Introduction
Prostate cancer (PCa) is one of the most common
types of cancer in men worldwide. In the United States, PCa is the
most frequent malignancy and the third common cause of
cancer-related death in males (1,2).
However, the etiology and pathological mechanisms of PCa require
extensive investigation, despite the current knowledge on the
several risk factors that may affect its origin and development
(3-5). In addition to genetic, biochemical
and metabolic events, biomechanical factors, such as the integrated
forces within tissues, also have an important role in tumor
development (6,7). Previous studies have shown that
pressure is significantly higher in human prostates with cancer
than in normal tissues (8),
prostate epithelial cells and stromal cells, and may confer
resistance to apoptosis (9,10).
Tumor-induced pressure in the bone microenvironment is also known
to promote PCa bone metastasis growth (11). Given that increased prostate
pressure may play a key role in the pathogenesis of PCa (8-11),
Piezo channels, which act as cell sensors and mechanotransduction
mediators, may have an important role in PCa development.
Piezo channels, including piezo type
mechanosensitive ion channel component 1 and 2 (Piezo1 and Piezo2),
were identified in 2010 as the long sought-after molecular carriers
of mechanically activated currents in many cells (12,13).
Piezo channels are essential for detecting external mechanical
stimuli, as well as for sensing mechanical forces within tissues
(such as lung expansion and blood flow) (14-18).
In addition to mechanotransduction, Piezo1 channels also have an
important role in cell neogenesis, survival, differentiation and
proliferation (13,18). For instance, Piezo1 channels
facilitate the migration and alignment of endothelial cells in
response to shear stress induced by blood flow, and disruption of
Piezo1 profoundly disturbs the developing vasculature (16,19).
In epithelial cells, the opening of Piezo1 channels induced by
mechanical stretching is followed by Ca2+ influx, which
activates the Ca2+-sensitive ERK1, in turn promoting
cell proliferation (20).
Proliferation and differentiation of stem cells in the fly midgut
are also triggered by Piezo1 channel activation (21). As a trefoil factor family 1
(TFF1)-binding protein, Piezo1 promotes TFF1-mediated migration and
invasion of gastric cancer cells (22). Given that Piezo1 significantly
promotes cell proliferation and migration, it was hypothesized that
Piezo1 and its downstream signaling pathways may have an important
role in the development of PCa.
In the present study, the expression level of Piezo1
channel was found to be significantly higher in DU145 and PC3 PCa
cell lines, as well as in human prostate malignant tumors compared
to non-malignant tissues. The downregulation of Piezo1 channel
suppressed the proliferation and migration of PCa cells, as well as
the growth prostate tumors inoculated in nude mice. Akt/mTOR
activation and acceleration of cell cycle progression may have been
responsible for Piezo1-induced progression of PCa. Taken together,
the present study strongly suggests that Piezo1 channels have a
crucial role in PCa development. Piezo1 could be a novel
therapeutic target in clinical treatment of PCa.
Materials and methods
Reagents and antibodies
Antibodies specific to Piezo1 (cat. no. 15939-1-AP),
β-actin (cat. no. 66009-1-Ig), CDK4 (cat. no. 11026-1-AP), cyclin
D1 (cat. no. 60186-1-Ig), and platelet and endothelial cell
adhesion molecule 1 (CD31; cat. no. 11265-1-AP) were purchased from
ProteinTech Group, Inc. (dilution, 1:1,000). Antibodies specific to
proliferating cell nuclear antigen (PCNA; cat. no. 2714-1) and
phosphorylated (p-)AKT (Ser473; cat. no. 2118-S) were purchased
from Epitomics (dilution, 1:500; Abcam). Antibodies specific to AKT
(cat. no. GB13011-2), PI3K (cat. no. GB13161) and Ki-67 (cat. no.
GB13030-2) were purchased from Wuhan Servicebio Technology Co.,
Ltd. (dilution, 1:500). The antibodies against p-mTOR (cat. no.
5536T) and mTOR (cat. no. 2972S) were purchased from Cell Signal
Technology, Inc. (dilution, 1:1,000). Antibodies specific to ERK
(cat. no. AF0155) and p-ERK (Thr202/Tyr204; cat. no. AF1015) were
purchased from Affinity Biosciences (dilution, 1:500). Lastly, the
mechanosensitive and stretch-activated ion channel inhibitor,
GsMTx4 (cat. no. ab141871), was obtained from Abcam.
Cell culture
The RWPE-1, PC3, and DU145 cell lines were purchased
from the Shanghai Institutes for Biological Sciences. RWPE-1 was
maintained in K-SFM medium (Thermo Fisher Scientific, Inc.). The
PC3 and DU145 cells were maintained in RPMI-1640 medium (Thermo
Fisher Scientific, Inc.) containing 10% FBS with 100 IU/ml
penicillin and 0.1 mg/ml streptomycin. Cell cultures were
maintained in a humidified atmosphere of 95% air and 5%
CO2 at 37˚C. All cell lines showed no signs of
mycoplasma contamination.
Establishment of Piezo1 knockdown
The target sequence of short hairpin RNA (shRNA) 1
and shRNA2 were, respectively, 5′-cccugugc auugauuaucccu-3′ and
5′-AGAAGAAGAUCGUCAAGUA-3′ (23).
The sequence of the non-targeting control shRNA is
5′-CCUAAGGUUAAGUCGCCCUC-3′. The lentiviral vector pLKO.1 (obtained
from Department of Pharmacology, Hebei Medical University) was used
to construct Piezo1 shRNA1, Piezo1 shRNA2 and control shRNA
lentivirus. The titer of lentivirus was up to 107 TU/ml, 500
µl lentivirus used to infect cells in 35 mm petri dishes.
Lentivirus-infected DU145 cells were screened with 1 µg/ml
puromycin to establish Piezo1 shRNA1 DU145, Piezo1 shRNA2 DU145 and
control shRNA DU145 cells.
Whole-cell patch clamp recording
Whole-cell patch clamp recordings were performed at
a room temperature of 22-24°C with 1×104 cells seeded on
coverslips. Coverslips with cultured cells were placed in a 0.5 ml
microchamber, mounted on the stage of an Olympus IX71 inverted
microscope (Olympus Corporation) and continuously perfused at 2
ml/min with bath solution. The bath solution contained 145 mM NaCl,
5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM
glucose and 10 mM HEPES, with an osmolarity 320 mOsm and pH 7.35.
The recording pipette solution contained 135 mM K-gluconate, 5 mM
KCl, 2.4 mM MgCl2, 0.5 mM CaCl2, 5 mM EGTA, 10 mM HEPES, 5 mM NaCl,
2 mM ATP and 0.33 mM NaGTP, with pH 7.35 and osmolarity 320 mOsm.
The recording electrodes were built from thin wall borosilicate
glass capillaries using a Flaming P-97 puller (Sutter Instrument
Company) and had resistances of 2-4 MΩ. The protocol used to study
Piezo mechanically activated (MA) currents in tumor cells was as
follows: The cells were held at −60 mV and cell membranes were
displaced by heat-polished glass probe. The probe, with a ~4
µm diameter tip, was positioned at an angle of 45° to the
dish surface and its movement was controlled by a Piezo-electric
device (Physik Instrumente, Ltd.). Cells were stimulated with a
series of mechanical stimuli in 1 µm increments to elicit
Piezo MA currents. The moving velocity of the probe was set at 0.5
µm/ms. Signals were recorded with an Axonpatch 700B
amplifier, filtered at 2 kHz and sampled at 5 kHz using pClamp 10.7
(Axon Instruments; Molecular Devices, LLC).
Cell proliferation assays
Cells were seeded at a density of 6×103
cells/well into a 96-well plate (150 µl/well). Cell
proliferation ability was examined using a Cell Proliferation Assay
(MTS; Promega Corporation), according to the manufacturer's
instructions. Colony formation assays were performed to monitor the
PCa cells cloning capability. Cells were seeded into a 6-well plate
at a density of 800 cells per well. After incubation for 12 days,
visible colonies were fixed with 100% methanol at room temperature
for 10 min, stained with 5 mg/ml crystal violet at room temperature
for 20 min, counted and normalized to control group. All
experiments were repeated at least three times.
Transwell assay
The lower chamber of Transwell® was
filled with medium containing 10% FBS. Cells were digested and
suspended in medium without serum and 4×103 cells were
placed in the upper chamber of a Transwell® (Corning,
Inc.). After a 48-h incubation, with or without GsMTx4 (4
µM), the cells were fixed with 4% formalin, at room
temperature for 10 min, then stained with 5 mg/ml crystal violet at
room temperature for 20 min. Five random visual fields were imaged,
and cells were counted. Experiments were repeated at least three
times.
Wound-healing assay
A linear wound was made using 200 µl pipette
tips across a culture of confluent cells. Cells (2×105
cells/well) were cultured in 12-well plates, and allowed to grow to
80-90% confluence. Then cells treated with or without GsMTx4 (4
µM) for 48 h, were imaged after incubation with serum-free
medium for 0, 24 or 48 h. The wound area was calculated at 0, 24
and 48 h after scratching using ImageJ 1.50i software (National
Institutes of Health) to assess the cell migration during wound
closure. Experiments were repeated at least three times.
Cell-cycle analysis
Cells (1×106 cells) were fixed with 70%
ethanol at 4°C overnight, treated with 100 µg/ml stock of
RNase and stained with 50 µg/ml of propidium iodide. The
stained cells were analyzed by flow cytometry (BD Biosciences), and
the results were analyzed using the Expo32 ADC analysis software
version 1.1C (BD Biosciences).
Calcium imaging
Cells (1×105) were loaded with 2
µM fluo-4-acetoxymethyl ester (fluo-4-AM; Molecular Probes;
Thermo Fisher Scientific, Inc.) at 37°C for 30 min. After loading,
the cells were washed three times with Dulbecco's PBS to remove the
extracellular dye, and then placed in a chamber mounted on the
stage of laser scanning confocal microscope (Leica TCS SP5; Leica
Microsystems GmbH). The cells were incubated with the same bath
solution as the patch clamp experiment. Fluo-4-AM loaded calcium
signals were excited at a wavelength of 488 nm, and the emission
fluorescence was measured at 530 nm. The calcium signals from the
Piezo1 channel induced by treatment with its agonist Yoda1 and the
mechanically induced calcium signals were measured. Yoda1 was
applied in bath solution at 1 µM to induce intracellular
calcium signals. The protocol for mechanical stimulation to induce
calcium signals was identical to the protocol used for recording
Piezo1 MA currents with whole-cell patch clamp and cells were
stimulated with a series of mechanical stimuli in 1 µm
increments to elicit calcium signals. Dynamic signals were recorded
at an interval of two seconds and normalized to the initial
fluorescence value. The area under curve (AUC), which means the
area between the curve and basal line axis along the time course,
was calculated with Origin 9.1 software (OriginLab).
Reverse transcription-quantitative PCR
(rt-qPCR)
Total RNA was extracted from the cells using RNAiso
Plus total RNA extraction reagent (Takara Bio, Inc.). cDNA was
synthesized using a PrimeScript RT reagent Kit with gDNA Eraser
(Takara Bio, Inc., Otsu, Japan). Genomic DNA is eliminated by
treatment with gDNA Eraser for 2 min at 42°C. The reaction
conditions were as follows: 37°C for 15 min, 85°C for 5 sec and 4°C
for termination. Subsequently, cDNA was and stored at −20°C. qPCR
was performed using a SYBR Premix Ex Taq Real-Time PCR Kit (Takara
Bio, Inc.). The reaction conditions were one cycle of initial
denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for
30 sec, 60°C for 30 sec. The PCR primer sequences were as follows:
Piezo1, forward 5′-ATGTTGCTCTACACCCTGACC-3′ and reverse
5′-CCAGCACACACATAGATCCAGT-3′; GAPDH, forward
5′-GGCATGGACTGTGGTCATGAG-3′ and reverse 5′-TGCACCACCAACTGCTTAGC-3′.
GAPDH was used as the internal reference gene. Each test was
performed in triplicate and the 2−ΔΔCq method was used
to calculate gene expression (24).
Determination of PI3K activity
The activity of PI3K was measured based on the
amount of NADH using the GENMED PI3K Assay Kit (Genmed Scientifics,
Inc.). The cells were harvested and processed according to
manufacturer's instructions. Protein concentration was determined
using the bicinchoninic acid protein assay kit (Beyotime Institute
of Biotechnology). The absorbance of the negative control and
samples was detected at a wavelength of 340 nm at 0 and 5 min using
a microplate reader (Thermo Fisher Scientific, Inc.). The amount of
NADH (µmol/min/mg) was calculated using the protein
concentration.
Western blot analysis
Total proteins extracted from cells using RIPA lysis
buffer (Beyotime Institute of Biotechnology). The concentrations of
proteins were detected using a bicinchoninic acid protein kit
(Beyotime Institute of Biotechnology). Protein samples (20
µg) were loaded onto 8% gels and separated by SDS-PAGE. The
resolved proteins were electrophoretically transferred to PVDF
membranes. To evaluate proteins levels, the blots were blocked with
5% non-fat milk in TBS with Tween-20 at room temperature for 1 h
and incubated with primary antibodies. β-actin was used as the
internal control. The blots were then probed with secondary
antibodies (IRDye 800CW goat anti-rabbit, cat. no. 926-32210; IRDye
800CW goat anti-mouse, cat. no. 926-32211; LI-COR Biosciences), and
the blots visualized using the Odyssey Fc System (LI-COR
Biosciences). Densitometry of the protein bands was performed using
ImageJ 1.50i software (National Institutes of Health). The
experiments were repeated at least three times.
In vivo xenograft tumor growth
Male Balb/c nu/nu mice (3-4 weeks of age; weight,
12.6-15.4 g; 7 mice/group; 21 mice in total) were purchased from
Beijing Vital River Laboratory Animal Technology Co., Ltd.
(Beijing, China) and kept at temperature (22-24°C) with a stable
humidity (55±15%) with free access to food/water in a 12 h/12 h
light/dark cycle, according to the guidelines of the local Animal
Care and Use Committee at Hebei Medical University. A total of
1×107 Piezo1 shRNA1 cells and 1×107 control
shRNA cells were injected subcutaneously into the right dorsonuchal
area of the nude mice, with seven animals being included in each
injection group. The tumors were measured every 7 days for 4 weeks,
and tumor size was calculated as follows: (a × b2)/2,
where a and b are the longest longitudinal and transverse
diameters, respectively. The endpoint of the experiments was the
28th day after injection. The mice were euthanized, and the tumors
were removed, weighed, imaged and fixed with 4% paraformaldehyde at
room temperature for 24 h. A total of 10 consecutive 4
µm-thick sections were prepared for hematoxylin-eosin (HE)
staining. The sections were stained with 0.2% hematoxylin staining
solution (Sangon Biotech Co., Ltd.) for 5 min at room temperature,
then stained with 0.5% eosin staining solution (Sangon Biotech Co.,
Ltd.) for 3 min at room temperature. Images were acquired using a
Leica microscope (Leica DM6000B; LAS V.4.3; Leica Microsystems,
GmbH).
Bioluminescence imaging in vivo To obtain
luciferase-labeled DU145 cells, 2×105 DU145 cells were
transfected with 10 µl LV-luc-puro lentivirus
(1×108 TU/ml) (Hanbio Biotechnology Co., Ltd.)
A total of 1×107 luciferase-labeled DU145
cells, obtained by transducing with LV-luc-puro lentiviral were
inoculated hypodermically into the right dorsonuchal area of nude
mice (nu/nu; male; 3-4 weeks of age; weight, 12.8-15.2 g; 5
mice/group; 25 mice in total; Vital River Laboratory Animal
Technology Co., Ltd.). The mice were housed at temperature
(22-24°C) with a stable humidity (55±15%) with free access to
food/water in a 12 h/12 h light/dark cycle.
AAV-Piezo1-shRNA1-DsRed2 and AAV-DsRed2 were manufactured by Hanbio
Biotechnology Co., Ltd., and the former was used to deliver
shRNA1-targeted Piezo1 to the tumor site. When tumor size reached
~100 mm3, which occurred around the 14th day, the mice
were randomly assigned to five groups. In four of the groups, mice
were separately treated with AAV-Piezo1-shRNA1-DsRed2, AAV-DsRed2,
saline or GsMTx4. The mice in the other group were left untreated
(blank). The injection of virus was administered on the 18th, 20th,
22nd, 24th and 28th days after tumor cell implantation. For each
injection, 17 µl of either AAV-Gluc-DsRed2 or AAV-S-TRAIL
(1012 virus particles/ml) were injected directly into
the tumor mass. Direct intra-tumor injections of GsMTx4 (12.5
µl at 400 pmol) or NaCl (12.5 µl) took place at the
18th, 20th, 22nd, 24th and 28th day after tumor cell
implantation.
Bioluminescence images were acquired with a Berthold
LB983 NC320 NightOwl System (Berthold Technologies GmbH & Co.
KG) and were used to serially monitor changes of tumor volume. The
mice were imaged for luciferase activity on days 17, 24, 31 and 38
after implantation of LV-luc-puro transduced DU145 cells. The mice
were intraperitoneally injected with D-luciferin (150 mg/kg body
weight), and the tumors were imaged 10 min after injection at a
20-sec exposure time. The study protocol was approved by Animal
Care and Use Committee at Hebei Medical University (Shijiazhuang,
China).
Immunohistochemical (IHC) staining
Human PCa tissue array (Wuhan Servicebio Technology
Co., Ltd.; cat. no. 1677234) contains 26 cases of paracarcinoma
tissues and 44 cases of carcinoma tissues with Gleason scores from
6 to 10 (25). One patient had
regional lymph node metastasis. All samples were from patients
within the age range of 28-87 years. Tissues were fixed in 4%
formalin at room temperature for 24 h and dehydrated with 75, 95
and 100% ethanol and xylene. Samples were embedded in paraffin and
sliced into 4 µm-thick sections. For IHC, sections were
deparaffinized in xylene, rehydrated in 100, 95 and 75% ethanol,
and distilled water, and then a microwave was used for antigen
retrieval. The sections were subsequently soaked in 0.3%
H2O2 to block the activities of endogenous
peroxidases and subsequently incubated with 10% goat serum (Sangon
Biotech Co., Ltd.) for 1 h to prevent the occurrence of
non-specific reactions, all at room temperature. Subsequently, the
sections were incubated with primary antibody at 4°C for 12 h.
Antibodies to Piezo1 (cat. no. 15939-1-AP; dilution, 1:250) and
CD31 (cat. no. 11265-1-AP; dilution, 1:1000) were purchased from
ProteinTech Group, Inc. Antibody specific to proliferating cell
nuclear antigen PCNA (cat. no. 2714-1; dilution, 1:200) was
purchased from Epitomics (dilution, 1:500; Abcam). Antibody
specific to Ki-67 (cat. no. GB13030-2; dilution, 1:300) was
purchased from Wuhan Servicebio Technology Co., Ltd. Immunostaining
was performed using the SP Immunohistochemical commercial assay kit
(cat. no. SP-900; Beijing Zhongshan Jinqiao Biotechnology Co.,
Ltd.) according to the manufacturer's instructions. They were then
incubated with kit's biotinylated secondary antibody at 37°C for 1
h, and streptavidin/peroxidase complex working solution for 1 h.
Peroxidase staining using a diaminobenzidine kit (Dako; Agilent
Technologies, Inc.). Nuclear couterstain were stained with 0.2%
hematoxylin staining solution (Sangon Biotech Co., Ltd.) for 1 min
at room temperature. After IHC staining, tissue specimen/samples
were scanned with Pannoramic MIDI (3DHISTECH, Ltd.) and analyzed
with a Pannoramic viewer (3DHISTECH, Ltd.). The percentage and
intensity of immunostaining were recorded, and the H-score was
calculated using the following formula: H score = ∑(PIxI) =
(percentage of cells of weak intensity x 1) + (percentage of cells
of moderate intensity x 2) + percentage of cells of strong
intensity x 3). The highest possible H-score is 300, and an
expression above the median was defined as high expression, while
an expression below the median was considered low expression
(26).
Statistical analysis
Data are presented as the mean ± SEM for the
indicated number of independently conducted experiments, and
analyzed with SPSS (SPSS, Inc.). Statistical significance was
evaluated using either a Student's t-test or a one-way analysis of
variance followed by Dunnett's post hoc test for multiple groups. A
χ2 test was used to analyze the human tissue arrays.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Piezo1 is upregulated in human PCa
tissues and cell lines
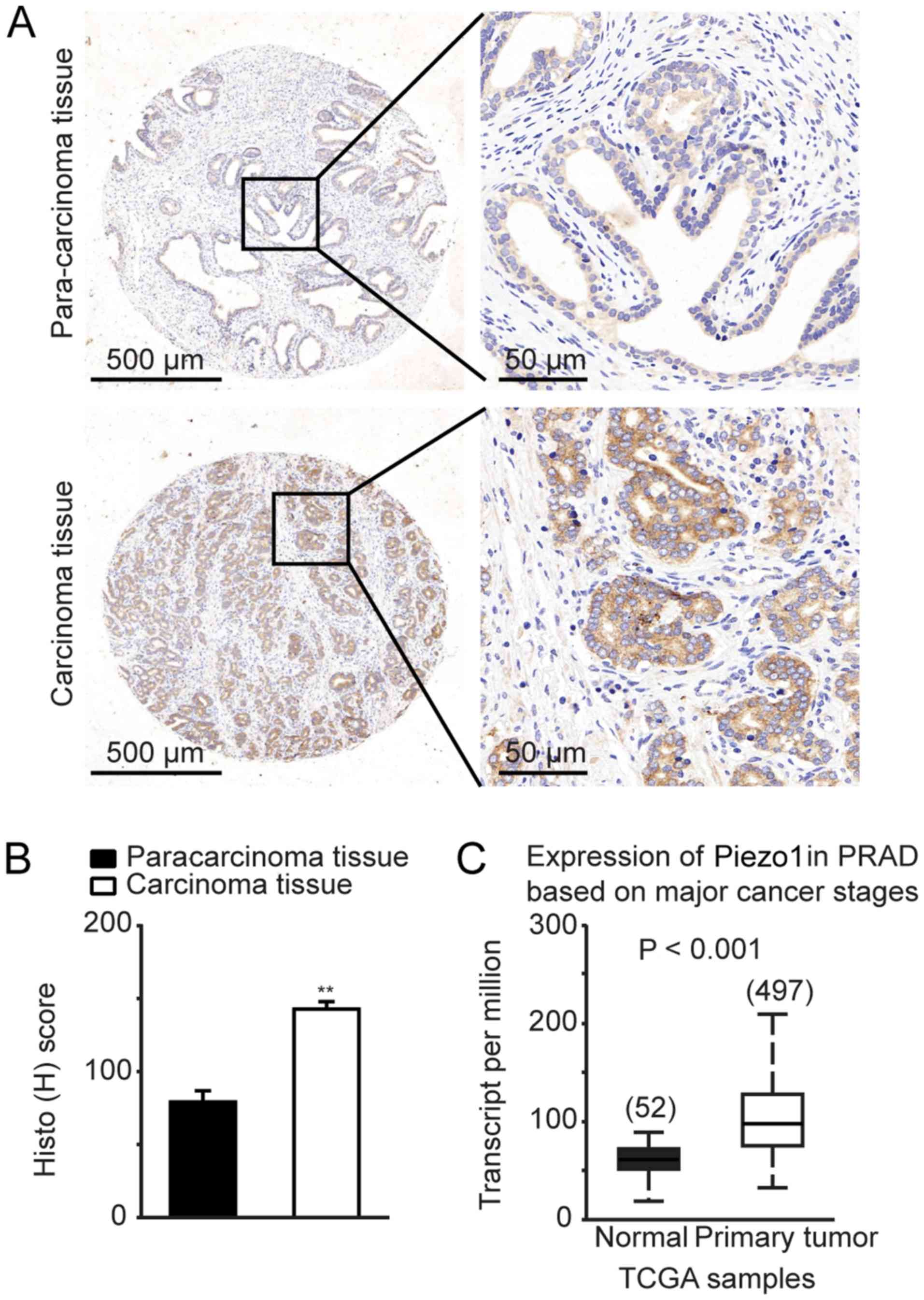
To determine the expression of Piezo1 channel in
human prostate tissues, human prostate tissue arrays were evaluated
by IHC staining. There were 44 cases of prostate carcinoma tissues
and 26 cases of benign tissues adjacent to PCa areas
(paracarcinoma). Piezo1 channel was highly expressed in PCa tissues
compared to paracarcinoma tissues (Fig. 1A), together with a significant
higher H-score for Piezo1 expression in prostate carcinoma tissues
(Fig. 1B).
Tissues array analysis indicated that the Piezo1
channel was upregulated in 31 out of 44 patients with PCa (Table I). However, only 4 out of 26 cases
of human prostate paracarcinoma tissues exhibited upregulation of
the Piezo1 channel, and the remaining 22 cases depicted
downregulation of Piezo1 (Table
I). Clinical evidence from the UALCAN (27) database demonstrated upregulation of
Piezo1, also known as FAM38A, in human PCa tissues (n=497), which
strongly supports the findings of the present study (Fig. 1C).
 | Table IExpression of Piezo1 channel in human
prostate carcinoma tissues and prostate paracarcinoma tissues. |
Table I
Expression of Piezo1 channel in human
prostate carcinoma tissues and prostate paracarcinoma tissues.
| Group | Number of
patients | Piezo1 expression
| P-value |
|---|
| Low | High |
|---|
| Paracarcinoma
tissue | 26 | 22 | 4 | 0.000008 |
| Carcinoma
tissue | 44 | 13 | 31 | |
Similar to the observation that the Piezo1 channel
is upregulated in human PCa tissues, the expression of Piezo1 at
the mRNA level was significantly higher in PC3 and DU145 PCa cell
lines than that in the normal prostate epithelial cell line RWPE-1.
The Piezo1 mRNA levels in the PC3 and DU145 cells were 6.5- and
2.8-fold higher than normal RWPE-1 cells, respectively (Fig. 2A). In addition, western blot
analysis revealed that the protein level of Piezo1 in the PC3 and
DU145 PCa cell lines increased 2.9- and 3.3-fold, respectively,
compared to that in RWPE-1 cells (Fig.
2B). To further characterize differences caused by Piezo1
channel downregulation in PCa cells compared with normal prostate
epithelial cells, patch clamp was performed to record the Piezo1 MA
currents (Fig. 2E and F). The
results showed that Piezo1 MA current densities in DU145 PCa cells
were ~10-fold higher than that in RWPE-1 cells at a displacement
stimulation of 9 µm (Fig. 2E
and F).
Lentiviral vectors expressing Piezo1 shRNA1, Piezo1
shRNA2 or control shRNA were constructed to knockdown the
expression of Piezo1 in DU145 PCa cells. After transfection with
Piezo1 shRNA1 or Piezo1 shRNA2, the mRNA levels of Piezo1 decreased
by 55.2% and 47.5%, respectively, compared to the control shRNA
(Fig. 2C). The protein expression
level of Piezo1 decreased by 52.1 and 50.7%, respectively, compared
to control shRNA (Fig. 2D). The
shRNA1-mediated Piezo1 knockdown also dramatically reduced MA
current densities in DU145 PCa cells (Fig. 2E and F). These results showed that
the Piezo1 channel is upregulated in human PCa tissues and cell
lines, suggesting that Piezo1 may have an important role in the
tumorigenesis of PCa.
Knockdown of Piezo1 channel expression or
inhibition of Piezo1 channel activity reduces the proliferation and
migration of PCa cells in vitro
To determine whether the Piezo1 channel has an
important role in PCa progression, its effect was evaluated on cell
proliferation and migration in vitro. The results of the MTS
assay revealed a significant decrease in the proliferation of DU145
PCa cells following Piezo1 shRNA1 or Piezo1 shRNA2 transfection
(P<0.05; Fig. 3A). The
antiproliferative effect of Piezo1 knockdown was also confirmed
with the colony formation assay on DU145 PCa cells. Colony
formation significantly decreased by 40.2% in the Piezo1 shRNA1
group and 36.7% in the Piezo1 shRNA2 group (Fig. 3B).
The wound-healing assay was performed to test the
effect of Piezo1 on wound closure/cell migration. As shown in
Fig. 4, Piezo1 knockdown by shRNA1
and shRNA2 reduced wound healing of DU145 PCa cells by 55.1 and
44.1%, respectively, at 48 h, while wound closure was complete in
the control shRNA group at 48 h (Fig.
4A). GsMTx4, a relatively specific inhibitor of Piezo1 channel
(28), also reduced wound healing
in DU145 PCa cells by 49.4 and 37.8% at 24 and 48 h, respectively
(Fig. 4B). These results indicated
that migration of PCa cells was reduced by both Piezo1 knockdown
and by Piezo1 channel inhibition. A Transwell® assay was
also performed using DU145 PCa cells. Cell migration was also
markedly reduced by 41.2, 38.5 and 44.2% following Piezo1 shRNA1
transfection, Piezo1 shRNA2 transfection and GsMTx4 treatment,
respectively (Fig. 4C and D).
Taken together, the knockdown of Piezo1 channel expression or its
inhibition resulted in a reduction of PCa cell proliferation and
migration, suggesting that Piezo1 may have oncogenic functions in
PCa.
Piezo1 knockdown or inhibition reduces
xenograft prostate tumor growth in vivo
To determine the role of Piezo1 channel in the
growth of prostate tumors and the effect of its inhibition during
tumor development, two experiments were designed.
DU145 PCa cells and cells expressing control shRNA
or Piezo1 shRNA1 were subcutaneously inoculated into
immunodeficient mice. The distinct tumor growth induced by
implanting DU145 PCa cells into nude mice is shown in Fig. 5A. Piezo1 shRNA silencing led to a
significant reduction in tumor growth (Fig. 5A-C). Both tumor volume and weight,
the latter measured after removal from mice, were reduced by Piezo1
shRNA silencing (Fig. 5A-C). The
tumor volume in the Piezo1 shRNA1 group was reduced by 61.6% by the
3rd week and 64.2% by the 4th week (Fig. 5A and B). Tumor weight in the Piezo1
shRNA1 group was decreased by 56.9% by the 4th week (Fig. 5A and C). The HE staining in
Fig. 5D illustrates tumor
development in the blank, control shRNA and Piezo1 shRNA1 groups.
Further IHC staining for Piezo1 indicated that its expression was
markedly reduced in the shRNA1 group (Fig. 5D). PCNA, which is a specific marker
indicating the proliferation potential of PCa cells (29), was significantly reduced in the
Piezo1 shRNA1 group (Fig. 5D).
Moreover, staining of tumor angiogenesis using anti-CD31, a
biomarker of endothelial cells (16), was significantly reduced in the
Piezo1 shRNA1 group (Fig. 5D).
Knockdown of Piezo1 caused a reduction in CD31 levels in prostate
tumor cells, which was consistent with the findings of a previous
study showing that Piezo1 knockout decreases the proliferation and
integration of endothelial cells (16). These data suggest that the Piezo1
channel may promote the growth of prostate tumors. The knockdown of
Piezo1 expression at the initial stages of PCa may have inhibited
the growth of the xenograft prostate tumor in vivo.
 | Figure 5Inhibition of prostate cancer
xenograft tumor growth by downregulation of Piezo1 in vivo.
(A) The left panel of image shows the nude mice carrying implanted
tumors grown from wild-type DU145 cells (blank), stable DU145 cells
infected with control shRNA and Piezo1 shRNA1. The right panel
shows the tumors isolated from mice of each group on the 28th day
of generation. (B) Tumor volume growth curve measured with calipers
every 7 days (n=7). (C) Measurements of tumor weights from nude
mice on the 28th day (n=7). (D) HE staining of xenograft tumors,
and immunostaining of Piezo1, PCNA and CD31 in wild-type DU145,
control shRNA DU145 and Piezo1 shRNA1 DU145 groups. The expression
of Piezo1, PCNA and CD31 were significantly decreased by Piezo1
shRNA1 interference. Scale bar, 50 µm. Data are presented as
the mean ± SEM. *P<0.05 and **P<0.01
vs. blank. shRNA, short hairpin RNA; Piezo1, piezo type
mechanosensitive ion channel component 1; HE, hematoxylin-eosin;
PCNA, proliferating cell nuclear antigen; CD31, platelet and
endothelial cell adhesion molecule 1. |
Additionally, to avoid the inaccuracies caused by
manual measurements of tumor volume, fluorescein-labeled DU145/Luc
PCa cells were used for bioluminescence imaging to observe tumor
growth in mice. Furthermore, to eliminate the possibility that
inhibition of prostate tumor growth was due to the non-specific
cell damage caused by Piezo1 shRNA silencing in the initial stage,
Piezo1 shRNA injections or GsMTx4 treatment were performed during
prostate tumor growth. After implantation of fluorescein-labeled
DU145/Luc PCa cells for 17 days, four out of five groups of mice
were treated with AAV-piezo1-shRNA1-DsRed2, AAV-DsRed2, saline or
GsMTx4 on the 18th, 20th, 22nd, 24th and 28th day, whereas mice in
another group were not treated (blank). On the 17th day, the photon
counts showed no significant differences among the five groups
(Fig. 6A and B). However, in the
three subsequent weeks, the increase of photon counts in the GsMTx4
group were significantly lower than that of the NaCl or blank
group, and the photon counts of the Piezo1 shRNA1 group were
dramatically lower than that of the control shRNA group (Fig. 6A and B). To verify whether the
bioluminescence imaging technology accurately reflects tumor size,
tumors were stripped and weighed on the 38th day. Consistent with
bioluminescence imaging results, tumor weight was markedly lower
following treatment with GsMTx4 or with Piezo1 shRNA1 injections
(Fig. 6C and D). Thus, both
bioluminescence images and tumor growth revealed that Piezo1
knockdown or the inhibition of Piezo1 channel activity
significantly suppressed prostate tumor growth. HE and IHC staining
revealed that Ki-67, a specific marker of cell proliferation
(29), was downregulated by Piezo1
shRNA treatment (Fig. S1).
 | Figure 6Inhibition of prostate cancer
xenograft tumor growth by downregulation of Piezo1 in vivo.
(A) Representative images of DU145/Luc xenograft tumor mice before
(day 17, implanted for 17 days) and after (days 24, 31, and 38,
implanted for 24, 31 and 38 days, respectively) treatment with
saline, GsMTx4, AAV-DsRed2 or AAV-piezo1-shRNA-DsRed2. (B) The
photon counts before (day 17) intratumoral treatment (top panel)
and the altered photon counts after (day 24, 31 and 38)
intratumoral treatment (botto panel). (C) Tumors were isolated from
nude DU145/Luc xenograft prostate tumor mice in each group on day
38. (D) Measurements of tumor weight from tumors collected from
nude DU145/Luc xenograft mice in each group on the day 38. Data are
presented as the mean ± SEM. *P<0.05. DU145/Luc,
luciferase-labeled DU145 cells; shRNA, short hairpin RNA; Piezo1,
piezo type mechanosensitive ion channel component 1; ns, not
significant. |
Taken together, these results suggest that the
Piezo1 knockdown and its inhibition may have suppressed tumor
growth, not only at the initial stage but also later in the
developmental process of PCa. All these data further illustrate
that Piezo1 is a potential treatment target for PCa.
Knockdown of Piezo1 expression inhibits
Yoda1- and mechanical stimulation-induced intracellular calcium
signals
Piezo1 channel is a mechanically activated cation
channel that allows Ca2+ to pass through and enter cells
(12,30). Moreover, Ca2+ is a
well-known modulator cancer cell proliferation, differentiation and
migration, and has an important role in tumorigenesis (31,32).
To determine the role of the Piezo1 channel in intracellular
calcium signaling, calcium imaging was performed in PCa DU145 cells
in control shRNA and Piezo1 shRNA1 group. The calcium levels
associated with the treatment with the Piezo1 channel activator
Yoda1 and the mechanically induced calcium signals were measured in
both groups. Mechanical stimulation-induced calcium signals in
Piezo1 shRNA1 DU145 cells were markedly lower than that in the
control shRNA DU145 cells. The AUC of the fluorescence curve in the
Piezo1 shRNA1 group was significantly smaller than that in the
control shRNA group (Fig. 7A and
B). Yoda1-induced calcium signals in Piezo1 shRNA1 DU145 cells
were also markedly lower than that in control shRNA DU145 cells.
The AUC of the fluorescence curve in the Piezo1 shRNA1 group was
significantly smaller than that in the control shRNA group
(Fig. 7C and D). These results
suggested that calcium signals are elicited via Piezo1
channel-mediated Ca2+ influx in PCa cells. Moreover,
Piezo1 knockdown significantly inhibits mechanical stimulation- or
Yoda1-elicited intracellular calcium signals.
Akt/mTOR signaling is involved in
downstream events of Piezo1 activation
The ERK and Akt/mTOR signaling pathways are major
molecular mechanisms involved in cell survival, proliferation,
motility and differentiation (33). Both pathways can be regulated by
intracellular Ca2+ signals (33-36).
To determine the molecular mechanisms underlying the
Piezo1-dependent PCa development, the involvement of ERK and/or
Akt/mTOR pathway in the downstream events of Piezo1 activation were
evaluated. The results of the GENMED PI3K kinase activity assay
showed that there were no significant differences in PI3K kinase
activity between the Piezo1 shRNA1 group and the control shRNA
group (Fig. 8A). Western blot
analysis showed that the level of PI3K, Akt and mTOR was not
changed, but the levels of p-Akt (Ser473) and p-mTOR (Ser2448)
markedly decreased in a PI3K-independent manner after knockdown of
Piezo1 channel by Piezo1 shRNA1 interference (Fig. 8B-I). The levels of p-Akt and p-mTOR
decreased by 34.6% and 37.6%, respectively (Fig. 8E and H). The ratios of p-Akt/Akt
and p-mTOR/mTOR were also markedly decreased after knockdown of
Piezo1 channel (Fig. 8F and I).
However, the expression of ERK1/2 and p-ERK1/2 (Thr202/Tyr204) did
not show any significant differences between the Piezo1 shRNA1 and
the control shRNA groups (Fig. 8J and
K). The ratio of p-ERK/ERK did not show any significant
difference after knockdown of Piezo1 channel (Fig. 8L). Taken together, these data
suggested that Piezo1 knockdown may have inhibited PCa cell
proliferation, migration and prostate tumor growth by blocking
Akt/mTOR phosphorylation.
 | Figure 8Downstream signals involved in Piezo1
channel activation in DU145 prostate cancer cells. (A) The activity
of PI3K was detected by GENMED PI3K Assay Kit based on the NADH
levels. (B) Representative western blot assay used to evaluate the
potential downstream signaling molecules associated with Piezo1
activation. Densitometry analysis of (C) PI3K, (D) Akt, (E) p-Akt,
(F) p-Akt/Akt, (G) mTOR, (H) p-mTOR, (I) pmTOR/mTOR, (J) ERK, (K)
p-ERK and (L) p-ERK/ERK. Data are presented as the mean ± SEM
(n=4). *P<0.05 and **P<0.01 vs.
control. shRNA, short hairpin RNA; Piezo1, piezo type
mecha-nosensitive ion channel component 1; p-, phosphorylated-. |
Knockdown of Piezo1 inhibits cell cycle
progression of PCa cells in vitro
In addition to regulating the above signaling
pathways, Ca2+ plays an important role throughout the
cell cycle and is especially important early in G1,
particularly for the G1/S and G2/M
transitions (31). Flow cytometry
analysis was performed to determine the role of Piezo1 in the cell
cycle progression of DU145 PCa cells. The results showed that the
proportion of cells in the G0/G1 phase
significantly increased, whereas cells in the S phase significantly
decreased in the Piezo1 shRNA1 group compared to the control shRNA
group (Fig. 9A and B). This result
indicated that inhibition of Piezo1 expression caused G1
phase arrest. CDK4, cyclin D1 and the cyclin D1-CDK4 complex are
the key effectors in regulation of cell cycle transition from
G1 to the S phase (31). Therefore, the expression levels of
cyclin D1 and CDK4 were evaluated by western blotting in DU145 PCa
cells. Compared with the control shRNA group, the expression of
CDK4 and cyclin D1 proteins considerably decreased with Piezo1
shRNA1 interference (Fig. 9C and
D). The expression of CDK4 and cyclin D1 decreased by 45.0 and
26.2%, respectively (Fig. 9C and
D). Taken together, these results indicated that the
downregulation of Piezo1 in DU145 PCa cells may have led to their
arrest at the G0/G1 phase by inhibiting the
expression of cyclin D1 and CDK4.
Discussion
The major findings of the present study are as
follows: i) Piezo1 is overexpressed in PCa cell lines and in human
PCa tissues; ii) downregulation of Piezo1 significantly reduced PCa
cell proliferation and migration in vitro, and inhibited
prostate tumor growth in vivo; iii) Piezo1-dependent
Ca2+ signals were generated in PCa cells; iv) Piezo1
downstream signaling may have involved Akt/mTOR, but not ERK1/2;
and v) Piezo1-dependent promotion of PCa cell transition from
G1 to S phase may be associated with PCa progression.
Based on these findings, upregulation of Piezo1 in PCa may mediate
an increase in Ca2+ signals. Subsequently, increased
intracellular Ca2+ may activate Akt/mTOR signaling
pathways, upregulating the expression of cyclin D1 and CDK4 and
promoting the assembly of the cyclin D1-CDK4 complex. These
cellular events may, therefore, have promoted PCa cell
proliferation and migration, leading to prostate tumor growth
(Fig. 10). The present results
have shown for the first time (to the best of our knowledge) that
the Piezo1 channel and its downstream signaling pathway may have an
important role in the tumorigenesis of human PCa. These findings
may also have several clinical implications. First, given that it
is overexpressed in PCa cells and tissues, Piezo1 may potentially
serve as a biomarker for the diagnosis and prognosis of PCa.
Second, both in vitro and in vivo studies indicate
that Piezo1 may potentially be used as a therapeutic target for
human PCa. Third, the development of small molecules that
selectively inhibit Piezo1 may be a useful pharmacological
intervention for the treatment of PCa or other cancers where Piezo1
is overexpressed.
Some studies have revealed that Piezo1 is implicated
in human cancer diseases. Piezo1 functions as a TFF1-binding
protein, promoting TFF1-mediated migration and invasion of gastric
cancer cells (22). The
overexpression of Piezo1, accompanied by an increased expression of
β1 integrin, also contributes to the migration of gastric cancer
cells (22). In addition, Piezo1
is overexpressed in malignant MCF-7 breast epithelial cancer cells.
Breast cancer patients with upregulated Piezo1 have higher hazard
ratios and shorter overall survival time (37). More recently, Chen et al
(38) reported that Piezo1 is
localized in focal adhesions and may activate integrin-focal
adhesion kinase signaling, regulating extracellular matrix
associated pathways and reinforcing tissue stiffness. In turn, a
stiffer mechanical microenvironment may lead to the upregulation of
Piezo1, further promoting glioma aggression. In accordance with
these studies, the present findings showed that Piezo1 expression
levels are relatively higher in human PCa tissues and cancer cells
compared with normal tissues and epithelial cells. High expression
of Piezo1 may have promoted the progression of PCa, although the
underlying signaling mechanisms are distinct from those described
in previous studies. However, the present results also contradict
previous findings: McHugh et al (39) described that depletion of the
Piezo1, which was localized to the endoplasmic reticulum,
inactivated β1 integrin affinity and reduced HeLa cell adhesion,
and its knockout promoted the migration of lung epithelial cells.
In addition, loss-of-function germline mutations in Piezo1 have
been identified in some patients with colorectal adenomatous
polyposis (40). Further research
into the association between Piezo1 and cancer is required.
Piezo1 channel mediates Ca2+ influx when
it receives mechanical stimulation (30,41).
Similar to these previous studies, the present experiments
demonstrated that activation of Piezo1 channel by mechanical
stimulation or Yoda1 treatment mediated Ca2+ influx in
PCa cells. Knocking down the expression of Piezo1 reduced the
calcium signals elicited by mechanical stimulation or the agonist
Yoda1. Ca2+ is a very important second messenger that
triggers various cellular biofunctions. The ERK and Akt/mTOR
signaling pathways play a key role in tumorigenesis, and their
activation and activity are regulated by intracellular
Ca2+ signals (33-36,42).
In the present study, the Akt/mTOR, but not ERK1/2, signaling
pathway was activated in DU145 PCa cells in a Piezo1-dependent
manner: Silencing Piezo1 significantly reduced the phosphorylation
levels of Akt and mTOR. Consistent with these findings, a previous
study showed that Piezo1 is required for the phosphorylation of Akt
in endothelial cells in response to shear stress induced by blood
flow (43). Akt is generally
activated by membrane phosphatidylinositol-(3,4,5)-P3, a substrate of PI3K
(33,44). However, in the present study,
Piezo1-mediated Akt activation was independent from PI3K activity,
as the knockdown of Piezo1 did not change the expression levels of
PI3K in DU145 PCa cells. Consistent with these results,
Ca2+ influx mediated by NMDA- or AMPA-type glutamate
receptors or voltage-gated Ca2+ channels, is also known
to activate Akt in a PI3K-independent manner (45-48).
The Piezo1-dependent activation of Akt may involve calmodulin (CaM)
and CaM-dependent protein kinase II (CaMKII), which are activated
by Ca2+. Ca2+/CaMKII activation of Akt plays
an important role in regulating cell survival and apoptosis
(35,36). Further research into whether
Ca2+/CaM/CaMKII signals are induced by Piezo1 activation
is required. However, the dynamic calcium signals recorded in the
present study is limited since it cannot accurately mimic the
intracellular calcium signals responding to the microenvironment of
cancerous tissues. Further research for measuring spontaneous
calcium events in PCa cells is required.
ERK1/2 can be activated by Ca2+ influx
produced by stretch-opened Piezo1 channels, which in turn promotes
epithelial cell proliferation (20). In dental pulp stem cells, ERK1/2
can be activated in a Piezo1-dependent manner by the mechanical
force of low-intensity pulsed ultrasound (49). However, in the present study, the
ERK does not appear to be involved in the downstream signaling
pathway of Piezo1 activation in DU145 PCa cells. Inhibiting the
expression of Piezo1 did not change the phosphorylation levels of
ERK1/2. The exact mechanisms underlying Piezo1-induced activation
of Akt/mTOR, but not ERK in DU145 PCa cells are not clear. A
negative feedback regulation between Akt and ERK pathways may
explain this phenomenon, especially since ERK activation can be
negatively regulated by Akt-mediated Raf phosphorylation, which is
the upstream activator of ERK (33,50).
Ca2+ plays a key role in cell cycle
regulation. The activation of cyclin D1 and CDK4, and the assembly
of cyclin D1-CDK4 complexes are essential for promoting cell cycle
transition from G1 to S phase (31). The present study showed that the
activation of cyclin D1 and CDK4 is suppressed, and the cell cycle
may, therefore, be arrested at G0/G1 phase
after Piezo1 knockdown in DU145 PCa cells. Piezo1-mediated
Ca2+ influx and its downstream signaling pathways may
increase the expression of cyclin D1 and CDK4, and the assembly of
cyclin D1-CDK4 complexes in PCa cells. Moreover, Piezo1-induced
activation of Akt may promote PCa cell transition from
G1 to S phase by activating cyclin D1, since Akt can
stabilize mature cyclin D1 (31,51).
Thus, the Piezo1 knockdown may have inactivated Akt, suppressed
cyclin D1 activation and arrested cells in G1 phase.
In the present study, Piezo1 shRNA only caused ~50%
knockdown of Piezo1 mRNA and proteins, but the Piezo1 MA current
densities were nearly abolished in the shRNA-treated PCa cells.
This result indicates that knockdown of Piezo1 monomer may have
markedly disturbed Piezo1 homotrimer assembly. Additionally, as the
MTS assay showed that knockdown of Piezo1 only induced a mild
suppression in cell viability, but it induced ~50% reduction on the
tumor size in the xenograft tumor growth experiment. One reason is
that cell proliferation conforms to an exponential growth pattern,
short-term cell viability observation in vitro cannot
accurately match long-term xenograft tumor growth in vivo,
the other possibility is that knockdown of Piezo1 may inhibit tumor
growth by inhibiting tumor angiogenesis since knockdown of Piezo1
significantly reduced the expression of vascular endothelial marker
CD31 in tumor tissues.
In summary, the present study found that the Piezo1
channel was upregulated in PCa cells, at the mRNA and protein
levels. The Piezo1 channel was also upregulated in human PCa
tissues. Piezo1-dependent activation of the Akt/mTOR signaling
pathway and acceleration of cell cycle progression may have
contributed to the tumorigenesis of PCa. Furthermore,
downregulation of Piezo1 may have suppressed the proliferation and
migration of PCa cells in vitro and inhibited prostate tumor
growth in vivo. The present study clearly indicates that the
Piezo1 channel has a crucial role in PCa tumorigenesis. Piezo1 may
also serve as a biomarker of PCa and could be used as a novel
therapeutic target in the treatment of human PCa.
Supplementary Materials
Funding
This work is supported by National Natural Science
Foundation of China (NSFC, 81571080 to ZJ, and 81573416 to WZ),
Natural Science Foundation of Hebei Province (H2015206240 to ZJ),
Science and technology research project of Hebei colleges
(ZD2017053 to ZJ), the Ministry of Education (Young Thousand Talent
Program to WZ) and High Talent Science Research Project of
Education Bureau Hebei Province (A2017010068 to WZ).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
ZJ and YZ designed the research. YH, CL, DZ, HM,
LH, QG, SW, YG performed the research. YH, WZ and ZJ analyzed the
data. ZJ, YH, WZ and YZ wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Hebei Medical University, and was performed in accordance with the
approved guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Fitzmaurice C, Allen C, Barber RM,
Barregard L, Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O,
Dandona R, Dandona L, et al: Regional, and national cancer
incidence, mortality, years of life lost, years lived with
disability, and disability-adjusted life-years for 32 cancer
groups, 1990 to 2015:. A systematic analysis for the global burden
of disease study JAMA Oncol. 3:524–548. 2017.
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bostwick DG, Burke HB, Djakiew D, Euling
S, Ho SM, Landolph J, Morrison H, Sonawane B, Shifflett T, Waters
DJ, et al: Human prostate cancer risk factors Cancer. 101(Suppl):
2371–2490. 2004.
|
|
4
|
DeMarzo AM, Nelson WG, Isaacs WB and
Epstein JI: Pathological and molecular aspects of prostate cancer.
Lancet. 361:955–964. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Howard N, Clementino M, Kim D, Wang L,
Verma A, Shi X, Zhang Z and DiPaola RS: New developments in
mechanisms of prostate cancer progression. Semin Cancer Biol. Sep
10–2018.(Epub ahead of print): S1044-579X(18)30079-8, 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Butcher DT, Alliston T and Weaver VM: A
tense situation: Forcing tumour progression. Nat Rev Cancer.
9:108–122. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu H, Mouw JK and Weaver VM: Forcing form
and function: Biomechanical regulation of tumor evolution. Trends
Cell Biol. 21:47–56. 2011. View Article : Google Scholar :
|
|
8
|
Hoyt K, Castaneda B, Zhang M, Nigwekar P,
di Sant'agnese PA, Joseph JV, Strang J, Rubens DJ and Parker KJ:
Tissue elasticity properties as biomarkers for prostate cancer.
Cancer Biomark. 4:213–225. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hegarty PK, Watson RW, Coffey RN, Webber
MM and Fitzpatrick JM: Effects of cyclic stretch on prostatic cells
in culture. J Urol. 168:2291–2295. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wadhera P: An introduction to acinar
pressures in BPH and prostate cancer. Nat Rev Urol. 10:358–366.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sottnik JL, Dai J, Zhang H, Campbell B and
Keller ET: Tumor-induced pressure in the bone microenvironment
causes osteocytes to promote the growth of prostate cancer bone
metastases. Cancer Res. 75:2151–2158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Coste B, Mathur J, Schmidt M, Earley TJ,
Ranade S, Petrus MJ, Dubin AE and Patapoutian A: Piezo1 and Piezo2
are essential components of distinct mechanically activated cation
channels. Science. 330:55–60. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu J, Lewis AH and Grandl J: Touch,
tension, and transduction - the function and regulation of Piezo
ion channels. Trends Biochem Sci. 42:57–71. 2017. View Article : Google Scholar
|
|
14
|
Ranade SS, Woo SH, Dubin AE, Moshourab RA,
Wetzel C, Petrus M, Mathur J, Bégay V, Coste B, Mainquist J, et al:
Piezo2 is the major transducer of mechanical forces for touch
sensation in mice. Nature. 516:121–125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ikeda R, Cha M, Ling J, Jia Z, Coyle D and
Gu JG: Merkel cells transduce and encode tactile stimuli to drive
Aβ-afferent impulses. Cell. 157:664–675. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Hou B, Tumova S, Muraki K, Bruns A,
Ludlow MJ, Sedo A, Hyman AJ, McKeown L, Young RS, et al: Piezo1
integration of vascular architecture with physiological force.
Nature. 515:279–282. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nonomura K, Woo SH, Chang RB, Gillich A,
Qiu Z, Francisco AG, Ranade SS, Liberles SD and Patapoutian A:
Piezo2 senses airway stretch and mediates lung inflation-induced
apnoea. Nature. 541:176–181. 2017. View Article : Google Scholar :
|
|
18
|
Murthy SE, Dubin AE and Patapoutian A:
Piezos thrive under pressure: Mechanically activated ion channels
in health and disease. Nat Rev Mol Cell Biol. 18:771–783. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ranade SS, Qiu Z, Woo SH, Hur SS, Murthy
SE, Cahalan SM, Xu J, Mathur J, Bandell M, Coste B, et al: Piezo1,
a mechanically activated ion channel, is required for vascular
development in mice. Proc Natl Acad Sci USA. 111:10347–10352. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gudipaty SA, Lindblom J, Loftus PD, Redd
MJ, Edes K, Davey CF, Krishnegowda V and Rosenblatt J: Mechanical
stretch triggers rapid epithelial cell division through Piezo1.
Nature. 543:118–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He L, Si G, Huang J, Samuel ADT and
Perrimon N: Mechanical regulation of stem-cell differentiation by
the stretch-activated Piezo channel. Nature. 555:103–106. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang XN, Lu YP, Liu JJ, Huang JK, Liu YP,
Xiao CX, Jazag A, Ren JL and Guleng B: Piezo1 is as a novel trefoil
factor family 1 binding protein that promotes gastric cancer cell
mobility in vitro. Dig Dis Sci. 59:1428–1435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang T, Chi S, Jiang F, Zhao Q and Xiao
B: A protein interaction mechanism for suppressing the
mechanosensitive Piezo channels. Nat Commun. 8:17972017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)). Method Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Shah RB and Zhou M: Recent advances in
prostate cancer pathology: Gleason grading and beyond. Pathol Int.
66:260–272. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Booy EP, Henson ES and Gibson SB:
Epidermal growth factor regulates Mcl-1 expression through the
MAPK-Elk-1 signalling pathway contributing to cell survival in
breast cancer. Oncogene. 30:2367–2378. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bae C, Sachs F and Gottlieb PA: The
mechanosensitive ion channel Piezo1 is inhibited by the peptide
GsMTx4. Biochemistry. 50:6295–6300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhong W, Peng J, He H, Wu D, Han Z, Bi X
and Dai Q: Ki-67 and PCNA expression in prostate cancer and benign
prostatic hyperplasia. Clin Invest Med. 31:E8–E15. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gnanasambandam R, Bae C, Gottlieb PA and
Sachs F: Ionic selectivity and permeation properties of human
PIEZO1 channels. PLoS One. 10:e01255032015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Roderick HL and Cook SJ: Ca2+
signalling checkpoints in cancer: Remodelling Ca2+ for
cancer cell proliferation and survival. Nat Rev Cancer. 8:361–375.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Monteith GR, Prevarskaya N and
Roberts-Thomson SJ: The calcium-cancer signalling nexus. Nat Rev
Cancer. 17:367–380. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mendoza MC, Er EE and Blenis J: The
Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends
Biochem Sci. 36:320–328. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Agell N, Bachs O, Rocamora N and
Villalonga P: Modulation of the Ras/Raf/MEK/ERK pathway by
Ca2+, and calmodulin. Cell Signal. 14:649–54. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gocher AM, Azabdaftari G, Euscher LM, Dai
S, Karacosta LG, Franke TF and Edelman AM: Akt activation by
Ca2+/calmodulin-dependent protein kinase kinase 2
(CaMKK2) in ovarian cancer cells. J Biol Chem. 292:14188–14204.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang R, Zhu Y, Dong X, Liu B, Zhang N,
Wang X, Liu L, Xu C, Huang S and Chen L: Celastrol Attenuates
Cadmium-Induced Neuronal Apoptosis via Inhibiting Ca2+
-CaMKII-Dependent Akt/mTOR Pathway. J Cell Physiol. 232:2145–2157.
2017. View Article : Google Scholar
|
|
37
|
Li C, Rezania S, Kammerer S, Sokolowski A,
Devaney T, Gorischek A, Jahn S, Hackl H, Groschner K, Windpassinger
C, et al: Piezo1 forms mechanosensitive ion channels in the human
MCF-7 breast cancer cell line. Sci Rep. 5:83642015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen X, Wanggou S, Bodalia A, Zhu M, Dong
W, Fan JJ, Yin WC, Min HK, Hu M, Draghici D, et al: A feedforward
mechanism mediated by mechanosensitive ion channel PIEZO1 and
tissue mechanics promotes glioma aggression. Neuron.
100:799–815.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
McHugh BJ, Murdoch A, Haslett C and Sethi
T: Loss of the integrin-activating transmembrane protein Fam38A
(Piezo1) promotes a switch to a reduced integrin-dependent mode of
cell migration. PLoS One. 7:e403462012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Spier I, Kerick M, Drichel D, Horpaopan S,
Altmüller J, Laner A, Holzapfel S, Peters S, Adam R, Zhao B, et al:
Exome sequencing identifies potential novel candidate genes in
patients with unexplained colorectal adenomatous polyposis. Fam
Cancer. 15:281–288. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Miyamoto T, Mochizuki T, Nakagomi H, Kira
S, Watanabe M, Takayama Y, Suzuki Y, Koizumi S, Takeda M and
Tominaga M: Functional role for Piezo1 in stretch-evoked
Ca2 influx and ATP release in urothelial cell cultures.
J Biol Chem. 289:16565–16575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Saxton RA and Sabatini DM: mTOR Signaling
in growth, metabolism, and disease. Cell. 168:960–976. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang S, Chennupati R, Kaur H, Iring A,
Wettschureck N and Offermanns S: Endothelial cation channel PIEZO1
controls blood pressure by mediating flow-induced ATP release. J
Clin Invest. 126:4527–4536. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vivanco I and Sawyers CL: The
phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat Rev
Cancer. 2:489–501. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yano S, Tokumitsu H and Soderling TR:
Calcium promotes cell survival through CaM-K kinase activation of
the protein-kinase-B pathway. Nature. 396:584–587. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ishiuchi S, Tsuzuki K, Yoshida Y, Yamada
N, Hagimura N, Okado H, Miwa A, Kurihara H, Nakazato Y, Tamura M,
et al: Blockage of Ca(2+)-permeable AMPA receptors suppresses
migration and induces apoptosis in human glioblastoma cells. Nat
Med. 8:971–978. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ishiuchi S, Yoshida Y, Sugawara K, Aihara
M, Ohtani T, Watanabe T, Saito N, Tsuzuki K, Okado H, Miwa A, et
al: Ca2+-permeable AMPA receptors regulate growth of
human glioblastoma via Akt activation. J Neurosci. 27:7987–8001.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Valerie NC, Dziegielewska B, Hosing AS,
Augustin E, Gray LS, Brautigan DL, Larner JM and Dziegielewski J:
Inhibition of T-type calcium channels disrupts Akt signaling and
promotes apoptosis in glioblastoma cells. Biochem Pharmacol.
85:888–897. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gao Q, Cooper PR, Walmsley AD and Scheven
BA: Role of Piezo channels in ultrasound-stimulated dental stem
cells. J Endod. 43:1130–1136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zimmermann S and Moelling K:
Phosphorylation and regulation of Raf by Akt (protein kinase B).
Science. 286:1741–1744. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Diehl JA, Cheng M, Roussel MF and Sherr
CJ: Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis
and subcellular localization. Genes Dev. 12:3499–3511. 1998.
View Article : Google Scholar : PubMed/NCBI
|