Introduction
Locally advanced rectal cancer is treated with
pre-operative chemoradiotherapy, followed by the resection of
rectal cancer (1,2). Pre-operative chemoradiotherapy (CRT)
increases the possibility of complete resection and reduces the
risk of local recurrence. However, radiation to the pelvic region
may cause adverse effects, such as urogenital and anal dysfunctions
(3,4). Recently, neoadjuvant chemotherapy
(NAC) without radiation to the pelvic region has also been applied
to locally advanced rectal cancer. However, approximately half of
NAC-treated cases exhibit poor responses to initial treatment
(5-7). Predicting the response to NAC from
clinical and pathological features of rectal cancer remains
difficult.
5-Fluorouracil (5FU) and its prodrugs,
fluoropyrimidine derivatives, are key chemotherapy drugs for
colorectal cancer. The 5FU agents are for adjuvant chemotherapy of
rectal cancer, and also for pre-operative CRT and NAC (1,2,5-9).
Previous studies reported that the expression levels of the target
enzyme thymidine synthase (TS) and the enzymes involved in
fluoropyrimidine metabolism are associated with the susceptibility
of colorectal cancer to 5FU (10-13).
The increased expression of TS (10,11)
and dihydro pyrimidine dehydrogenase, which is a degradation enzyme
of 5FU (10-13), and decreased expression of orotate
phosphoribosyl transferase, which is a metabolic enzyme that
convert 5FU into its active form (13), are suggestive for the resistance to
5FU. Previously, the association of resistance to 5FU with genetic
features or non-coding RNAs has been reported. Micro-satellite
instability status was also associated with the susceptibility to
5FU (14). Additional reports have
indicated a link between microRNAs and long non-coding RNAs
(lncRNAs) with the susceptibility of colorectal carcinoma cells to
5FU (15-22).
LncRNAs are RNAs that >200 base pairs in length
and serve significant roles in various biological functions, such
as gene expression, protein expression and chromatin status
(23). It has been noted that
lncRNAs are involved in the proliferation, invasion and metastasis
of carcinomas at various sites (24). Furthermore, an association between
lncRNAs and the resistance to chemotherapeutic agents was reported
in carcinomas (25). However, it
is unclear which lncRNA is useful for the prediction of the
susceptibility to 5FU.
In the present study, we examined the expression
profile of lncRNAs in 5FU-susceptible and 5FU-resistant cultured
colorectal cancer cells using PCR array analysis and identified
possible candidate lncRNAs, H19 and urothelial cancer associated 1
(UCA1), by bioinformatic analysis. The expression levels of H19 and
UCA1 and their alterations were examined in paired biopsy and
resected specimens. Alterations in expression levels, rather than
the basal expression levels of these lncRNAs, were suggested to be
predictive for the susceptibility to NAC.
Materials and methods
Cell lines and culture
HCT116, DLD-1 and SW480 human colorectal carcinoma
cell lines were employed in this study. HCT116 cells were obtained
from the RIKEN BioResource Research Center; DLD-1 cells were
obtained from Taiho Pharmaceutical Co., Ltd. SW480 cells were
obtained from the American Type Culture Collection. To generate
5FU-resistant cell lines, the parental cells (designated as
HCT116/p, DLD-1/p, and SW480/p) were repeatedly treated with 5FU
(Sigma-Aldrich; Merck KGaA) at 37°C for 120 h. The concentration of
5FU started at 0.2 µM and was increased to 2, 5 and 10
µM and then by 10 µM until 100 µM (26). After each treatment step, cells
were cultured in medium without 5FU until the cells proliferated to
80% confluency. The treated cells were designated as HCT116/5fu,
DLD-1/5fu, and SW480/5fu. DLD-1/5fu cells were kindly provided by
Taiho Pharmaceutical. The cells were cultured in RPMI 1640 (Thermo
Fisher Scientific, Inc.) with 10% fetal bovine serum (Nichirei
Bioscience Inc.).
5FU susceptibility assay
Cells were plated on 6-well plates at
1.5×105 cells/well, and cultured in medium without 5FU
or with 1, 2, 5 or 10 µM 5FU at 37°C for 48 h. Then, the
number of living cells was counted using a Cell Counter, model R1
(Olympus Corporation). The experiments were performed in
triplicate. Survival was expressed as the percentage of living
cells treated with 5FU, as calculated by dividing the mean numbers
of living cells treated with 5FU by the mean numbers of living
cells without 5FU. The half maximal-inhibitory concentration
(IC50) was calculated using GraphPad Prism, version 7
(GraphPad Software, Inc.).
Cell proliferation assay
Cells were plated in 6-well plates at
1.5×105 cells/well in medium with or without 10
µM 5FU at 37°C until 96 h. At 24, 48, 72 and 96 h, the
numbers of living cells were counted using a Cell Counter, model
R1.
RNA extraction from cultured cells
Cells were plated on 60-mm dishes at
2.5×105 cells/well, and cultured with or without 10
µM 5FU at 37°C for 48 h. After a wash with phosphate
buffered saline (PBS), total RNA was extracted using TRIzol (Thermo
Fisher Scientific, Inc.) according to the recommended protocol. The
concentration of total RNA was measured.
Comprehensive analysis of lncRNA
expression
DLD-1/p and DLD-1/5fu cells were cultured for 48 h
without 5FU. Total RNA was extracted from these cells using TRIzol
(Thermo Fisher Scientific, Inc.). Complementary DNA (cDNA) was
synthesized from 1,000 ng total RNA using the RT2 First
Strand Kit (Qiagen, Inc.) according to the manufacturer's
instructions. The expression profile of lncRNAs was obtained using
the RT2 lncRNA PCR Human Cancer PathwayFinder (Qiagen,
Inc.). The PCR mixture was a 25 µl solution containing 1X
RT2 SYBR Green ROX qPCR Mastermix (Qiagen, Inc.), cDNA
synthesized from 8.5 ng total RNA, and primers. The expression of a
total of 84 lncRNAs and 5 control housekeeping genes was analyzed;
the list of analyzed lncRNAs is available at the manufacturer's
website (cat. no. LAHS-002Z). PCR was performed using a 7500
Real-Time PCR System (Thermo Fisher Scientific, Inc.). The reaction
was initiated with incubation at 95°C for 10 min, followed by 60
cycles of incubation at 95°C for 15 sec and at 60°C for 1 min. The
expression levels of lncRNAs were standardized with levels of the
housekeeping genes. The expression levels of lncRNAs in DLD-1/5fu
cells were calculated as fold-change relative to the levels in
DLD-1/p cells by the 2-ΔΔCq method (27) and expressed as base-2 logarithm of
fold-change.
Pathway analysis of lncRNA
Pathways of the lncRNAs were analyzed by Ingenuity
Pathway Analysis (IPA; Qiagen, Inc.). The scores of networks were
calculated by the numbers of focus molecules.
Estimation of doubling time of cultured
cells
Cells were plated on 6-well plates at
1.5×105 cells/well and cultured with or without 10
µM 5FU. At 0, 24, 48, 72 and 96 h after plating, the cell
numbers were counted with Cell Counter, model R1. The doubling time
was estimated from the linear phase of growth curve of cultured
cells.
Cell cycle assay
Cells were plated on 100 mm dishes at
7.5×105 cells/well and cultured with or without 10
µM 5FU. After 48 h, the cells were collected and counted
using a Cell Counter, model R1. One million cells were transferred
to 1.5 ml tube, and the cells were fixed in 1 ml of 70% ethanol at
-20°C for 3 h. The cells were then washed once with PBS and
resuspended in 200 µl of Cell Cycle reagents of Muse Cell
Cycle Kit (cat. no. MCH 100106, Luminex Japan Corporation Ltd.).
The cells were incubated for 30 min at room temperature in the
dark. Cell cycle phases were determined by Muse Cell Analyzer
(Luminex Japan Corporation Ltd.). Approximately 10,000 cells were
analyzed, and the number of cells at cell cycle phases of G0/G1, S
and G2/M were counted. The cell cycle phases were expressed as the
percentage of cells at these phases.
Western blot analysis
Cells were plated on 100 mm dishes at
7.5×105 cells/well and cultured with or without 10
µM 5FU for 48 h. After a wash with PBS, the cells were lysed
in 0.5% SDS/50 mM Tris-HCl (pH 7.6) and sonicated for 10 min.
Protein samples were loaded onto 5-20% SDS-PAGE gradient gels and
transferred to a polyvinylidene difluo-ride membrane. After
blocking of the membrane with 5% skim milk in Tris-buffered
saline/0.5% Tween 20 at room temperature for 30 min, the membrane
was incubated with primary antibodies at 4°C overnight:
Anti-retinoblastoma (Rb) antibody (cat. no. 9309; 1:2,000; Cell
Signaling Technology, Inc.), anti-p27kip1 antibody (cat. no.
610242; 1:5,000; Becton, Dickinson and Company), and anti-β-actin
(cat. no. A5316; 1:10,000; Sigma-Aldrich; Merck KGaA). After
washing the membrane with a buffer containing 250 mM Tris-HCl/150
mM NaCl/0.1% TritonX, the membrane was then incubated with
horseradish peroxidase-labeled anti-mouse IgG antibody (cat. no.
A106PU; 1:10,000; American Qualex Scientific Products) at room
temperature for 60 min. Peroxidase activity was detected with
chemiluminescence using SuperSignal West Dura Extended Duration
Substrate (Thermo Fisher Scientific, Inc.). The bands were
quantified using Quantity One Software version 4. 6. 2 (Bio-Rad
Laboratories, Inc.).
Rectal cancer cases
Cases of rectal cancer treated with NAC between
January 2012 and April 2017 were retrieved from the records of the
Department of Gastrointestinal and Hepato-Biliary-Pancreatic
Surgery, Nippon Medical School Hospital. A total of 35 cases of
rectal cancer, pre-operatively diagnosed as stage cT3 or cT4
without distant metastasis and treated with six cycles of
combination therapy with 5FU, leucovorin, and oxaliplatin, were
identified. Molecular targeting drugs were not used and
radiotherapy was not performed. After 1 month after the end of
chemotherapy, the tumor was resected.
Histological subtypes, depth of the tumor invasion
and histological therapeutic responses were evaluated in the
resected tissue by two investigators in accordance with the
criteria of the Japanese Classification of Colorectal Carcinoma
(28): Grade 0, no denaturation
and necrosis of the cancer; Grade 1a, denaturation and necrosis in
less than one-third of the cancer tissue; Grade 1b, denaturation
and necrosis in one-third or more to less than two-thirds of the
cancer tissue; Grade 2, denaturation, necrosis and loss in
two-thirds or more of the cancer tissue; and Grade 3, no residual
cancer cells.
Two patients with Grade 3 response were excluded
from the study as no cancer cells were identified in the resected
tissue. Another two patients from whom biopsy specimens were not
available were also excluded. Finally, 31 patients with rectal
cancer who received NAC were studied. The present study was
approved by the Ethics Committee of Nippon Medical School Hospital
(approval no. 29-03-909). Written informed consent was obtained
from all patients.
RNA extraction from biopsy and resected
cancer tissues
Total RNA was extracted from formalin-fixed,
paraffin-embedded tissue of biopsy specimens taken before NAC and
resected tissue specimens taken after NAC treatment using RNeasy
FFPE Kit (Qiagen, Inc.). Five slices of 10-µm-thick sections
of biopsy tissue were deparaffinized using xylene and ethanol in
1.5 ml tubes and dried. Three slices of 10-µm-thick sections
of resected tissue were deparaffinized, hydrated, and stained with
hematoxylin at room temperature for 30 sec. Cancer tissue was
dissected out, collected and then transferred to 1.5 ml tubes, and
dried. Tissues were incubated at 56°C overnight in 240 µl of
Buffer PKD provided with the kit. Total RNA was extracted according
to the manufacturer's protocol, and the concentration of total RNA
was determined by spectrophotometry.
Reverse transcription and quantitative
PCR
cDNA was reverse-transcribed from total RNA using
the SuperScript VILO cDNA Synthesis Kit (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. One microgram
of total RNA extracted from cultured cells and 250 ng total RNA
extracted from biopsy specimens and resected tissues were used as
templates.
Quantitative PCR was performed in a 20-µl
reaction mixture containing 1X TaqMan Fast Universal PCR Master Mix
(Thermo Fisher Scientific, Inc.), 1X TaqMan primers and probes;
synthesized cDNA was then reverse transcribed. The TaqMan primers
and probes were as follows: H19 (Hs00262142), UCA1 (Hs01909129),
and 18S ribosome RNA (rRNA) (Hs03928990) (all from Thermo Fisher
Scientific, Inc.). The reaction was initiated with incubation at
95°C for 20 sec, followed by 40 cycles of incubation at 95°C for 1
sec and at 60°C for 20 sec. cDNA that was reverse transcribed from
a mixture of total RNA extracted from DLD-1/p, SW480/p, and
HCT116/p cells were used for standardization. Alterations in
fluorescence were monitored by the Step One Plus Real-Time PCR
System (Thermo Fisher Scientific, Inc.). The expression levels of
H19 and UCA1 were standardized with those of 18S rRNA. The
expression levels were calculated by the 2-ΔΔCq method
(27) and expressed as fold-change
relative to the level of the mixture of total RNA extracted from
DLD-1/p, SW480/p, and HCT116/p cells. The fold-changes in the
expression levels of 5FU-treated cultured cells compared with
untreated cultured cells were expressed as 5FU-treated/untreated
ratio. The fold-changes in resected specimens compared with biopsy
specimens were calculated as resection/biopsy ratio.
Statistical analysis
Data are expressed as the mean ± standard deviation.
χ2 and Mann-Whiney U tests, and two-way ANOVA with
Sidak's post-hoc test were performed using GraphPad Prism, version
7 (GraphPad Software, Inc.). Hierarchical clustering was conducted
using JMP software version 13.2 (SAS Institute Japan, Ltd.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
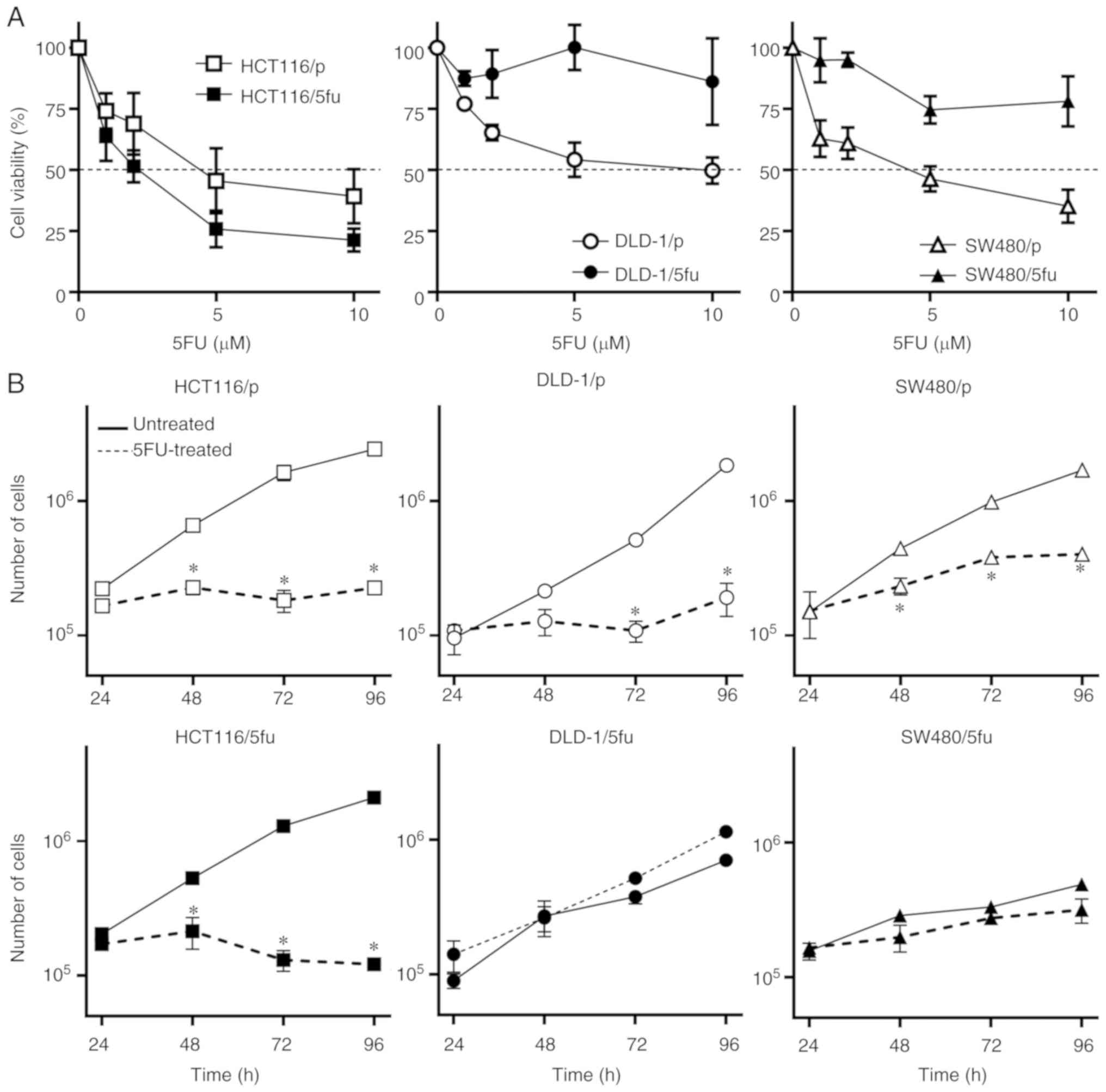
Susceptibility of cultured cells to
5FU
We treated a panel of parental and 5FU-resistant
colorectal carcinoma cell lines with various concentrations of 5FU
and determined the IC50 values (Fig. 1A). The IC50 values of
the parental cells HCT116/p, DLD-1/p, and SW480/p were 4.5, 6.4 and
3.5 µM, respectively. The IC50 values of
HCT116/5fu, DLD-1/5fu, and SW480/5fu cells were 2.0, 103.8 and 26.2
µM, respectively (data not shown).
We next examined the proliferation of cell lines
treated with 10 µM 5FU. The proliferation of all three
parental cell lines HCT116/p, DLD-1/p and SW480/p was significantly
inhibited by 10 µM 5FU compared with the untreated cells
(Fig. 1B). The proliferation of
DLD-1/5fu and SW480/5fu cells were not notably inhibited by 5FU,
whereas suppressed proliferation was exhibited by HCT116/5fu cells
following treatment with 5FU.
Collectively, these results indicated that the three
parental cell lines and HCT116/5fu cells were susceptible to 5FU,
while the DLD-1/5fu and SW480/5fu cell lines appeared to be
resistant to 5FU.
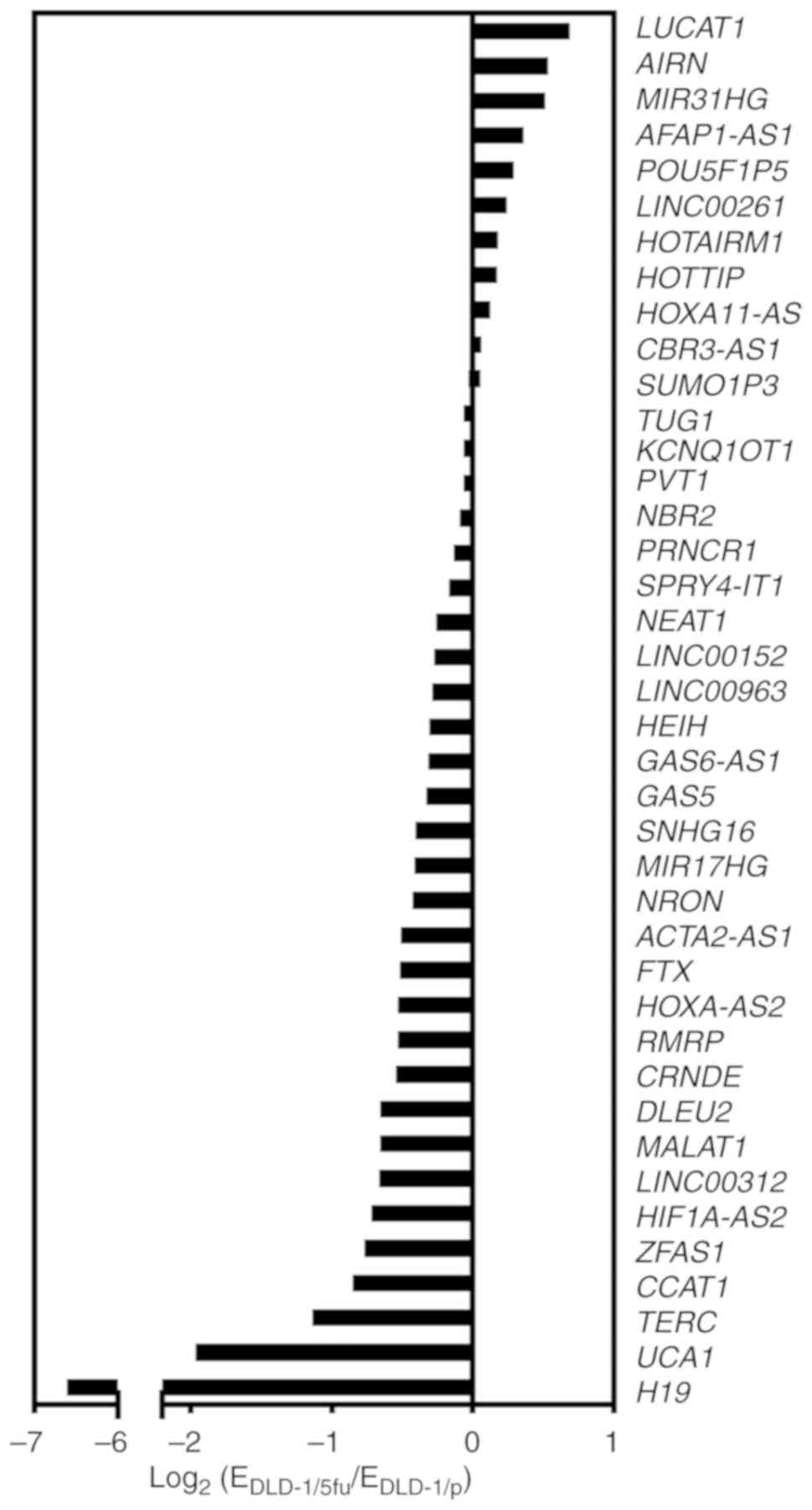
Comprehensive and bioinformatics analysis
of lncRNA expression in 5FU-resistant cells
We next performed a comprehensive analysis of the
expression profile of lncRNAs in the 5FU-resistant line DLD-1/5fu
compared with DLD-1/p parental cells. Of the 84 lncRNAs from the
expression array, 40 lncRNAs were considered to be suitable for the
analysis as the threshold cycle was <30 cycles in both DLD-1/p
and DLD-1/5fu sets. Among the 40 lncRNAs, 11 lncRNAs were
upregulated and 29 lncRNAs were downregulated in DLD-1/5fu cells
compared with DLD-1/p cells (Fig.
2). No lncRNAs were upregulated by >2-fold. Among the
downregu-lated lncRNAs, telomerase RNA component (TERC), UCA1, and
H19 were decreased less than half.
Five networks were identified by IPA (Table I). Networks 1-3 contained 10
lncRNAs. UCA1 was categorized in network 1, and H19 and TERC were
categorized in network 2. Pathway analysis revealed numerous
connections of H19 with other molecules (Fig. 3). Although UCA1 showed fewer
connections, UCA1 had connections with molecules in other networks.
Only a few connections were observed with TERC. We thus suspected
that H19 and UCA1 may be key molecules involved in the
susceptibility of cultured cells to 5FU.
 | Table INetworks identified by Ingenuity
Pathway Analysis. |
Table I
Networks identified by Ingenuity
Pathway Analysis.
| ID | Molecules in
network | Score | Focus
molecules | Top diseases and
function |
|---|
| 1 | Akt, AR, COL2A1,
corticosterone, CRNDEa, CTNNB1,
EGFR, FTXa, GAS5a, Gsk3, HMGA1, HNRNPA2B1,
HOXA11-ASa, IGF2, IL1,
LINC00963a, MIR17HGa, MIR31HGa, MMP14, MTDH, PARP1, PDGF BB, PGR,
PRNCR1a, progesterone, Rb,
S100A8, SMARCA4, SOX2, SOX9, TWIST2, UCA1a, ZBTB7A, ZEB1, ZFAS1a | 22 | 10 | Cellular movement,
cell cycle, cellular development |
| 2 | CDKN3, COL2A1,
corticosterone, CTCF, Cyclin D, NMT3L, FGF2, Gsk3, GSK3B,
H19a, H2BFM, HOTAIRM1a, hydrogen peroxide, IGF2,
KCNQ1OT1a, LINC00261a, MALAT1a, Map3k7, NBR2a, PDGF BB, PGR, POMC, PRKAA, Rb,
RMRPa, Srebp, SUMO1P3a, TERCa, TGFB1, TNF, tretinoin, TUG1a, TWIST2, ZBTB7A, ZFP57 | 22 | 10 | Cellular
development, cellular growth and prolif eration, cellular
movement |
| 3 | AFAP1-AS1a, CASP8, CBR3-AS1a, CCAT1a, CCND1, CD28, CD3, CDC73, CHEK1,
corticosterone, CYTORa,
DLEU2a, DNMT3A, ELAVL1, Gsk3,
HMGA1, HNRNPU, HOXA-AS2a, IGF1R,
IGF2, IGF2BP3, MYC, NEAT1a,
NOS2, PARP1, PDGF BB, PI3K (complex), PVT1a, Rb, RHOA, S100A8, SNHG16a, SOX9, SPRY4-IT1a, TWIST2 | 22 | 10 | Cell cycle,
cellular movement, cell death and survival |
| 4 | HOTTIPa, HOXA13 | 3 | 1 | Cell death and
survival, developmental disorder, hereditary disorder |
| 5 | LRRK2, NFATC2,
NRONa | 2 | 1 | Cellular
development, cellular growth and, proliferation nervous system
development and function |
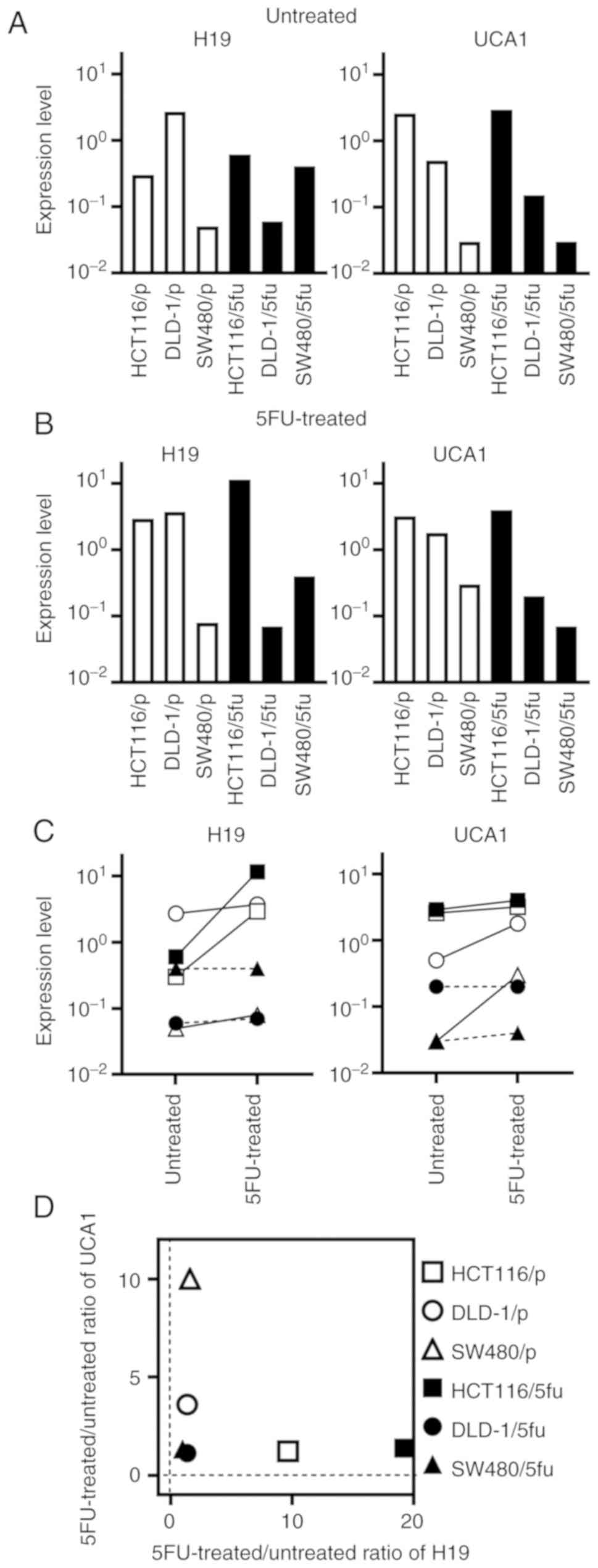
H19 and UCA1 expression in cultured
cells
We next examined the expression levels of H19 and
UCA1 in cultured cells. The expression levels of H19 and UCA1
markedly varied among the untreated cultured cells (Fig. 4A) and among the 5FU-treated
cultured cells (Fig. 4B). The
expression level of H19 was changed >10-fold in HCT116/p and
HCT116/5fu cells by treatment with 5FU, whereas the change of H19
in other cultured cells were <2-fold (Fig. 4C). The expression level of UCA1 was
changed >3-fold in DLD-1/p and SW480/p cells by treatment with
5FU, whereas the change was less apparent in other cultured
cells.
The association of between the 5FU-treated/untreated
ratios of H19 and UCA1 expression and cell lines is presented in
Fig. 4D. The ratios were low in
the 5FU-resistant cell lines DLD-1/5fu and SW480/5fu. Conversely,
the ratio of H19 or UCA1 expression was increased in the three
parental cultured cells and HCT116/5fu cells, which were
susceptible to 5FU.
Doubling time and cell cycle phases of
cultured cells
With 5FU treatment, the doubling time was
significantly increased in the parental cultured cells of HCT116/p,
DLD-1/p and SW480/p compared with their untreated counterparts
(Table II). The doubling time of
untreated HCT116/5fu cells was comparable with untreated parental
cells, and the time was significantly increased following treatment
with 5FU. The doubling time of untreated DLD-1/5fu and SW480/5fu
was significantly increased than that of their parental cells.
Treatment with 5FU did not affect the doubling time.
 | Table IIEffect of 5FU on doubling time of
cultured cells. |
Table II
Effect of 5FU on doubling time of
cultured cells.
| Cell line | Doubling time (h)
| Fold change |
|---|
| Untreated | 5FU-treated |
|---|
| HCT116/p | 20±5 | 200±46b | 10.0 |
| DLD-1/p | 16±2 | 155±27b | 9.6 |
| SW480/p | 22±2 | 67±4b | 3.0 |
| HCT116/5fu | 24±1 | 301±17b | 12.5 |
| DLD-1/5fu | 24±4a | 25±4 | 1.0 |
| SW480/5fu | 68±3a | 73±13 | 1.1 |
The cell cycle phase was determined with a Muse Cell
Analyzer (Luminex Japan Corporation Ltd.). The cell cycle phases
were expressed as percentage of the number of cells at the phases
in total number of cells analyzed (Fig. S1). Following treatment with 5FU,
the number of cells in S phase appeared to be increased for SW480/p
and SW480/5fu (Fig. 5A). The
change in cell cycle phases was subtle in HCT116/p, DLD-1/p,
HCT116/5fu and DLD-1/5fu cells.
Rb and p27kip1 expression in cultured
cells
The expression of Rb and p27kip1, which are involved
in the cell cycle, and are target molecules of H19 and UCA1. The
expression of Rb was notably decreased in DLD-1/p and SW480/p cells
in response to 5FU; Rb expression was also reduced in HCT116/p and
HCT116/5fu cells (Fig. 5B). In
contrast, the expression levels of Rb appeared to be unchanged in
DLD-1/5fu and SW480/5fu cells treated with 5FU. The expression of
p27kip1 was down-regulated in three parental cells and HCT116/5fu
cells, which were susceptible to 5FU, whereas the expression was
notably unchanged in DLD-1/5fu and SW480/5fu cells, which were
resistant to 5FU.
Clinicopathological features of rectal
cancer cases
The clinicopathological features of the 31 patients
with rectal cancer included in this study were summarized in
Table III. Among the 31 cases,
the numbers of cases with Grade 0, Grade 1a, Grade 1b and Grade 2
were 0, 15, 8 and 8, respectively. Cases with therapeutic responses
of Grade 1b and 2 were designated as 'favorable response' cases,
and cases with therapeutic responses of Grade 0 and 1a were
designated as 'poor response' cases. There was no significant
difference in the clinicopathological features between the two
groups.
 | Table IIIClinicopathological features of
rectal cancer cases. |
Table III
Clinicopathological features of
rectal cancer cases.
| Characteristic | Response
| P-value |
|---|
| Favorable
(n=16) | Poor (n=15) |
|---|
| Age, years | 64 (39-72) | 66 (46-76) | 0.178 |
| Sex | | | 0.354 |
| Male | 13 | 10 | |
| Female | 3 | 5 | |
| Clinical depth of
tumor invasiona | | | 0.325 |
| cT3 | 15 | 15 | |
| cT4 | 1 | 0 | |
| Pathological depth
of tumor invasiona | | | 0.573 |
| pTis | 1 | 0 | |
| pT1 | 0 | 0 | |
| pT2 | 6 | 5 | |
| pT3 | 9 | 10 | |
| pT4 | 0 | 0 | |
| Tumor
histology | | | 0.376 |
| Well
differentiated | 6 | 8 | |
| Moderately
differentiated | 10 | 7 | |
| Histological
therapeutic response | | | <0.001 |
| Grade 0 | 0 | 0 | |
| Grade 1a | 0 | 15 | |
| Grade 1b | 8 | 0 | |
| Grade 2 | 8 | 0 | |
| Hierarchical
analysis | | | 0.045 |
| Cluster 1 | 6 | 11 | |
| Cluster 2+3 | 10 | 4 | |
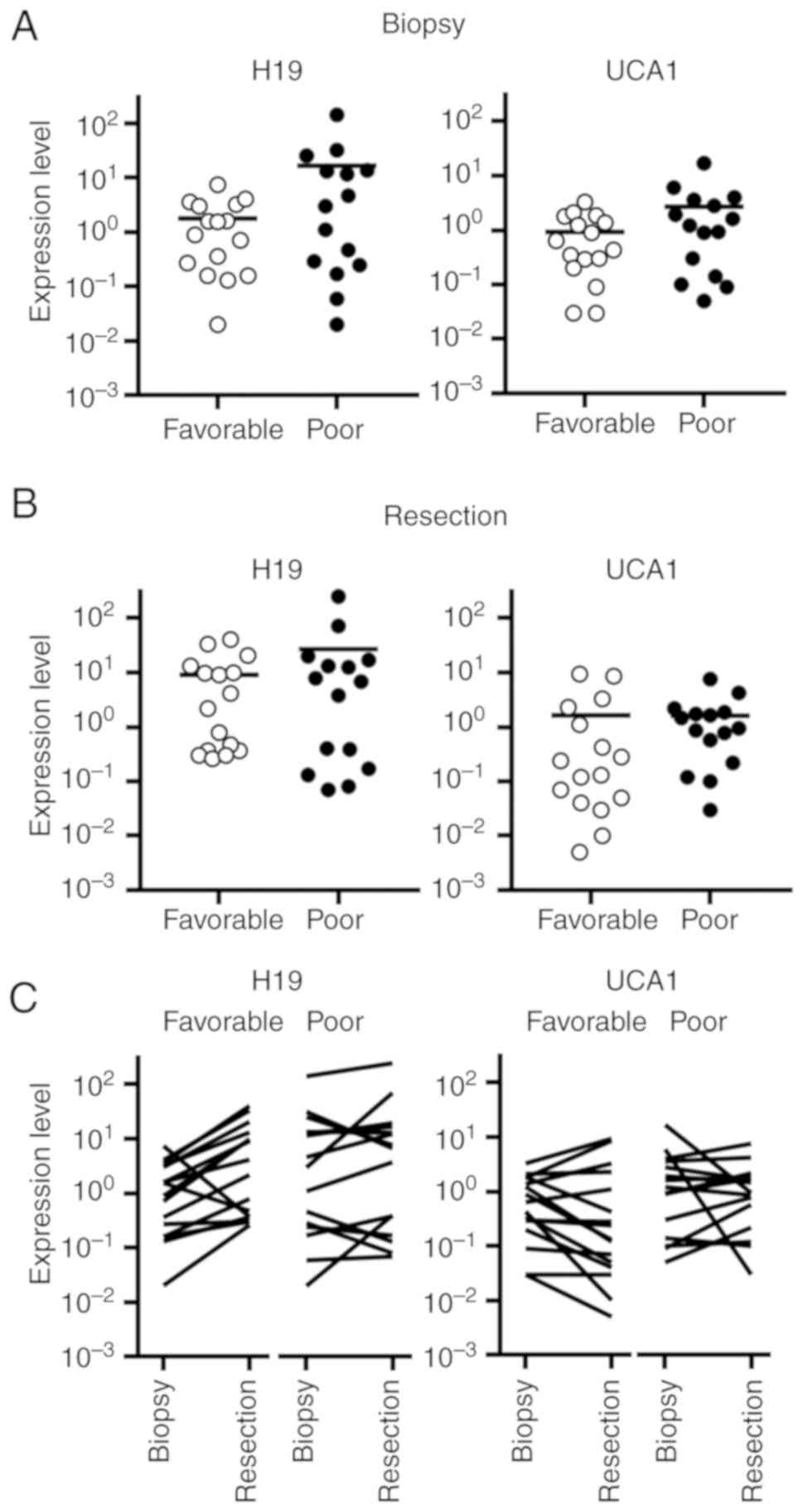
Expression of H19 and UCA1 in rectal
cancer
The expression levels of H19 and UCA1 in the biopsy
specimens of the cases with poor response were higher than those
with favorable responses (Fig.
6A). In the resected tissue specimens, H19 and UCA1 expression
levels appeared to be comparable between the cases with poor and
favorable responses (Fig. 6B).
The expression level of H19 was altered >2-fold
in 12 of 16 cases (75%) with favorable response and in 5 of 15
cases (33%) with poor response (Fig.
6C). Alterations in the expression levels of UCA1 markedly
varied in cases with favorable response as well as those with poor
response.
The association between the resection/biopsy ratios
of H19 and UCA1 was shown in Fig.
6D. Cases with poor response tended to be located in the area
in which both ratios were low. By hierarchical clustering, three
major clusters were separated. Cluster 1 included the cases in
which both ratios were low. Cases that showed a higher ratio of H19
were separated into cluster 2, and the cases that showed a higher
ratio of UCA1 were separated into cluster 3. In cluster 1, 11 of
the 17 cases exhibited a poor response (65%), whereas in clusters 2
and 3, 4 of the 14 cases were linked to a poor response (29%)
(P=0.045, Table III).
Discussion
Several studies have examined the association
between lncRNAs and the susceptibility to 5FU in colorectal cancer
(15,16,18-22).
However, a specific lncRNA that may be reliable for the prediction
of susceptibility to 5FU has not yet been identified. In the
current study, we examined the expression levels of 84 lncRNAs in
5FU-susceptible and 5FU-resistant cultured cells using PCR array
analysis. Bioinformatic analysis revealed H19 and UCA1 as candidate
molecules that may be involved in the susceptibility to 5FU. A
previous study reported differentially expressed lncRNAs in
5FU-resistant cultured colorectal carcinoma cells compared with
5FU-susceptible cultured cells (15). Although the cultured cells were
different from the cells used in the present study, H19 and UCA1
were included as differentially expressed lncRNAs. In addition,
recent studies reported the association of H19 and UCA1 with the
susceptibility to 5FU (16,22).
In the present study, the expression levels of H19
and UCA1 varied largely in 5FU-treated and untreated cultured
cells, and their expression levels were notably higher in
5FU-susceptible cells compared with 5FU-resistant cells. In
contrast with our findings, high H19 levels in 5FU-resistant cells
were detected (22). This
discrepancy may be due to the difference in the cultured cells used
in this study and our investigation. Notably, the
5FU-treated/untreated ratios appeared to be more closely associated
with the susceptibility compared with the expression levels in
untreated or 5FU-treated cells. The 5FU-treated/untreated ratios of
H19 and UCA1 were low in 5FU-resistant cells, whereas the ratios
were high in 5FU-susceptible cells. To the best of our knowledge,
we are the first to report on the association between alterations
in the expression levels of lncRNAs in 5FU-treated cultured cells
with the susceptibility to 5FU.
In human cases of rectal cancer treated with NAC,
there was no significant difference in the expression levels of H19
and UCA1 between cases with favorable and poor responses both in
biopsy tissue and resected tissue. The cases with low
resection/biopsy ratios of H19 and UCA1 tended to exhibit a poor
response to NAC. These results appear to be consistent with that in
cultured cells. We proposed that changes in H19 or UCA1 expression
in response to 5FU, rather than the stable expression before or
after exposure to 5FU, may be useful for the prediction of the
susceptibility to 5FU-based NAC in human rectal cancer. However, a
small patient cohort was examined in the present study; alterations
in the expression levels of H19 and UCA1 in response to NAC should
to be verified in a larger number of patients.
In 5FU-susceptible cultured cells, cell
proliferation was inhibited by 5FU. The doubling time was increased
due to treatment with 5FU in 5FU-susceptible cells. In
5FU-resistant cells, the doubling time was significantly longer
than their parental cells, and the doubling time was markedly
unchanged by treatment with 5FU. The cell cycle phases appeared to
be comparable in cultured cells. The inhibition in the
proliferation in 5FU-susceptible cells due to impaired cell cycle
progression was observed following inhibition of thymidylate
synthase and DNA synthesis (10,29).
The expression levels of Rb and p27kip1 were decreased in
5FU-susceptible cells treated with 5FU, whereas their expressions
were unchanged in 5FU-resistant cells. Previous studies showed that
elevated expression of H19 and UCA1 suppressed the expressions of
Rb and p27kip1, respectively (30,31).
Thus, we speculate that the reduction in Rb and p27kip1 levels may
be, in part, caused by the upregulation of H19 and UCA1. These
alterations in expression may represent the response to the
impaired cell cycle caused by 5FU. Another study also showed that
the expression levels of lncRNAs were upregulated in response to
treatment with various chemical agents in human pluripotent stem
cells (32). Elevations of H19 and
UCA1 expression may also be linked to the cellular stress response.
The precise mechanism underlying the elevation of H19 and UCA1 in
response to 5FU requires further investigation.
Based on the results of present study, it is not
plausible to predict the response to NAC by the quantitation of H19
and UCA1 only in biopsy specimens taken before NAC in human cases
of rectal cancer. In vitro analyses demonstrated that the
change of expression levels of H19 and UCA1 was noted in
5FU-susceptible cultured cells even at 48 h after the initiation of
treatment with 5FU. It is thus expected that the response is
predicted by the quantitation of H19 and UCA1 in paired biopsy
specimens taken before NAC and early after the initiation of NAC.
The early prediction of response to 5FU-based NAC may aid the
identification of cases with poor responses, and facilitate the
administration of intensive treatment or radiotherapy.
Supplementary Data
Acknowledgments
The authors thank Dr Shin-ichi Horike and Dr Makiko
Meguro-Horike, at the Advanced Science Research Center, Kanazawa
University, for their constructive suggestions. The authors thank
Ms. Kiyoko Kawahara, Mr. Takenori Fujii, Mr. Kiyoshi Teduka, Ms.
Yoko Kawamoto and Ms. Taeko Kitamura, Department of Integrated
Diagnostic Pathology, Nippon Medical School for their skillful
assistance.
Abbreviations:
|
5FU
|
5-fluorouracil
|
|
IC50
|
half maximal-inhibitory
concentration
|
|
IPA
|
Ingenuity Pathway Analysis
|
|
lncRNA
|
long non-coding RNA
|
|
NAC
|
neoadjuvant chemotherapy
|
|
PBS
|
phosphate buffered saline
|
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YY, TS, and RW were involved in the conception and
design of the study. YY, KI, and MK performed the histological and
biochemical experiments. MK, TY, and HY acquired the clinical data.
YY prepared the figures. YY, TS, and RW drafted the manuscript. HY
and ZN reviewed and edited the manuscript. ZN supervised the
experiments and writing the manuscript and gave the final approval
of manuscript to be published. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
This study was conducted according to the
Declaration of Helsinki and the Japanese Society of Pathology. The
study was approved by the Ethics Committee of Nippon Medical School
Hospital (approval no. 29-03-909). Written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rödel C, Martus P, Papadoupolos T, Füzesi
L, Klimpfinger M, Fietkau R, Liersch T, Hohenberger W, Raab R,
Sauer R and Wittekind C: Prognostic significance of tumor
regression after preoperative chemoradiotherapy for rectal cancer.
J Clin Oncol. 23:8688–8696. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beddy D, Hyland JM, Winter DC, Lim C,
White A, Moriarty M, Armstrong J, Fennelly D, Gibbons D and Sheahan
K: A simplified tumor regression grade correlates with survival in
locally advanced rectal carcinoma treated with neoadjuvant
chemoradiotherapy. Ann Surg Oncol. 15:3471–3477. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marijnen CA, van de Velde CJ, Putter H,
van den Brink M, Maas CP, Martijn H, Rutten HJ, Wiggers T,
Kranenbarg EK, Leer JW and Stiggelbout AM: Impact of short-term
preoperative radiotherapy on health-related quality of life and
sexual functioning in primary rectal cancer: Report of a
multicenter randomized trial. J Clin Oncol. 23:1847–1858. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parc Y, Zutshi M, Zalinski S, Ruppert R,
Fürst A and Fazio VW: Preoperative radiotherapy is associated with
worse functional results after coloanal anastomosis for rectal
cancer. Dis Colon Rectum. 52:2004–2014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hasegawa S, Goto S, Matsumoto T, Hida K,
Kawada K, Matsusue R, Yamaguchi T, Nishitai R, Manaka D, Kato S, et
al: A multicenter phase 2 study on the feasibility and efficacy of
neoadjuvant chemotherapy without radiotherapy for locally advanced
rectal cancer. Ann Surg Oncol. 24:3587–3595. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koike J, Funahashi K, Yoshimatsu K,
Yokomizo H, Kan H, Yamada T, Ishida H, Ishibashi K, Saida Y,
Enomoto T, et al: Efficacy and safety of neoadjuvant chemotherapy
with oxali-platin, 5-fluorouracil, and levofolinate for T3 or T4
stage II/III rectal cancer: The FACT trial. Cancer Chemother
Pharmacol. 79:519–525. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Koizumi M, Yamada T, Shinji S, Yokoyama Y,
Takahashi G, Iwai T, Takeda K, Hara K, Ohta K, Uchida E and Yoshida
H: Feasibility of neoadjuvant FOLFOX therapy without radiotherapy
for baseline resectable rectal cancer. In Vivo. 32:937–943. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hong YS, Nam BH, Kim KP, Kim JE, Park SJ,
Park YS, Park JO, Kim SY, Kim TY, Kim JH, et al: Oxaliplatin,
fluorouracil, and leucovorin versus fluorouracil and leucovorin as
adjuvant chemotherapy for locally advanced rectal cancer after
preoperative chemoradiotherapy (ADORE): An open-label, multicentre,
phase 2, randomised controlled trial. Lancet Oncol. 15:1245–1253.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oki E, Murata A, Yoshida K, Maeda K,
Ikejiri K, Munemoto Y, Sasaki K, Matsuda C, Kotake M, Suenaga T, et
al: A randomized phase III trial comparing S-1 versus UFT as
adjuvant chemotherapy for stage II/III rectal cancer (JFMC35-C1:
ACTS-RC). Ann Oncol. 27:1266–1272. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Soong R, Shah N, Salto-Tellez M, Tai BC,
Soo RA, Han HC, Ng SS, Tan WL, Zeps N, Joseph D, et al: Prognostic
significance of thymidylate synthase, dihydropyrimidine
dehydrogenase and thymidine phosphorylase protein expression in
colorectal cancer patients treated with or without
5-fluorouracil-based chemotherapy. Ann Oncol. 19:915–919. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salonga D, Danenberg KD, Johnson M,
Metzger R, Groshen S, Tsao-Wei DD, Lenz HJ, Leichman CG, Leichman
L, Diasio RB, et al: Colorectal tumors responding to 5-fluorouracil
have low gene expression levels of dihydropyrimidine dehydrogenase,
thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res.
6:1322–1327. 2000.PubMed/NCBI
|
|
13
|
Tokunaga Y, Sasaki H and Saito T: Clinical
role of orotate phosphoribosyl transferase and dihydropyrimidine
dehydrogenase in colorectal cancer treated with postoperative
fluoropyrimidine. Surgery. 141:346–353. 2006. View Article : Google Scholar
|
|
14
|
Ribic CM, Sargent DJ, Moore MJ, Thibodeau
SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R,
Shepherd LE, et al: Tumor microsatellite-instability status as a
predictor of benefit from fluorouracil-based adjuvant chemotherapy
for colon cancer. N Eng J Med. 349:247–257. 2003. View Article : Google Scholar
|
|
15
|
Lee H, Kim C, Ku JL, Kim W, Yoon SK, Kuh
HJ, Lee JH, Nam SW and Lee EK and Lee EK: A long non-coding RNA
snaR contributes to 5-fluorouracil resistance in human colon cancer
cells. Mol Cells. 37:540–546. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu
Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell
proliferation and 5-fluorouracil resistance in colorectal cancer by
inhibiting miR-204-5p. Sci Rep. 6:238922016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie T, Huang M, Wang Y, Wang L, Chen C and
Chu X: MicroRNAs as regulators, biomarkers and therapeutic targets
in the drug resistance of colorectal cancer. Cell Physiol Biochem.
40:62–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bian Z, Zhang J, Li M, Feng Y, Yao S, Song
M, Qi X, Fei B, Yin Y, Hua D and Huang Z: Long non-coding RNA
LINC00152 promotes cell proliferation, metastasis, and confers 5-FU
resistance in colorectal cancer by inhibiting miR-139-5p.
Oncogenesis. 6:3952017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li P, Zhang X, Wang L, Du L, Yang Y, Liu
T, Li C and Wang C: lncRNA HOTAIR Contributes to 5FU resistance
through suppressing miR-218 and activating NF-κB/TS signaling in
colorectal cancer. Mol Ther Nucleic Acids. 8:356–369. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li J, Li X, Cen C, Ai X, Lin C and Hu G:
The long non-coding RNA ENST00000547547 reduces 5-fluorouracil
resistance of colorectal cancer cells via competitive binding to
microRNA-31. Oncol Rep. 39:217–226. 2018.
|
|
21
|
Qiao L, Liu X, Tang Y, Zhao Z, Zhang J and
Liu H: Knockdown of long non-coding RNA prostate cancer-associated
ncRNA transcript 1 inhibits multidrug resistance and
c-Myc-dependent aggressiveness in colorectal cancer Caco-2 and
HT-29 cells. Mol Cell Biochem. 441:99–108. 2018. View Article : Google Scholar
|
|
22
|
Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang
R, Jin Y, Zou C, Chen Y, Wang G, et al: Long non-coding RNA H19
confers 5-Fu resistance in colorectal cancer by promoting
SIRT1-mediated autophagy. Cell Death Dis. 9:11492018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sanchez Calle A, Kawamura Y, Yamamoto Y,
Takeshita F and Ochiya T: Emerging roles of long non-coding RNA in
cancer. Cancer Sci. 109:2093–2100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng H, Zhang J, Shi J, Guo Z, He C, Ding
L, Tang JH and Hou Y: Role of long non-coding RNA in tumor drug
resistance. Tumour Biol. 37:11623–11631. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Murakami Y, Kazuno H, Emura T, Tsujimoto
H, Suzuki N and Fukushima M: Different mechanisms of acquired
resistance to fluorinated pyrimidines in human colorectal cancer
cells. Int J Oncol. 17:277–283. 2000.PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Japanese Society for Cancer of the Colon
and Rectum: Japanese Classification of Colorectal, Appendiceal, and
Anal Carcinoma. 9th Edition. Tokyo: Kanehara; 2018
|
|
29
|
Yoshikawa R, Kusunoki M, Yanagi H, Noda M,
Furuyama JI, Yamamura T and Hashimoto-Tamaoki T: Dual antitumor
effects of 5-fluorouracil on the cell cycle in colorectal carcinoma
cells: A novel target mechanism concept for pharmacokinetic
modulating chemotherapy. Cancer Res. 61:1029–1037. 2001.PubMed/NCBI
|
|
30
|
Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung
JJ and Kwok TT: Oncofetal H19-derived miR-675 regulates tumor
suppressor RB in human colorectal cancer. Carcinogenesis.
31:350–358. 2010. View Article : Google Scholar
|
|
31
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tani H, Onuma Y, Ito Y and Torimura M:
Long non-coding RNAs as surrogate indicators for chemical stress
responses in human-induced pluripotent stem cells. PLoS One.
9:e1062822014. View Article : Google Scholar : PubMed/NCBI
|