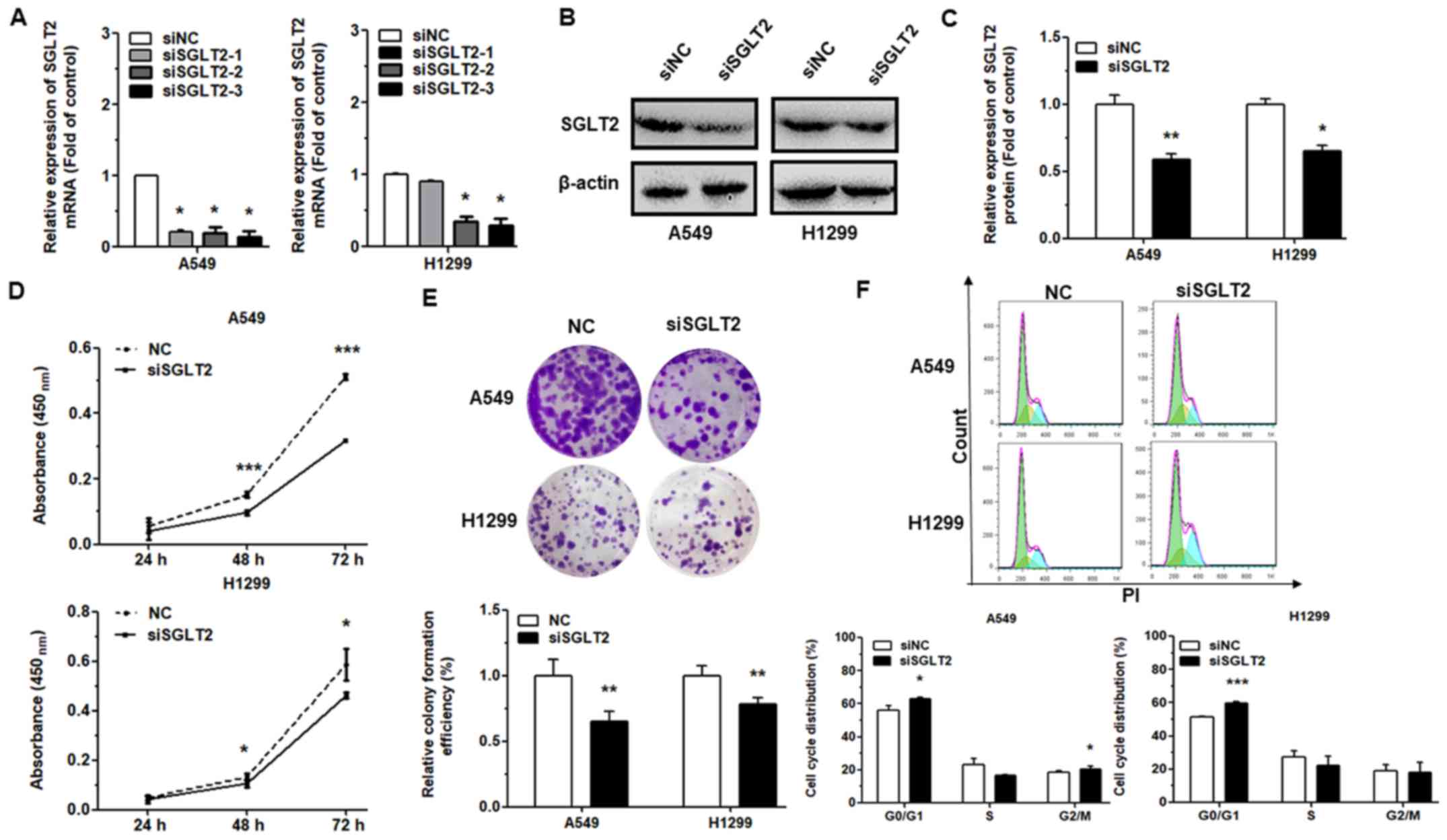
Effects of the inhibition of SGLT2 on
cell proliferation and cell cycle progression
To determine whether the biological effects of
miR-296-5p may be attributed to the direct targeting of
SGLT2, we silenced its expression using siRNA and detected
alterations in cell proliferation and cell cycle progression, as
previously described (26,34). Transfection of the NSCLC cells with
SGLT2-directed siRNA suppressed the SGLT2 mRNA (Fig. 5A) and protein (Fig. 5B and C) levels, as compared to the
control. Moreover, the results of CCK-8 and cell colony assays
revealed that the proliferative capacity of the NSCLC cells was
significantly downregulated at 72 h following transfection with
siSGLT2 (Fig. 5D and E). We also
found that at 48 h following SGLT2 knockdown, the proportion
of cells in the G0/G1 phase increased as compared with the control
(Fig. 5F). Taken together, our
data highlight that the observed effects of miR-296-5p on cell
proliferation and cell cycle progression are mediated by the
targeting of SGLT2.
Discussion
Lung cancer does not have an exact cause, as some
environmental factors, such as tobacco, radon, asbestos and other
industrial carcinogens may increase the risk of developing lung
cancer (8,35,36).
NSCLC has a high incidence and high mortality, and has become the
focus of research in recent years (37). However, the specific mechanisms of
NSCLC remain unclear, and further investigations are required into
this matter. The dysregulation of miRNAs has been linked to the
development of various types of human cancer. Recently, increasing
evidence supports the notion that the aberrant expression of miRNAs
plays critical roles in NSCLC occurrence and progression (38,39)
miRNAs can function as oncogenes or tumor suppressors (40,41).
Our previous studies confirmed that miRNAs, such as miR-34a
(32), miR-486-5p (42) and miR-181a-5p(43) can function as tumor suppressor
genes, while miR-18a-5p (28) and
miR-150 (44) can function as
oncogenes in NSCLC.
Previous findings have established miR-296-5p as a
tumor suppressor through its ability to suppress cancer cells in
different types of cancer (45-48).
However, the roles of miR-296-5p in lung cancer tumorigenesis and
the underlying mechanisms have not yet been completely reported.
Therefore, in this study, we investigated the potential functions
of miR-296-5p in the development and progression of NSCLC. Our
results revealed that miR-296-5p was downregulated in the majority
of the NSCLC patient samples examined and in several NSCLC cell
lines. Moreover, we found that the low expression of miR-296-5p was
related to tumor size according to case information analysis after
measuring the expression of miR-296-5p in 46 pairs of patients by
RT-qPCR. When the tumor is small, the expression of miR-296-5p is
relatively high and in larger tumors, the expression of miR-296-5p
relatively low, which is consistent with the role of miR-296-5p as
a tumor suppressor in lung cancer. In addition, our data confirmed
miR-296-5p as a tumor suppressor in lung cancer and that it
directly targets SGLT2. In the future, we aim to further
investigate the roles of miR-296-5p in suppressing invasiveness and
the cell migratory capability in vitro and in
vivo.
The findings of this study, and other studies shed
some light into the role of miR-296-5p in regulating cancer. It has
also been found that miR-296-5p plays an important role in diabetes
mellitus (DM), which is regulated by SGLT2 (24,49-52).
This study confirmed that miR-296-5p can suppress DM by targeting
SGLT2 (data not shown). Therefore, miR-296 may have more
functions than previously anticipated.
In conclusion, the present study demonstrates that
miR-296-5p functions as a tumor suppressor in NSCLC by targeting
SGLT2. Our findings provide a further understanding of the
potential mechanisms through which miRNAs affect the development
and oncogenesis of NSCLC. The findings of this study may aid in the
development of more effective treatment strategies for NSCLC.
Moreover, miR-296-5p may prove to be a useful prognostic marker for
NSCLC.
Funding
This study was funded by the National Natural
Science Foundation (grant no. 81601887).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
XIAOTIAN Z and XINJU Z conceived the experiments; XL
developed the methodology; PQ and HW analyzed and interpreted the
data; ZM and YC were involved in the conception and design of the
study and edited the manuscript. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
The use of patient samples was approved by the
Ethics Committee of Shanghai Hospital and consent was obtained from
all the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors acknowledge Miss Fatemeh Alsadat Jafari
Sheshtamad (Mashhad University of Medical Science, Mashhad, Iran)
for the critical reading of the manuscript. The authors would also
like to thank her for her valuable comments and suggestions and for
the language editing of the article.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu C, Li S, Chen T, Hu H, Ding C, Xu Z,
Chen J, Liu Z, Lei Z, Zhang HT, et al: miR-296-5p suppresses cell
viability by directly targeting PLK1 in non-small cell lung cancer.
Oncol Rep. 35:497–503. 2016. View Article : Google Scholar
|
|
4
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the Global
Burden of Disease Study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jain RK and Chen H: Spotlight on
brigatinib and its potential in the treatment of patients with
metastatic ALK-positive non-small cell lung cancer who are
resistant or intolerant to crizotinib. Lung Cancer (Auckl).
8:169–177. 2017.
|
|
6
|
Goffin J, Lacchetti C, Ellis PM, Ung YC
and Evans WK; Lung Cancer Disease Site Group of Cancer Care
Ontario’s Program in Evidence-Based Care: First-line systemic
chemotherapy in the treatment of advanced non-small cell lung
cancer: A systematic review. J Thorac Oncol. 5:260–274. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao Y, Gao F, Ma JL, Sun WZ and Song LP:
The potential clinical applications and prospects of microRNAs in
lung cancer. OncoTargets Ther. 7:901–906. 2014. View Article : Google Scholar
|
|
8
|
Kanwal M, Ding XJ and Cao Y: Familial risk
for lung cancer. Oncol Lett. 13:535–542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ye Y, Li SL and Wang SY: Construction and
analysis of mRNA, miRNA, lncRNA, and TF regulatory networks reveal
the key genes associated with prostate cancer. PLoS One.
13:e01980552018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Morales S, De Mayo T, Gulppi FA,
Gonzalez-Hormazabal P, Carrasco V, Reyes JM, Gómez F, Waugh E and
Jara L: Genetic variants in pre-miR-146a, pre-miR-499,
pre-miR-125a, pre-miR-605, and pri-miR-182 are associated with
breast cancer Ssusceptibility in a South American Population. Genes
(Basel). 9:92018. View Article : Google Scholar
|
|
11
|
Tian W, Du Y, Ma Y, Gu L, Zhou J and Deng
D: MALAT1-miR663a negative feedback loop in colon cancer cell
functions through direct miRNA-lncRNA binding. Cell Death Dis.
9:8572018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dua K, Hansbro NG, Foster PS and Hansbro
PM: MicroRNAs as therapeutics for future drug delivery systems in
treatment of lung diseases. Drug Deliv Transl Res. 7:168–178. 2017.
View Article : Google Scholar
|
|
13
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar
|
|
14
|
Bartels CL and Tsongalis GJ: MicroRNAs:
Novel biomarkers for human cancer. Clin Chem. 55:623–631. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bowen T, Jenkins RH and Fraser DJ:
MicroRNAs, transforming growth factor beta-1, and tissue fibrosis.
J Pathol. 229:274–285. 2013. View Article : Google Scholar
|
|
16
|
Fu Q, Song X, Liu Z, Deng X, Luo R, Ge C,
Li R, Li Z, Zhao M, Chen Y, et al: miRomics and proteomics reveal a
miR-296-3p/ PRKCA/FAK/Ras/c-Myc feedback loop modulated by HDGF/
DDX5/β-catenin complex in lung adenocarcinoma. Clin Cancer Res.
23:6336–6350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vaira V, Faversani A, Dohi T, Montorsi M,
Augello C, Gatti S, Coggi G, Altieri DC and Bosari S: miR-296
regulation of a cell polarity-cell plasticity module controls tumor
progression. Oncogene. 31:27–38. 2012. View Article : Google Scholar :
|
|
18
|
Savi F, Forno I, Faversani A, Luciani A,
Caldiera S, Gatti S, Foa P, Ricca D, Bulfamante G, Vaira V, et al:
miR-296/Scribble axis is deregulated in human breast cancer and
miR-296 restoration reduces tumour growth in vivo. Clin Sci (Lond).
127:233–242. 2014. View Article : Google Scholar
|
|
19
|
Barbagallo D, Piro S, Condorelli AG,
Mascali LG, Urbano F, Parrinello N, Monello A, Statello L, Ragusa
M, Rabuazzo AM, et al: miR-296-3p, miR-298-5p and their downstream
networks are causally involved in the higher resistance of
mammalian pancreatic α cells to cytokine-induced apoptosis as
compared to β cells. BMC Genomics. 14:622013. View Article : Google Scholar
|
|
20
|
Carreras-Badosa G, Bonmatí A, Ortega FJ,
Mercader JM, Guindo-Martínez M, Torrents D, Prats-Puig A,
Martinez-Calcerrada JM, de Zegher F, Ibáñez L, et al: Dysregulation
of placental miRNA in maternal obesity is associated with pre- and
postnatal growth. J Clin Endocrinol Metab. 102:2584–2594. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Togliatto G, Dentelli P, Rosso A, Lombardo
G, Gili M, Gallo S, Gai C, Solini A, Camussi G and Brizzi MF:
PDGF-BB carried by endothelial cell-derived extracellular vesicles
reduces vascular smooth muscle cell apoptosis in diabetes.
Diabetes. 67:704–716. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shivapurkar N, Mikhail S, Navarro R, Bai
W, Marshall J, Hwang J, Pishvaian M, Wellstein A and He AR:
Decrease in blood miR-296 predicts chemotherapy resistance and poor
clinical outcome in patients receiving systemic chemotherapy for
metastatic colon cancer. Int J Colorectal Dis. 28:8872013.
View Article : Google Scholar
|
|
23
|
Wang L, Bo X, Zheng Q, Xiao X, Wu L and Li
B: miR-296 inhibits proliferation and induces apoptosis by
targeting FGFR1 in human hepatocellular carcinoma. FEBS Lett.
590:4252–4262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gallo LA, Wright EM and Vallon V: Probing
SGLT2 as a therapeutic target for diabetes: Basic physiology and
consequences. Diab Vasc Dis Res. 12:78–89. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scafoglio C, Hirayama BA, Kepe V, Liu J,
Ghezzi C, Satyamurthy N, Moatamed NA, Huang J, Koepsell H, Barrio
JR, et al: Functional expression of sodium-glucose transporters in
cancer. Proc Natl Acad Sci USA. 112:E4111–E4119. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ishikawa N, Oguri T, Isobe T, Fujitaka K
and Kohno N: SGLT gene expression in primary lung cancers and their
metastatic lesions. Jpn J Cancer Res. 92:874–879. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yao Y, Shao J, Wu J, Zhang Q, Wang J, Xiao
D and Huang F: The functional variant in the 3′UTR of PTPRT with
the risk of esophageal squamous cell carcinoma in a Chinese
population. Cell Physiol Biochem. 36:306–314. 2015. View Article : Google Scholar
|
|
28
|
Liang C, Zhang X, Wang HM, Liu XM, Zhang
XJ, Zheng B, Qian GR and Ma ZL: MicroRNA-18a-5p functions as an
oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis.
8:e27642017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
30
|
Shao Y, Sun Q, Liu X, Wang P, Wu R and Ma
Z: tRF-Leu-CAG promotes cell proliferation and cell cycle in
non-small cell lung cancer. Chem Biol Drug Des. 90:730–738. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang B, Ma Z, Li X, Zhang C, Shao Y, Liu
Z, Li Y and Jin Y: Tanshinones suppress non-small cell lung cancer
through up-regulating miR-137. Acta Biochim Biophys Sin (Shanghai).
48:768–770. 2016. View Article : Google Scholar
|
|
32
|
Li YL, Liu XM, Zhang CY, Zhou JB, Shao Y,
Liang C, Wang HM, Hua ZY, Lu SD and Ma ZL: MicroRNA-34a/EGFR axis
plays pivotal roles in lung tumorigenesis. Oncogenesis. 6:e3722017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang P, Liu X, Shao Y, Wang H, Liang C,
Han B and Ma Z: MicroRNA-107–5p suppresses non-small cell lung
cancer by directly targeting oncogene epidermal growth factor
receptor. Oncotarget. 8:57012–57023. 2017.PubMed/NCBI
|
|
34
|
Hine J, Paterson H, Abrol E, Russell-Jones
D and Herring R: SGLT inhibition and euglycaemic diabetic
ketoacidosis. Lancet Diabetes Endocrinol. 3:503–504. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Brinker TJ, Alfitian J, Seeger W,
Groneberg DA, von Kalle C, Enk AH, Herth FJF, Kreuter M, Bauer CM,
Gatzka M, et al: A face-aging smoking prevention/cessation
intervention for nursery school students in Germany: An
appearance-focused interventional study. Int J Environ Res Public
Health. 15:152018. View Article : Google Scholar
|
|
36
|
Ma H, Chen X, Hu H, Li B, Ying X, Zhou C,
Zhong J, Zhao G and Duan S: Hypermethylation of MDFI promoter with
NSCLC is specific for females, non-smokers and people younger than
65. Oncol Lett. 15:9017–9024. 2018.PubMed/NCBI
|
|
37
|
Cheong HT, Xu F, Choy CT, Hui CWC, Mok TSK
and Wong CH: Upregulation of Bcl2 in NSCLC with acquired resistance
to EGFR-TKI. Oncol Lett. 15:901–907. 2018.PubMed/NCBI
|
|
38
|
Romero-Cordoba SL, Salido-Guadarrama I,
Rodriguez-Dorantes M and Hidalgo-Miranda A: miRNA biogenesis:
Biological impact in the development of cancer. Cancer Biol Ther.
15:1444–1455. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tutar Y: miRNA and cancer; computational
and experimental approaches. Curr Pharm Biotechnol. 15:4292014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ji W, Sun B and Su C: Targeting microRNAs
in cancer gene therapy. Genes (Basel). 8:82017. View Article : Google Scholar
|
|
41
|
Ebrahimi A and Sadroddiny E: MicroRNAs in
lung diseases: Recent findings and their pathophysiological
implications. Pulm Pharmacol Ther. 34:55–63. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shao Y, Shen YQ, Li YL, Liang C, Zhang BJ,
Lu SD, He YY, Wang P, Sun QL, Jin YX, et al: Direct repression of
the oncogene CDK4 by the tumor suppressor miR-486-5p in non-small
cell lung cancer. Oncotarget. 7:34011–34021. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ma Z, Qiu X, Wang D, Li Y, Zhang B, Yuan
T, Wei J, Zhao B, Zhao X, Lou J, et al: miR-181a-5p inhibits cell
proliferation and migration by targeting Kras in non-small cell
lung cancer A549 cells. Acta Biochim Biophys Sin (Shanghai).
47:630–638. 2015. View Article : Google Scholar
|
|
44
|
Wang DT, Ma ZL, Li YL, Wang YQ, Zhao BT,
Wei JL, Qi X, Zhao XT and Jin YX: miR-150, p53 protein and relevant
miRNAs consist of a regulatory network in NSCLC tumorigenesis.
Oncol Rep. 30:492–498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lee H, Shin CH, Kim HR, Choi KH and Kim
HH: MicroRNA-296-5p promotes invasiveness through downregulation of
nerve growth factor receptor and caspase-8. Mol Cells. 40:254–261.
2017. View Article : Google Scholar :
|
|
46
|
Lee KH, Lin FC, Hsu TI, Lin JT, Guo JH,
Tsai CH, Lee YC, Lee YC, Chen CL, Hsiao M, et al: MicroRNA-296-5p
(miR-296-5p) functions as a tumor suppressor in prostate cancer by
directly targeting Pin1. Biochim Biophys Acta. 1843:2055–2066.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li T, Lu YY, Zhao XD, Guo HQ, Liu CH, Li
H, Zhou L, Han YN, Wu KC, Nie YZ, et al: MicroRNA-296-5p increases
proliferation in gastric cancer through repression of
Caudal-related homeobox 1. Oncogene. 33:783–793. 2014. View Article : Google Scholar
|
|
48
|
Maia D, de Carvalho AC, Horst MA, Carvalho
AL, Scapulatempo-Neto C and Vettore AL: Expression of miR-296-5p as
predictive marker for radiotherapy resistance in early-stage
laryngeal carcinoma. J Transl Med. 13:2622015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Watanabe A, Choe S, Chaptal V, Rosenberg
JM, Wright EM, Grabe M and Abramson J: The mechanism of sodium and
substrate release from the binding pocket of vSGLT. Nature.
468:988–991. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cramer SC, Pardridge WM, Hirayama BA and
Wright EM: Colocalization of GLUT2 glucose transporter,
sodium/glucose cotransporter, and gamma-glutamyl transpeptidase in
rat kidney with double-peroxidase immunocytochemistry. Diabetes.
41:766–770. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Vallon V, Rose M, Gerasimova M, Satriano
J, Platt KA, Koepsell H, Cunard R, Sharma K, Thomson SC and Rieg T:
Knockout of Na-glucose transporter SGLT2 attenuates hyperglycemia
and glomerular hyperfiltration but not kidney growth or injury in
diabetes mellitus. Am J Physiol Renal Physiol. 304:F156–F167. 2013.
View Article : Google Scholar :
|
|
52
|
Chao EC and Henry RR: SGLT2 inhibition--a
novel strategy for diabetes treatment. Nat Rev Drug Discov.
9:551–559. 2010. View Article : Google Scholar : PubMed/NCBI
|