Introduction
Colorectal cancer (CRC) is the third most common
type of cancer worldwide and the fourth leading cause of
cancer-related mortality, exceeded by lung cancer, liver cancer and
gastric cancer (1). The
occurrence of CRC is the result of the gradual accumulation of
genetic and epigenetic alterations, which lead to homeostasis
dysfunction and neoplastic transformation (2). Age, genetic and environmental
factors are widely involved in the initiation of CRC; other
recognized risk factors include inflammatory bowel disease,
obesity, a sedentary lifestyle, a history of abdominal radiation
and acromegaly (3). Despite
recent advancements being made in screening strategies and
effective treatments, the prognosis of patients with advanced CRC
remains poor. Furthermore, the latest molecular targeted agents
seem to be active only for metastatic CRC, and they exponentially
increase the cost of CRC treatment (4). Therefore, methods for the
determination of potent diagnostic and prognostic biomarkers are
urgently required for the effective intervention of CRC.
Long non-coding RNAs (lncRNAs) are a class of
non-coding RNA transcripts that are >200 nucleotides in length
and lack protein-coding ability (5). Dysregulated lncRNA expression is
closely related to the pathogenesis of cancer, metabolic disorders
and cardiovascular diseases (6).
lncRNAs are implicated in tumorigenesis through different types of
molecular mechanisms, and certain regulatory factors often lead to
the abnormal expression of lncRNAs in CRC, thus causing malignant
transformation (7). In addition,
the association of abnormally regulated lncRNAs with clinical
outcomes suggests the potential of lncRNAs as effective diagnostic
and prognostic predictors and therapeutic targets for CRC (8). Nuclear paraspeckle assembly
transcript 1 (NEAT1) is an emerging lncRNA located at nuclear
paraspeckles (9). The aberrant
overexpression of NEAT1 occurs in solid tumors, which is typically
responsible for the poor survival of patients (10). NEAT1 drives carcinogenesis and
progression by regulating the expression of genes involved in
cancer cell growth, migration and invasion, as well as
epithelial-mesenchymal transition (EMT) and chemotherapeutic
resistance (11). Notably, lncRNA
NEAT1 knockdown has been shown to enhance 5-fluorouracil
sensitivity in patients with CRC by attenuating autophagy (12). Although NEAT1 is generally viewed
as a diagnostic and prognostic marker for CRC (13), the specific mechanisms of action
of NEAT1 in CRC remain unclear.
Thus, the present study aimed to determine the
lncRNA NEAT1 interacting partners and elucidate the molecular
mechanisms underlying the oncogenic functions of NEAT1 in CRC. The
findings presented herein may provide a novel theoretical basis for
the management of CRC.
Materials and methods
Ethics statement
The use of ovarian tissues was approved by the
Ethics Committee of the Second Affiliated Hospital of Soochow
University. Informed consent was signed by each eligible
participant. All animal experiments are approved by the Ethics
Committee of the Second Affiliated Hospital of Soochow University
(S.No. 20190523b116). All experimental procedures were implemented
on the Ethical Guidelines for the study of experimental pain in
conscious animals.
Bioinformatics analysis
lncRNA NEAT1-related diseases were searched through
the lncdisease database (14).
The differential expression of NEAT1 and Cul4A in CRC samples
(n=286) and normal samples (n=41) collected by TCGA were searched
through the UALCAN cancer database (http://ualcan.path.uab.edu/analysis.html) (15). The differential expression of
Cul4A in CRC samples (n=97) and normal samples (n=100) collected by
CPTAC were searched through the UALCAN cancer database (http://ualcan.path.uab.edu/analysis.html) (15). The cellular localization of NEAT1
was predicted through LncMAP database (http://bio-bigdata.hrbmu.edu.cn/LncMAP/) (16). The NEAT1-related
lncRNA-transcription factor-gene regulatory network in CRC was
searched through LncMAP database (http://bio-bigdata.hrbmu.edu.cn/LncMAP/).
Tissue samples
The present study recruited 55 patients with CRC (30
males and 25 females) aged 27-49 years, from December, 2018 to
December, 2019, at the Second Affiliated Hospital of Soochow
University. All cancer tissues were collected via surgical tumor
resection, and the adjacent non-cancerous tissues were used as
negative controls (NCs). For the experiment, the cancer tissues and
normal tissues of each patient were matched. The inclusion criteria
were as follows: Confirmed CRC by pathology, compliance with
surgical indications, no tumor-specific therapy before operation,
complete clinicopathological data available and informed consent
provided. The exclusion criteria were as follows: Patients with
other tumors, gastrointestinal dysfunction, autoimmune diseases, or
infectious diseases. The tissue fragments were refrigerated in
liquid nitrogen immediately after dissection and stored at
-80°C.
Cells and cell culture
The human epithelial cell line, NCM460 (CC-YM02142,
Shanghai Enzyme Research Biotechnology Co., Ltd.) and the HCT116
human CRC cell line (CCL-247™, American Type Culture Collection)
were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin (100 U/ml)/streptomycin (0.1
mg/ml) (Invitrogen; Thermo Fisher Scientific, Inc.) in a humidified
incubator at 37°C with 5% CO2. The cells were used in
subsequent experiments when they reached 60% confluency.
Cell treatment
Small interfering RNA (siRNA) targeting lncRNA NEAT1
(si-NEAT1-1, si-NEAT1-2 and si-NEAT1-3) and its NC (si-NC, Shanghai
GenePharma Co., Ltd.) were transfected into the HCT116 cells,
respectively at a final concentration of 50 nM. siRNA targeting
E2F1 and its NC (Sangon Biotech, Co., Ltd.) termed si-E2F1 and
si-NC, respectively, were transfected into the HCT116 cells.
si-NEAT1-1 was transfected with si-KDM5A and its NC (Sangon
Biotech, Co., Ltd.) or pcDNA-Cul4A and its pcDNA-NC (Sangon
Biotech, Co., Ltd.) into HCT116 cells, and termed si-NEAT1 +
si-KDM5A, si-NEAT1 + si-NC, si-NEAT1 + pc-Cul4A and si-NEAT1 +
pc-NC, respectively. All transfections (2 µl siRNA or 1,000
ng plasmid) were conducted using Lipofectamine 2000®
(Invitrogen; Thermo Fisher Scientific, Inc.). Untreated HCT116
cells were used as the blank group. CPI-455 is a specific pan-KDM5A
inhibitor, and the IC50 of KDM5A is 10 nM (17). HCT116 cells were treated with 15
µmol/l CPI-455 (CAS 1628208-23-0, Topscience Co., Ltd.) or
phosphate-buffered saline (PBS) for 48 h (18), and termed PBS and CPI-455.
Subsequent experiments were carried out after 24 h.
Colony formation assay
The 2X RPMI-1640 medium containing 20% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and 2×103 HCT116 cells
were mixed with an equal volume of 0.7% agarose (Sigma-Aldrich,
Merck KGaA). The mixture was immediately placed into six-well
plates (Thermo Fisher Scientific Inc.) containing 0.5% agarose
substrate made from 1X RPMI-1640 medium supplemented with 10% FBS
and cultured at 37°C with 5% CO2 for 10 days. The medium
was refreshed every three days. The medium was removed and the
cells were washed with PBS and fixed with 4% paraformaldehyde for
20 min. The paraformaldehyde was then removed and the cells were
stained with 0.2% crystal violet (Beyotime Institute of
Biotechnology, Shanghai, China) at 37°C for 5 min. The colonies
were analyzed using ImageJ software v1.8.0 (National Institutes of
Health).
Terminal deoxynucleotidyl transferase
(TdT)-mediated dUTP nick-end labeling (TUNEL) assay
TUNEL assay was performed using an apoptosis
detection kit (KeyGEN Biotech Corp., Ltd.). The specific operations
were as follows: The cells were fixed with 4% polyformaldehyde for
20 min, washed with PBS, and treated with 100 µl protease K
(20 g/ml) was at room temperature for 20 min. The cells were then
washed with PBS for 5 min, immersed in 4% paraformaldehyde for 5
min, and then supplemented with 100 µl DNase I buffer for 5
min. After the liquid was removed, the cells were detached with 100
µl DNase I (200 U/ml) for 10 min, washed with deionized
water 4 times and PBS for 5 min, cultured with 10 µl
equilibration solution in a wet box for 10 min, and supplemented
with 100 µl TUNEL reaction mixture solution. After applying
the sealing film, the cells were reacted at 37°C for 1 h in the
dark in a wet box and immersed in 20X SSC for 15 min to terminate
the reaction. TUNEL-positive HCT116 cells in five random fields of
vision of each well were observed and counted under a fluorescence
microscope (Olympus Corporation) to calculate the proportion of
TUNEL-positive cells.
Transwell assay
HCT116 cells (2×105) were seeded into
Matrigel-coated (for invasion detection) or uncoated (for migration
detection) apical chambers. HCT116 cells were suspended in
serum-free RPMI1640 medium and seeded into the apical chamber with
2×105 cells per well. RPMI-1640 medium containing 10%
FBS was added to the basolateral chamber. Following 24 h of
incubation at 37°C with 5% CO2, the non-migrated or
non-invasive cells were scraped off using a cotton swab. The cells
at the bottom of the chamber were fixed with methanol
(Sigma-Aldrich, Merck KGaA) for 10 min and stained with 0.5%
crystal violet at room temperature for 20 min. Five visual fields
were selected and randomly photographed using an inverted
microscope (Nikon Corporation).
RNA-fluorescence in situ hybridization
(FISH)
RNA-FISH was used to determine the localization of
lncRNA NEAT1 in the cells. The DNA oligo probe of NEAT1
(FAM-labeled) was purchased from Shanghai GenePharma Co., Ltd..
Subsequently, 1×105 HCT116 cells were seeded into
24-well plates, and the medium was removed after 24 h. Following
three washes with PBS, the cells were fixed with paraformaldehyde
and pre-hybridized with PBS containing 0.5% Triton X-100. The cells
were then subjected to hybridization buffer with NEAT1 probe at 4°C
overnight. The nuclei were stained with
4′,6-diamidino-2-phenylindole at room temperature for 5 min
(Beyotime Institute of Biotechnology). Images were captured under a
Leica SP5 confocal microscope (Leica Microsystems GmbH).
Nuclear/cytosol fractionation assay
The cytoplasmic and nuclear extracts were obtained
using the NE-PER nuclear and cytoplasmic extraction kit (Thermo
Fisher Scientific, Inc.). NEAT1 expression in the nucleoplasm
extract was then detected by reverse transcription-quantitative PCR
(RT-qPCR) as described below.
RNA immunoprecipitation (RIP)
Anti-E2F1 (5 µg/mg, ab179445, Abcam) was used
for RIP assay with immunoglobulin G (IgG) (ab172730, Abcam) as the
control. RIP was performed in accordance with the instructions of
the Magna RIP™ RNA-binding protein immunoprecipitation kit (Merck
KGaA). The isolated RNAs were purified, and the co-precipitated
RNAs were detected by RT-qPCR as described below.
RNA pull-down assay
Biotin-labeled lncRNA NEAT1 and NC (Sangon Biotech,
Co., Ltd.) were incubated with HCT116 cell lysate then supplemented
with streptavidin-coated magnetic beads (Life Technologies; Thermo
Fisher Scientific, Inc.). The biotin-conjugated RNA complex was
used for the RNA pull-down assay. The expression of E2F1 was
detected by RT-qPCR as described below.
Xenograft model of CRC using nude
mice
A xenograft model of CRC was established in nude
mice aged four to six weeks. Nude mice were purchased from Shanghai
SLAC Laboratory Animal Co., Ltd. and kept in isolation cages. The
humane endpoint used in the animal experiments was when the tumor
growth burden was >10% of the body weight of the animal, the
average tumor diameter was >20 mm, or the tumor metastasized or
grew rapidly to ulcer, causing infection or necrosis. The mice were
divided into two groups (12 mice per group) as follows: The
si-NEAT1 group (0.2 ml of PBS containing si-NEAT1-1-treated
2×106 HCT116 cells subcutaneously injected into the
right armpit of the mice) and the si-NC group (0.2 ml of PBS
containing si-NC-treated 2×106 HCT116 cells
subcutaneously injected into the right armpit of the mice). The
mice were monitored every day. The tumor volume was measured every
three days using the following formula: Volume = (length ×
width2)/2. The nude mice were euthanized by an
intraperitoneal injection of pentobarbital sodium (≥100 mg/kg) at
21 days after the injection. It was observed that the animals had
no spontaneous breathing for 2-3 min without blinking reflex, which
confirmed the death of the animals. The tumors of six mice in each
group were excised for immunohistochemistry, and those from the
other six mice were used for RT-qPCR.
Immunohistochemistry
After dewaxing, dehydration and antigen repair, the
tissue sections were blocked with goat serum (Beyotime Institute of
Biotechnology) for 20 min, and the sheep serum was discarded. The
tissue sections were cultured overnight with the primary antibody
(1:200, ab16667, Abcam) at 4°C. The tissue sections were then
cultured with the secondary antibody (1:2,000, ab205718, Abcam) and
developed using DAB (ZSGB-Bio Co., Ltd.). The nuclei were
counterstained with 15% hematoxylin (Beyotime Institute of
Biotechnology), followed by observation under a microscope (CKX41,
Olympus Corporation).
RT-qPCR
Total RNA was extracted from the cells and tissues
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific Inc.). Subsequently, 1 µg RNA was reverse
transcribed into cDNA using PrimerScript RT master mix (Takara
Biotechnology Co., Ltd.). RT-qPCR was performed using the
Quantitative SYBR-Green PCR kit (Qiagen GmbH) and the 7500 Fast
Real Time PCR System (Applied Biosystems; Thermo Fisher Scientific
Inc.). The reaction conditions were as follows: Pre-denaturation at
95°C for 5 min, and then 40 cycles of denaturation at 95°C for 10
sec, annealing at 65°C for 20 sec, and extension at 72°C for 30
sec. The primers used are presented in Table I. The data were analyzed using the
2-ΔΔCq method (19).
 | Table IPrimer sequences used for
RT-qPCR. |
Table I
Primer sequences used for
RT-qPCR.
| Primer | Sequence
(5′-3′) |
|---|
| NEAT1 (H) | F:
ATGGAGCCCCGTGACCTCTCACCT
R: CTAGACCTGCCATTTCTCACACAC |
| KDM5A (H) | F:
ATGGCGGGCGTGGGGCCGGGGGG
R: CTAACTGGTCTCTTTAAGATCCTC |
| E2F1 (H) | F:
ATGGCCTTGGCCGGGGCCCCTGCG
R: TCAGAAATCCAGGGGGGTGAGGTC |
| Cul4A (H) | F:
ATGGCGGACGAGGCCCCGCGGAA
R: TCAGGCCACGTAGTGGTACTGA |
| NEAT1 (M) | F:
ATGGGGGTAGAGGCGTTCGACTGC
R: CATATCTGGTGCCAAAAGTATTA |
| KDM5A (M) | F:
ATGGCGTCCGTGGGCCCGGGGGGCT
R: CTAACTGGTCTCTTTAAGATCCT |
| Cul4A (M) | F:
ATGGCGGACGAGGGCCCTCGGA
R: TCATGCCACGTAGTGGTACTGA |
| U6 (H) | F:
CGCTTCGGCAGCACATATAC
R: AATATGGAACGCTTCACGA |
| GAPDH (H) | F:
ATGGTTTACATGTTCCAATATG
R: TTACTCCTTGGAGGCCATGTGG |
| U6 (M) | F:
GTGCTCGCTTCGGCAGCACATATA
R: AATATGGAACGCTTCACGAATT |
| GAPDH (M) | F:
ATGCTGCCCTTACCCCGGGGTCC
R: TTACTCCTTGGAGGCCATGTAGGC |
Western blot analysis
Total protein was isolated from HCT116 cells using
radio-immunoprecipitation assay buffer (Duanhuan Biotechnology Co.,
Ltd.), and the protein concentration was determined using the
bicinchoninic acid (BCA) kit (Duanhuan Biotechnology Co., Ltd.).
The protein was separated on 12% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes. The membranes were blocked with
PBS containing 5% skim milk at room temperature for 2 h. The
membranes were then incubated with the primary antibodies, tubulin
(0.5 µg/ml, ab59680), GAPDH (1:2,500, ab9485), Wnt3a
(1:1,000, ab219412), β-catenin (1:500, ab68183) and H3K4me3
(1:1,000, ab213224) (all from Abcam) at 4°C overnight. The
membranes were then incubated with the secondary antibody (1:2,000,
ab205718, Abcam) at room temperature for 2 h and visualized using a
chemiluminescence reagent (EMD Millipore). The gray value of each
band was quantified using ImageJ software v1.8.0 (National
Institutes of Health), with β-actin as the internal control.
Statistical analysis
Data analysis was performed using SPSS 21.0 (IBM,
Inc.). Data are expressed as the mean ± standard deviation. An
unpaired t-test was adopted for comparisons between two groups.
One-way or two-way analysis of variance was employed for
comparisons among multiple groups, following Sidak's multiple
comparisons test or Tukey's multiple comparisons test. P<0.05
was considered to indicate a statistically significant
difference.
Results
lncRNA NEAT1 is highly expressed in
CRC
The pathological mechanisms of CRC are complex with
respect to an abnormal lncRNA expression (20-22). The implication of lncRNAs in CRC
has become increasingly clear (23). The lncRNA disease database
(http://www.cuilab.cn/lncRNAdisease)
(14) revealed that lncRNA NEAT1
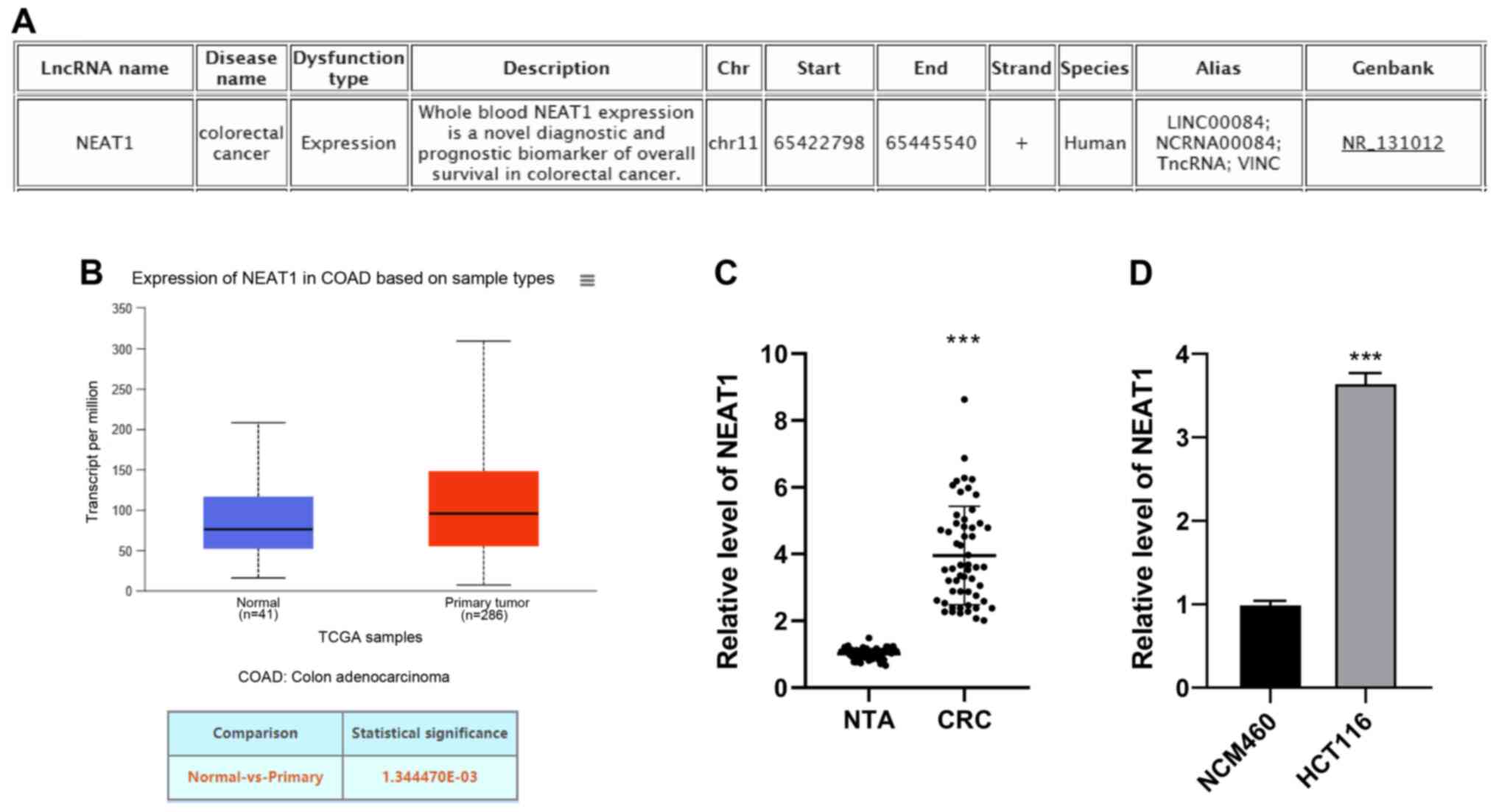
is associated with CRC (Fig. 1A).
lncRNA NEAT1 expression in CRC was predicted through TCGA analysis
(http://ualcan.path.uab.edu/analysis.html) (15), and the results revealed that the
mRNA expression of NEAT1 was upregulated in CRC (Fig. 1B). Additionally, NEAT1 was
overexpressed in CRC tissues and cells (Fig. 1C and D). The median relative
expression of NEAT1 (3.6) in 55 patients with CRC was used as the
cut-off point. The patients were divided into the NEAT1 low
expression group (27 cases) and high expression group (28 cases). A
high expression of NEAT1 was positively associated with the CEA
level (P=0.015), tumor size (P=0.031) and tumor-node-metastasis
(TNM) stage (P=0.029), as shown in Table II. Taken together, these findings
demonstrated that lncRNA NEAT1 was highly expressed in CRC, and may
thus be related to the occurrence and development of CRC.
 | Table IIAssociation between NEAT1 expression
and clinicopathological features of patients with colorectal
cancer. |
Table II
Association between NEAT1 expression
and clinicopathological features of patients with colorectal
cancer.
| Clinicopathological
feature | No. of patients
(n=55) | Expression of NEAT1
| χ2 | P-value |
|---|
| Low (n=27) | High (n=28) |
|---|
| Age (years) | | | | | |
| <36 | 27 | 12 | 15 | 0.458 | 0.593 |
| ≥36 | 28 | 15 | 13 | | |
| Sex | | | | | |
| Male | 30 | 16 | 14 | 0.475 | 0.591 |
| Female | 25 | 11 | 14 | | |
| Tumor location | | | | | |
| Rectum | 25 | 13 | 12 | 0.155 | 0.798 |
| Colon | 30 | 14 | 16 | | |
| CEA level | | | | | |
| ≤5 ng/ml | 25 | 17 | 8 | 6.557 | 0.015 |
| >5 ng/ml | 30 | 10 | 20 | | |
| Lymph node
metastasis | | | | | |
| Absent | 29 | 15 | 14 | 0.170 | 0.789 |
| Present | 26 | 12 | 14 | | |
| Tumor size | | | | | |
| ≤5 cm | 28 | 18 | 10 | 5.269 | 0.031 |
| >5 cm | 27 | 9 | 18 | | |
| Distant
metastasis | | | | | |
| Absent | 34 | 20 | 14 | 3.375 | 0.097 |
| Present | 21 | 7 | 14 | | |
| TNM stage | | | | | |
| Stage I-II | 22 | 15 | 7 | 5.347 | 0.029 |
| Stage III-IV | 33 | 12 | 21 | | |
Silencing of lncRNA NEAT1 suppresses the
malignant behaviors of HCT116 cells
To determine the role of lncRNA NEAT1 in CRC,
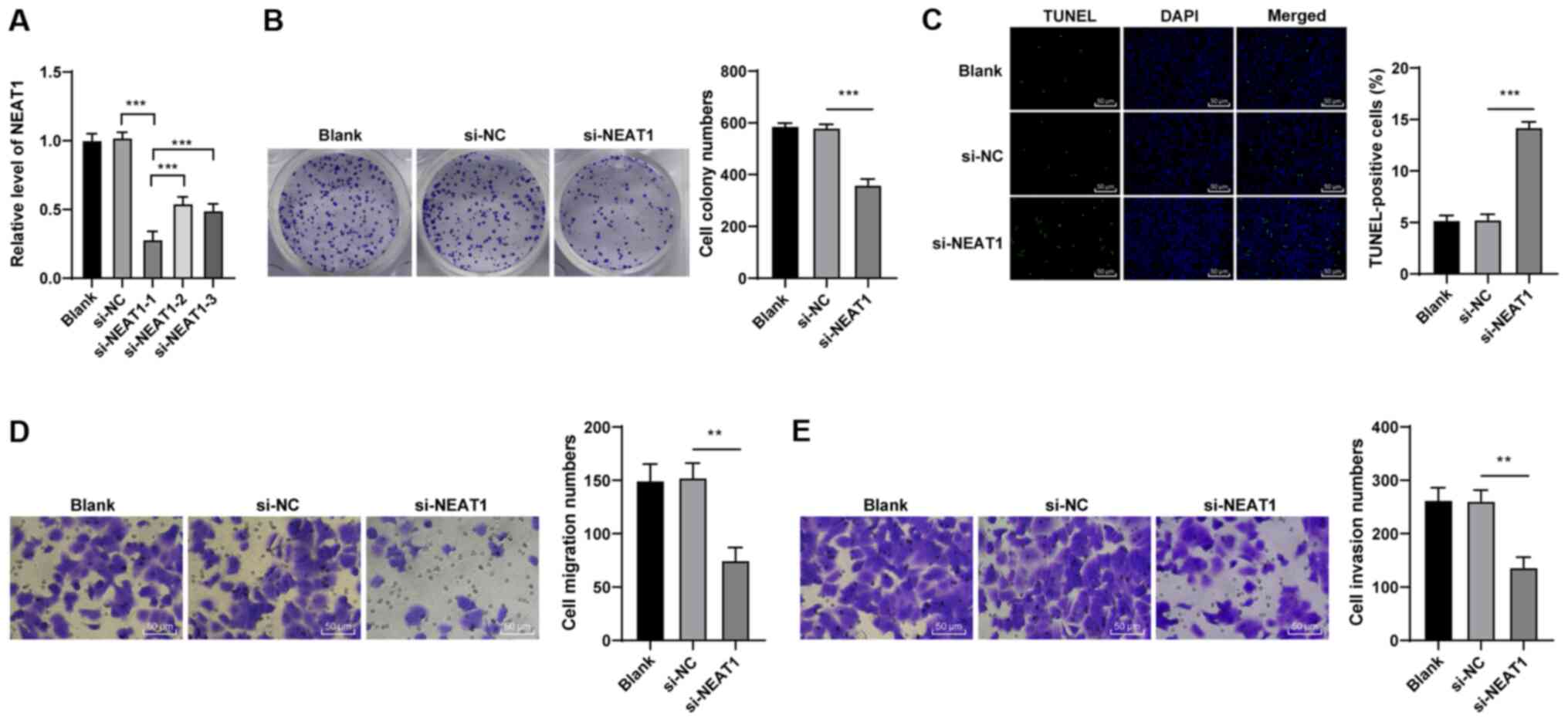
si-NEAT1-1, si-NEAT1-2 and si-NEAT1-3 were transfected into HCT116
cells, and the transfection efficiency was verified by RT-qPCR
(Fig. 2A). si-NEAT1-1 exhibited
the highest transfection efficiency (P<0.001); thus, si-NEAT1-1
was used in subsequent experiments. Subsequently, the malignant
behaviors of the HCT116 cells transfected with si-NEAT1 were
evaluated. The si-NEAT1-transfected cells exhibited a suppressed
colony formation ability (P<0.01; Fig. 2B) and an increased apoptosis
(P<0.01; Fig. 2C), compared
with the si-NC-transfected cells. Additionally, si-NEAT1
transfection suppressed cell migration and invasion (all P<0.01;
Fig. 2D and E). In brief, these
findings demonstrated that the downregulation of NEAT1 suppressed
the malignant behaviors of HCT116 cells.
lncRNA NEAT1 inhibits KDM5A expression by
binding to E2F1
The aforementioned experiments confirmed that
si-NEAT1 inhibited the malignant behaviors of HCT116 cells. The
present study then explored the downstream molecular mechanisms of
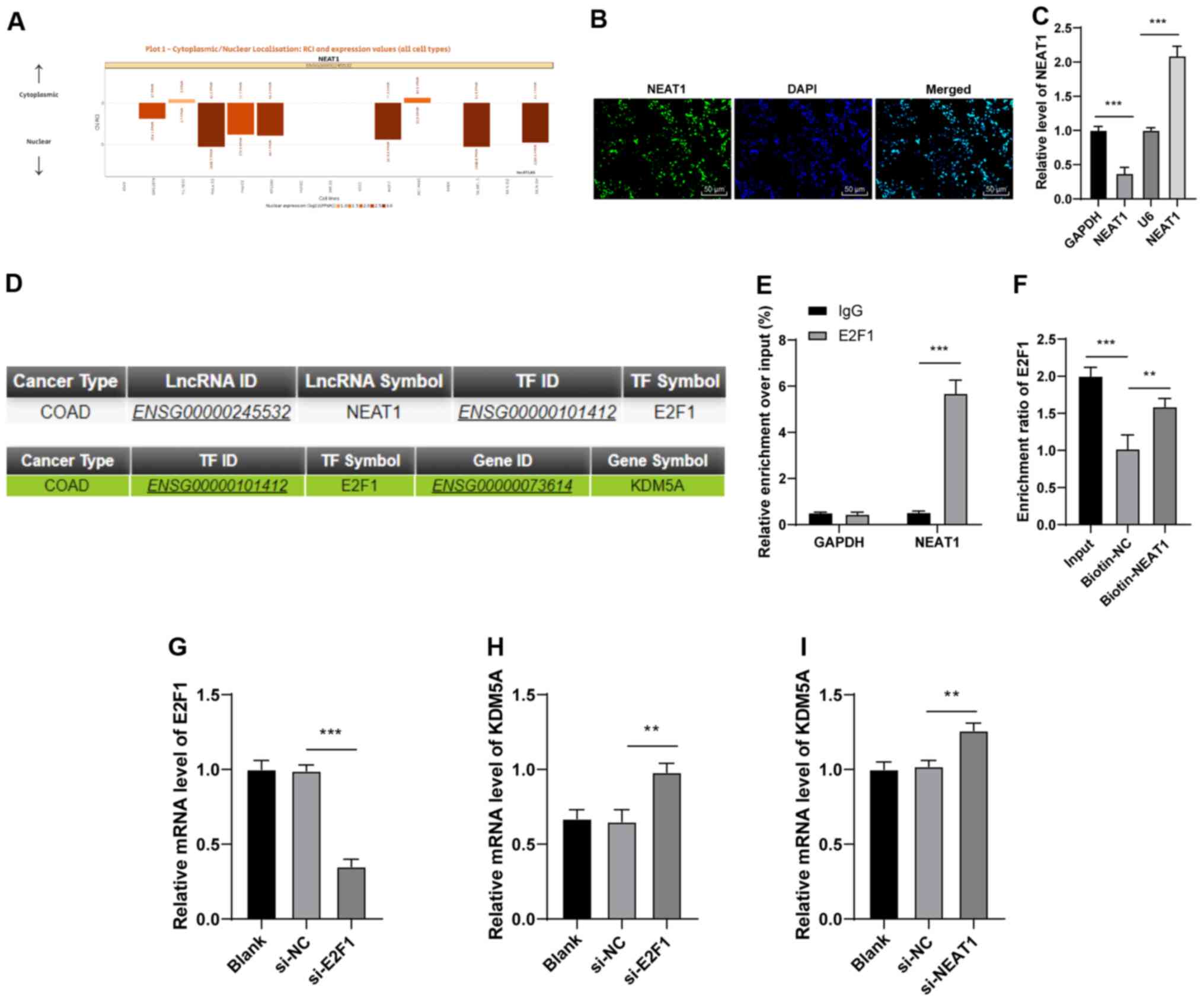
NEAT1 in CRC. First, NEAT1 was predicted to be mainly located in
the nucleus of HCT116 cells, through LncATLAS database (http://lncatlas.crg.eu/) (24) (Fig.
3A). RNA-FISH and nuclear/cytosol fractionation assay confirmed
the localization of lncRNA NEAT1 (Fig. 3B and C). Emerging evidence
suggests that NEAT1 may bind to transcription factors to regulate
the expression of downstream genes (25,26). Hence, the present study predicted
the transcription factors and downstream genes of NEAT1 by using
LncMAP database (http://bio-bigdata.hrbmu.edu.cn/LncMAP/) (16). The results demonstrated that NEAT1
bound to the transcription factor E2F1 (Fig. 3D), and E2F1 bound to the
downstream gene, KDM5A (Fig. 3D).
The results of the RIP assay revealed that the enrichment of the
NEAT1 co-precipitation group was notably increased compared with
that of the IgG co-precipitation group (P<0.01; Fig. 3E). The results of the RNA
pull-down assay revealed that the enrichment of the biotin-NEAT1
group was higher than that of the biotin-NC group (P<0.01;
Fig. 3F). These results suggested
that there was a binding association between NEAT1 and E2F1. To
verify the role of E2F1 in the regulation of KDM5A by NEAT1, the
HCT116 cells were transfected with si-E2F1 (P<0.001; Fig. 3G), and the results revealed that
KDM5A expression was notably promoted in the si-E2F1-transfected
HCT116 cells (P<0.01; Fig.
3H). Moreover, KDM5A expression was elevated in the
si-NEAT1-transfected HCT116 cells (P<0.01; Fig. 3I). Thus, these data indicated that
lncRNA NEAT1 could bind to E2F1 to inhibit KDM5A expression.
Downregulation of KDM5A reverses the
inhibitory effects of NEAT1 silencing on the malignant behaviors of
HCT116 cells
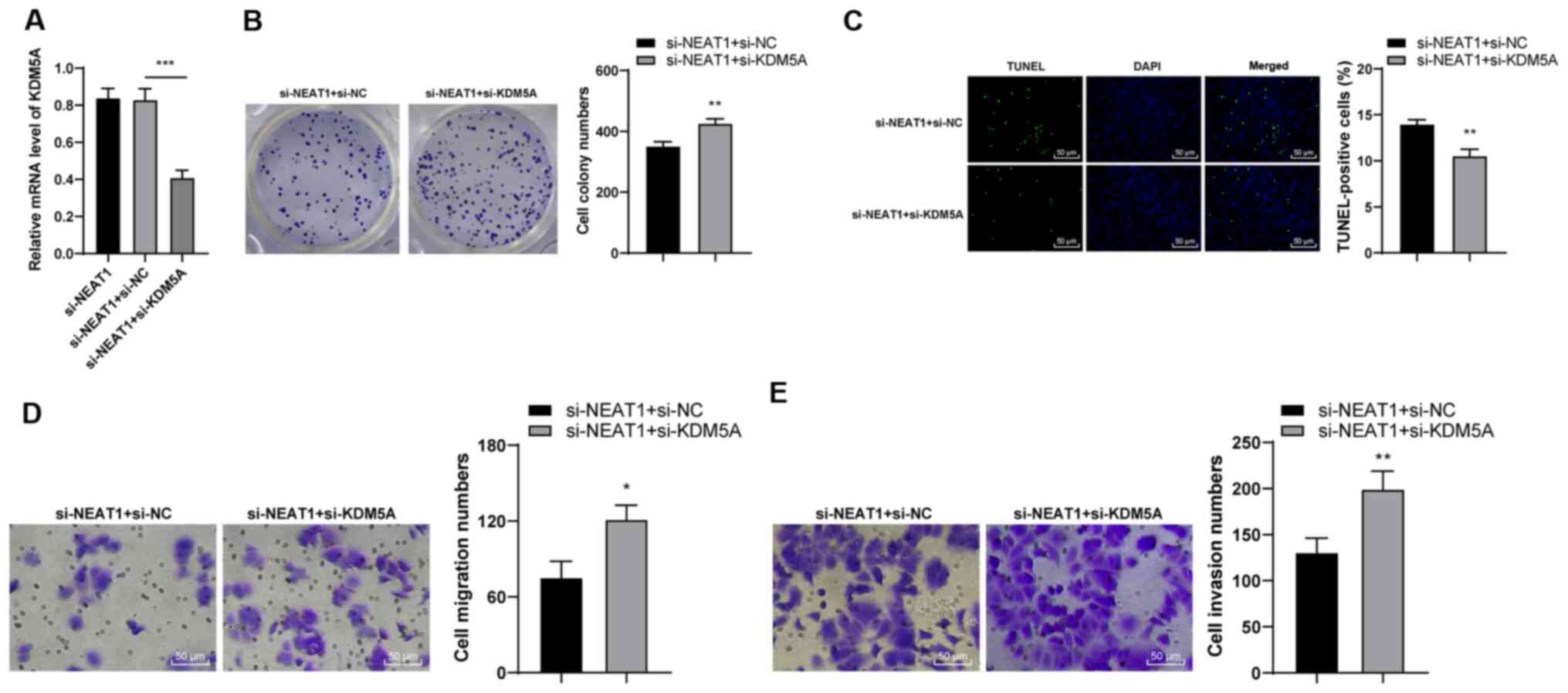
To explore the role of KDM5A in CRC, KDM5A
expression was silenced in si-NEAT1-transfected cells (P<0.001;
Fig. 4A) and observed that colony
formation was enhanced (P<0.01; Fig. 4B) and apoptosis was inhibited
(P<0.01; Fig. 4C). In
addition, cell migration and invasion were enhanced in the si-NEAT1
+ si-KDM5A-transfected cells (P<0.05; Fig. 4D and E). Taken together, these
findings demonstrated that the downregulation of KDM5A reversed the
inhibitory effects of NEAT1 silencing on the malignant behaviors of
HCT116 cells.
KDM5A inhibits Cul4A expression via the
demethylation of H3K4me3
KDM5A is associated with transcriptional regulation
due to its capacity to catalyze the removal of methyl groups from
H3K4me3 (27). KDM5A regulates
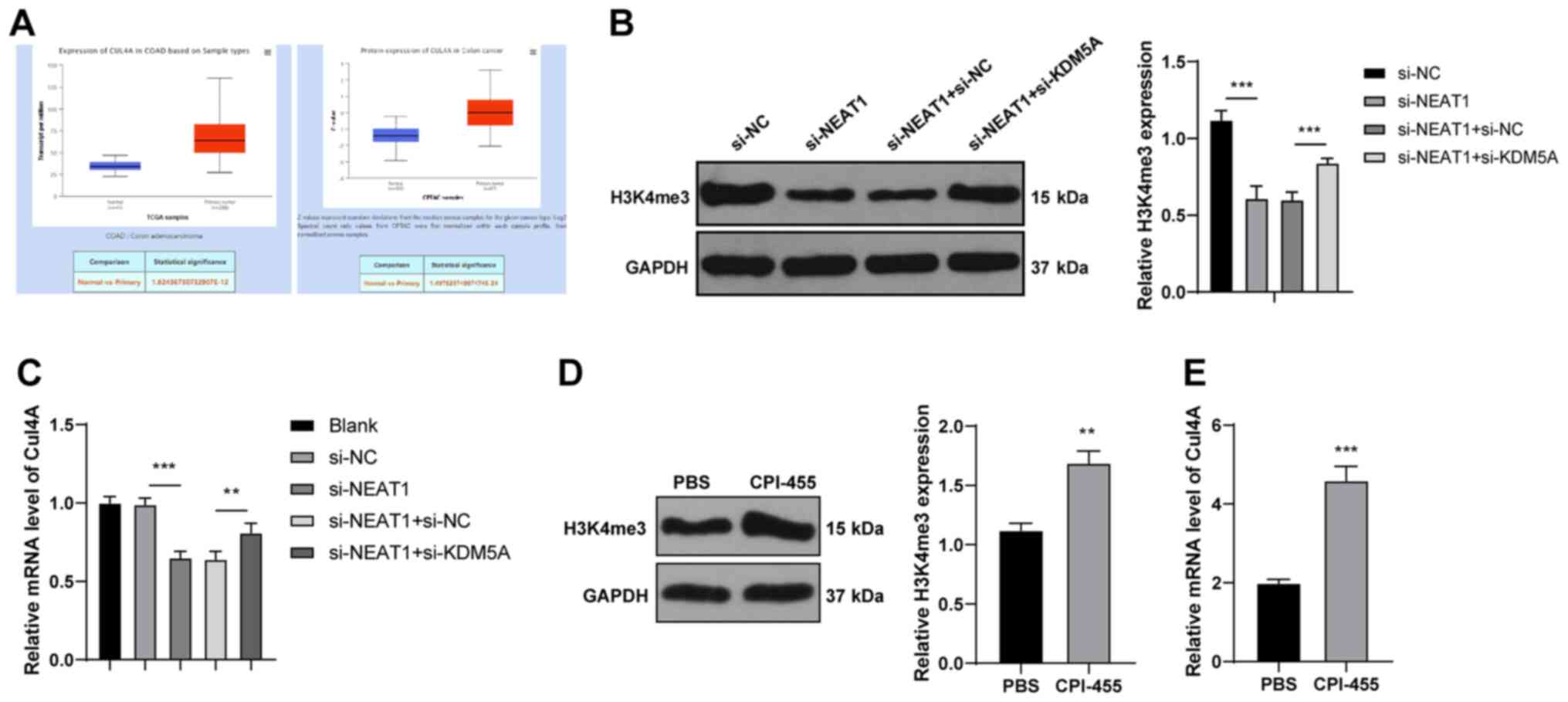
demethylation in the promoter of Cul4A (28). TCGA analysis (http://ualcan.path.uab.edu/analysis.html) (15) revealed that Cul4A was highly
expressed in CRC (Fig. 5A). It
was hypothesized that KDM5A may affect Cul4A expression by
regulating H3K4me3; thus, H3K4me3 methylation and Cul4A expression
were detected in the HCT116 cells. Compared with those of the
si-NC-transfected cells, the methylation level of H3K4me3 and Cul4A
expression were decreased in the si-NEAT1-transfected cells, but
were increased in the si-NEAT1 + si-KDM5A-transfected cells
(P<0.001; Fig. 5B and C),
suggesting that KDM5A regulated Cul4A expression via H3K4me3.
Subsequently, the level of H3K4me3 in HCT116 cells was enhanced by
CPI-455 treatment (P<0.01; Fig.
5D) and it was found that Cul4A expression was promoted with
the increase in the H3K4me3 levels (P<0.001; Fig. 5E). These results suggest that
KDM5A inhibits H3K4me3 methylation in the promoter of Cul4A, thus
inhibiting Cul4A expression.
Overexpression of Cul4A promotes the
proliferation and migration of si-NEAT1-transfected HCT116
cells
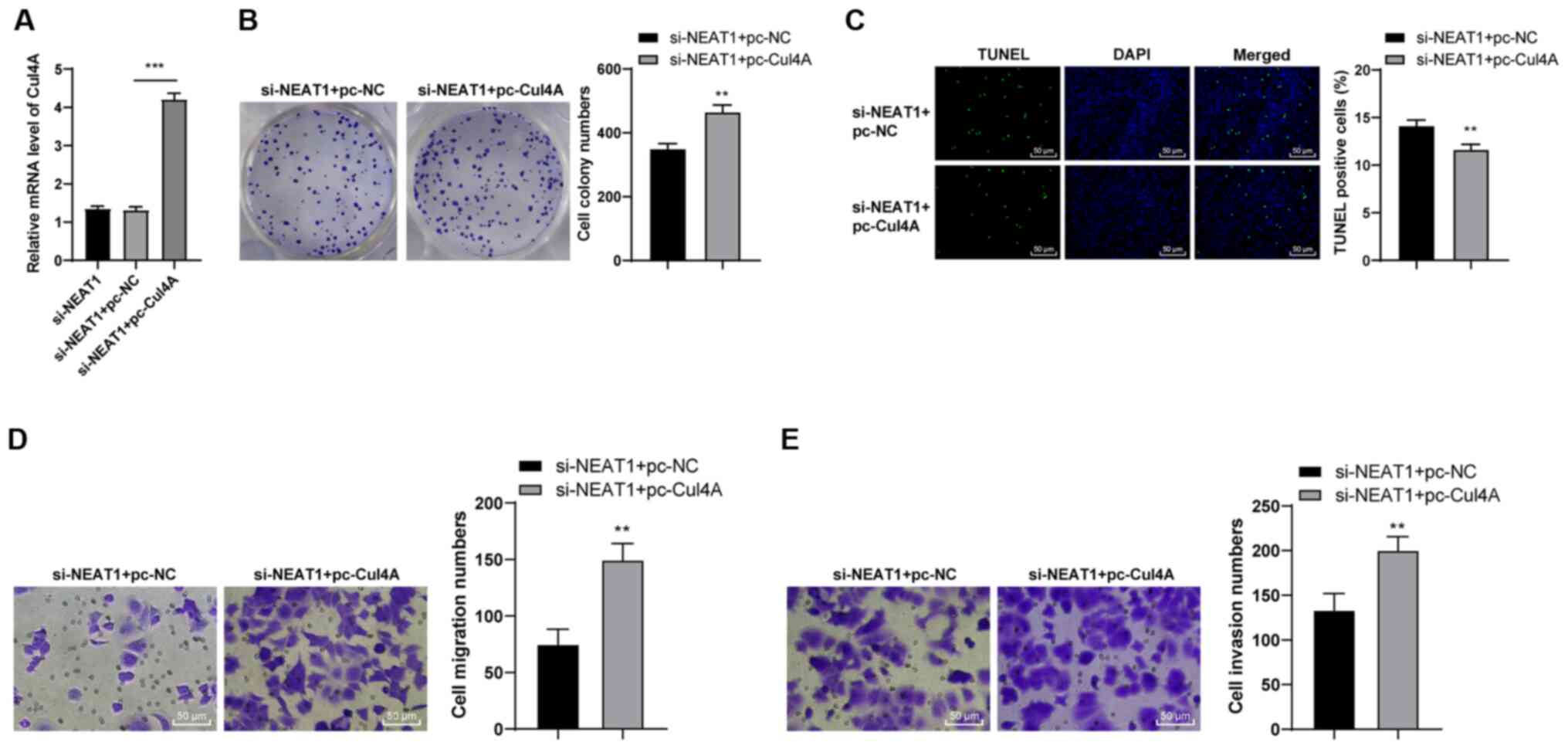
To verify the role of Cul4A in CRC, joint
experiments were designed. The si-NEAT1-transfected HCT116 cells
were transfected with pc-Cul4A (P<0.001; Fig. 6A), and the malignant behaviors of
the HCT116 cells were examined. Compared with the si-NEAT1 + pc-NC
cells, the si-NEAT1 + Cul4A cells exhibited a notably increased
colony formation ability (P<0.01; Fig. 6B) and a reduced apoptosis
(P<0.01; Fig. 6C). In
addition, an enhanced Cul4A expression promoted cell migration and
invasion (all P<0.01; Fig. 6D and
E). These results suggested that the overexpression of Cul4A
facilitated the malignant behavior of si-NEAT1-transfected HCT116
cells.
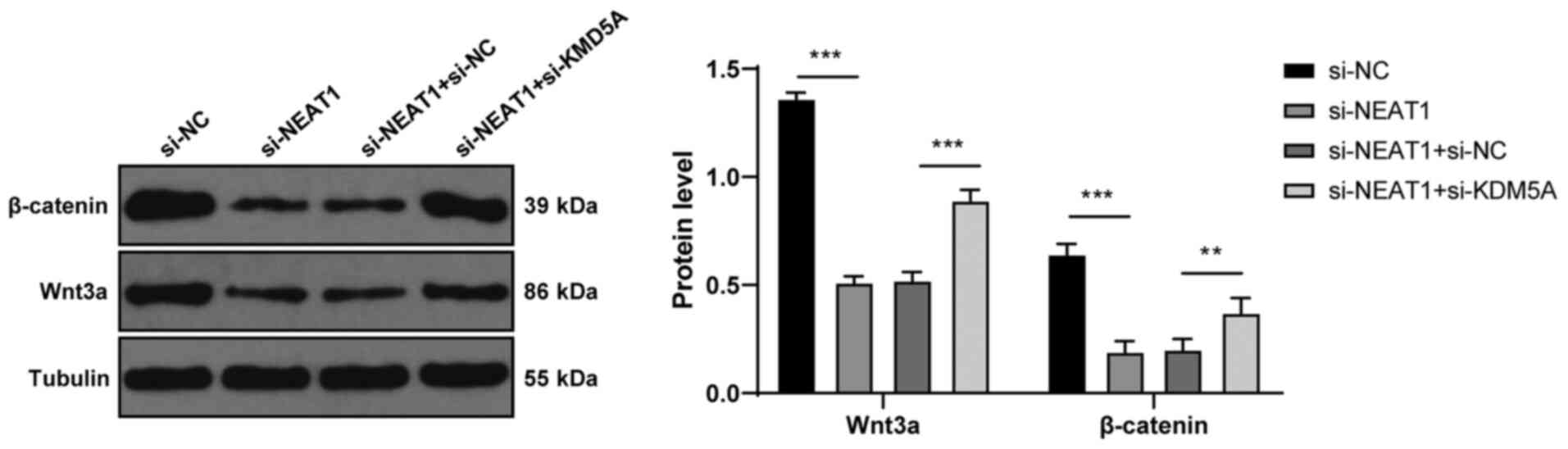
lncRNA NEAT1 activates the Wnt pathway
via KDM5A/Cul4A
The Wnt pathway plays a vital role in CRC (29,30), and there exists a regulatory
association between Cul4A and this pathway (31). Hence, the present study detected
proteins related to the Wnt pathway. Compared with the
si-NC-transfected cells, the si-NEAT1-transfected cells exhibited a
decreased expression of Wnt3a and β-catenin (P<0.001; Fig. 7), while si-KDM5A transfection
partially increased the expression of Wnt3a and β-catenin (all
P<0.01; Fig. 7). Thus, lncRNA
NEAT1 activated the Wnt pathway via KDM5A/Cul4A.
lncRNA NEAT1 promotes CRC via KDM5A/Cul4A
in vivo
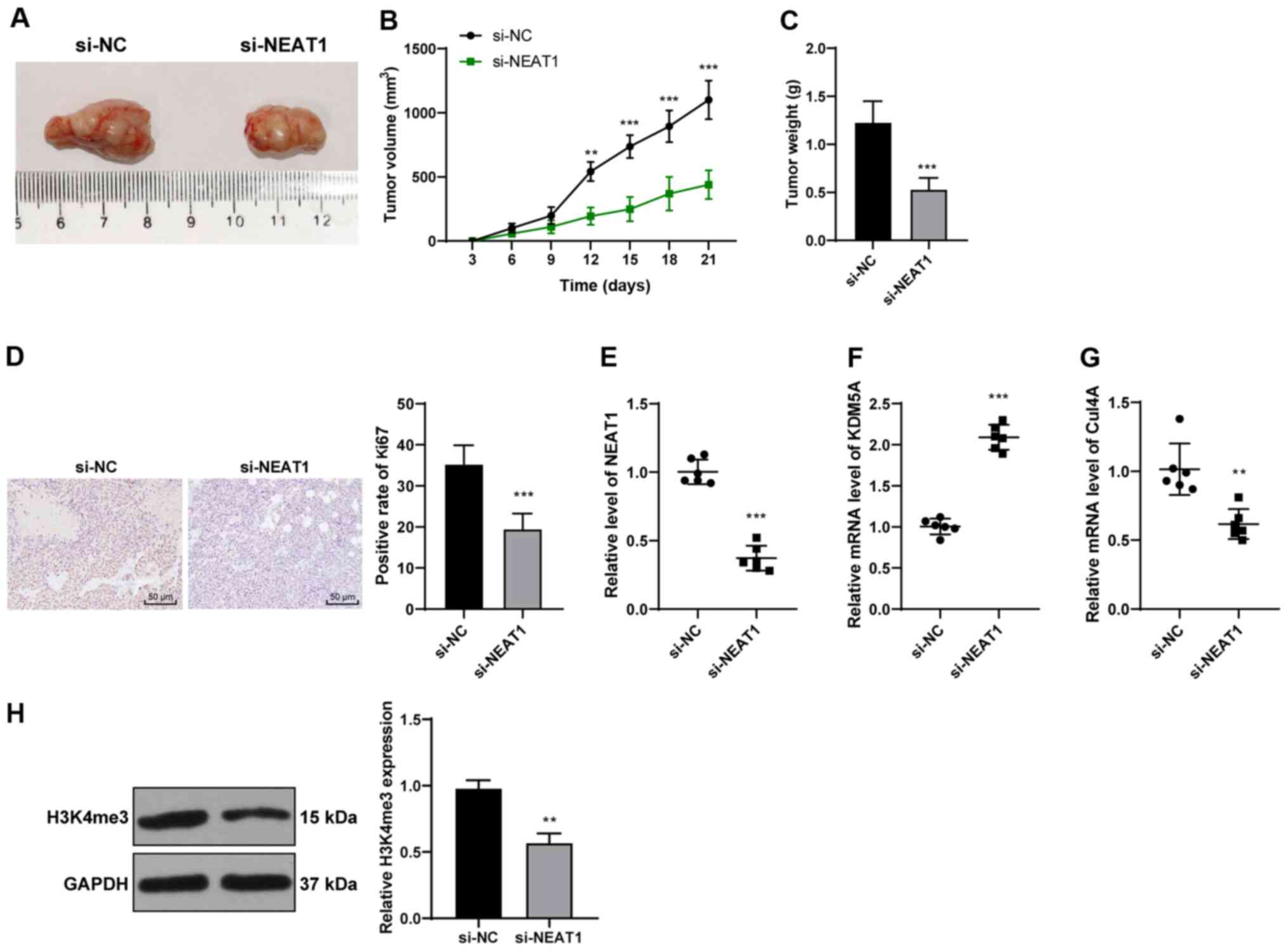
To verify the effects of lncRNA NEAT1 in
vivo, a xenograft model of CRC was established by injecting
HCT116 cells into the right armpits of mice (Fig. 8A). Tumor growth was inhibited
(P<0.01; Fig. 8B and Table SI) and tumor weight was reduced
in the mice injected with si-NEAT1-transfected cells (P<0.001;
Fig. 8C). Ki67 is a marker of
reactive proliferation (32).
Immunohistochemistry revealed that the injection of
si-NEAT1-transfected cells led to a decrease in the Ki67-positive
rate (P<0.001; Fig. 8D).
Compared with the si-NC group, the si-NEAT1 group exhibited a
notably decreased NEAT1 expression, an increased KDM5A expression,
and decreased H3K4me3 levels and Cul4A expression (all P<0.001;
Fig. 8E-H). Thus, the silencing
of lncRNA NEAT1 expression promoted KDM5A expression and inhibited
Cul4A expression, thus, supprssing the development of CRC in
vivo.
Discussion
CRC mostly develops from benign polyps to distant
metastasis and, consequently, early diagnosis and intervention are
essential for the long-term survival of patients (33). Recently, the biological
characteristics of lncRNAs and novel gene therapies of CRC have
attracted considerable attention (34). The present study elucidated the
potential mechanisms of action of lncRNA NEAT1 in facilitating CRC
progression.
With the advancement of tumor molecular biology, the
role of lncRNAs in the progression of CRC has been increasingly
investigated (35). lncRNA NEAT1
was found to be associated with CRC through the LncRNADisease
database (14). The aberrant
NEAT1 expression has been documented in CRC, in which its elevated
expression is related to poor outcomes (10). In the present study, NEAT1 was
predicted expression by TCGA analysis and it was found that NEAT1
was upregulated in CRC. Additionally, NEAT1 was highly expressed in
cancer tissues and cells. An elevated NEAT1 expression contributes
to poor differentiation, metastasis, and TNM stage (36). In the present study, HCT116 cells
were then transfected with si-NEAT1 to further determine the role
of NEAT1 in CRC. The si-NEAT1-transfected cells exhibited a
suppressed colony formation ability, increased apoptosis, and a
suppressed cell migration and invasion. The knockdown of NEAT1
impaired the proliferation and migration of CRC cells, thus
suppressing CRC progression (37). Thus, the silencing of NEAT1
suppressed the malignant behaviors of HCT116 cells.
The potential mechanisms of lncRNA NEAT1 in CRC were
then explored. The mechanisms of lncRNAs may be related to the
extensive subcellular localization in cells (38). NEAT1 was found to be mainly
located in the nucleus of HCT116 cells. NEAT1 may regulate the
expression of downstream genes by binding to transcription factors.
The database predicted that NEAT1 binds to transcription factor
E2F1, and E2F1 binds to KDM5A. E2F1, a crucial molecule implicated
in cell proliferation and apoptosis, is overexpressed in a broad
range of human cancers, including CRC (39,40). An elevated E2F1 expression in
cancer cells can induce metastasis and invasion (41), as well as enhance the
aggressiveness of CRC (42). KDMs
are enzymes of dimethyl-lysine residues in histones, and KDM5
specifically removes methylation from H3K4me1/2/3, which is a
hallmark associated with the action of transcription (27). The dysregulation of KDM5A
contributes to the pathogenesis of lung and gastric cancers
(43). However, currently,
knowledge regarding the specific role of KDM5A in CRC is limited.
The present study confirmed the binding association between NEAT1
and E2F1. KDM5A expression was notably promoted in the si-E2F1- and
si-NEAT1-transfected HCT116 cells. The present study is the first,
to the best of our knowledge, to reveal that NEAT1 binds to E2F1 to
inhibit KDM5A expression. To verify the role of KDM5A in CRC, KDM5A
expression was silenced in si-NEAT1-transfected cells. The results
revealed that the downregulation of KDM5A reversed the inhibitory
effects of NEAT1 silencing on the malignant behaviors of HCT116
cells.
Subsequently, the downstream mechanism of KDM5D in
CRC was investigated. KDM5A can catalyze the removal of methyl
groups from H3K4me3 (27). KDM5D
has been shown to suppress EMT in gastric cancer through the
demethylation of the Cul4A promoter (28). Accordingly, it was hypothesized
that KDM5A may affect Cul4A expression by regulating H3K4me3. Cul4A
participates in a variety of critical cell functions, including
apoptosis, cell cycle progression, genomic stability and histone
modification (44). Cul4A
amplification or overexpression can be observed in several human
malignancies, including breast, prostate and lung cancer (45-47). Importantly, Cul4A facilitates the
proliferation and metastasis of CRC cells by modulating H3K4
trimethylation in EMT (48). The
present study demonstrated that H3K4me3 methylation and Cul4A
expression were inhibited in the si-NEAT1-transfected cells,
whereas they were enhanced in the si-NEAT1 + si-KDM5A-transfected
cells, suggesting that KDM5A regulated Cul4A expression via
H3K4me3. H3K4me3 was then overexpressed in HCT116 cells by CPI-455
treatment and it was found that Cul4A expression was promoted with
an increase in the H3K4me3 levels. These results suggest that KDM5A
inhibits H3K4me3 methylation in the promoter of Cul4A, thus
inhibiting Cul4A expression. Joint experiments were conducted to
verify the role of Cul4A in CRC. The upregulation of Cul4A
facilitated the malignant behavior of si-NEAT1-treated HCT116
cells. Consistently, Li et al demonstrated that the
knockdown of Cul4A notably suppressed the progression of EMT and
metastasis of colon cancer cells in vitro (49).
Thereafter, the present study determined the
downstream pathway regulated by Cul4A. The Wnt signaling pathway is
a critical cascade closely related to cancer progression. In
particular, the role of the Wnt pathway in tumorigenesis has been
prominently described in CRC (50). The Wnt pathway controls β-catenin
levels for signal transduction through phosphorylation and
ubiquitin-mediated degradation (51). Cul4A is a novel Wnt target gene
that physically interacts with p27 in Wnt-responsive cells
(52). Proteins related to the
Wnt pathway were then detected. The expression of Wnt3a and
β-catenin in the si-NEAT1-transfected cells was notably decreased,
whereas it was partially increased following si-KDM5A transfection.
Zhang et al demonstrated that NEAT1 activatesd the Wnt
pathway to facilitate CRC progression and metastasis (38). Moreover, in vivo
experiments confirmed that the silencing of lncRNA NEAT1 promoted
KDM5A expression and inhibited Cul4A expression, thus suppressing
the development of CRC. Briefly, lncRNA NEAT1 activated the Wnt
pathway via KDM5A/Cul4A.
In conclusion, the present study demonstrated that
lncRNA NEAT1 bound to E2F1 to inhibit KDM5A expression, promote
Cul4A expression and activate the Wnt pathway, thereby facilitating
the progression of CRC. However, the competing endogenous RNA
mechanism of lncRNA NEAT1 in CRC remains to be elucidated.
Additionally, the present study merely demonstrated that the Wnt
pathway was activated; however, its function remains unclear. In
future, the authors aim to verify the specific mechanisms of the
Wnt pathway in CRC, and explore the feasibility and safety of NEAT1
as an entry point for CRC treatment.
Supplementary Data
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
XS and KZ were involved in the conceptualization.
WW, YW, GC, ZYe, LX, LG and ZYa were involved in data validation,
research, the provision of resources, data reviewing and manuscript
writing. LX and LG were involved in the reviewing and editing of
the manuscript. XS and ZYe confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The usage of ovarian tissues was approved by the
Ethics Committee of the Second Affiliated Hospital of Soochow
University. Informed consent was signed by each eligible
participant. All animal experiments are approved by the Ethics
Committee of the Second Affiliated Hospital of Soochow University
(S.No. 20190523b116). All experimental procedures were implemented
on the Ethical Guidelines for the study of experimental pain in
conscious animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by funds from the Education and
Health Foundation of Suzhou City (grant no. SYSD2019107), and
Suzhou Medical Team Introduction Program (grant no.
SZYJTD201804).
References
|
1
|
Mármol I, Sánchez-de-Diego C, Pradilla
Dieste A, Cerrada E and Rodriguez Yoldi MJ: Colorectal carcinoma: A
general overview and future perspectives in colorectal cancer. Int
J Mol Sci. 18:1972017. View Article : Google Scholar :
|
|
2
|
Abbaszadegan MR and Moghbeli M: Genetic
and molecular origins of colorectal cancer among the Iranians: An
update. Diagn Pathol. 13:972018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging biomarkers.
Gastroenterology. 149:1204–1225.e12. 2015. View Article : Google Scholar :
|
|
5
|
Cheng J, Chen J, Zhang X, Mei H, Wang F
and Cai Z: Overexpression of CRNDE promotes the progression of
bladder cancer. Biomed Pharmacother. 99:638–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar :
|
|
7
|
Wei L, Wang X, Lv L, Zheng Y, Zhang N and
Yang M: The emerging role of noncoding RNAs in colorectal cancer
chemoresistance. Cell Oncol (Dordr). 42:757–768. 2019. View Article : Google Scholar
|
|
8
|
Xu MD, Qi P and Du X: Long non-coding RNAs
in colorectal cancer: Implications for pathogenesis and clinical
application. Mod Pathol. 27:1310–1320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu X, Li Z, Zheng H, Chan MT and Wu WK:
NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif.
50:e123292017. View Article : Google Scholar
|
|
11
|
Dong P, Xiong Y, Yue J, Hanley SJB,
Kobayashi N, Todo Y and Watari H: Long non-coding RNA NEAT1: A
novel target for diagnosis and therapy in human tumors. Front
Genet. 9:4712018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu F, Ai FY, Zhang DC, Tian L, Yang ZY
and Liu SJ: LncRNA NEAT1 knockdown attenuates autophagy to elevate
5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer
Med. 9:1079–1091. 2020. View Article : Google Scholar
|
|
13
|
Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F,
Zhuo C, Zheng Y, Li B, Wang Z and Xu Y: Nuclear-enriched abundant
transcript 1 as a diagnostic and prognostic biomarker in colorectal
cancer. Mol Cancer. 14:1912015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41(Database Issue): D983–D986. 2013. View Article : Google Scholar :
|
|
15
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Li L, Wang Z, Pan T, Sahni N, Jin X,
Wang G, Li J, Zheng X, Zhang Y, et al: LncMAP: Pan-cancer atlas of
long noncoding RNA-mediated transcriptional network perturbations.
Nucleic Acids Res. 46:1113–1123. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vinogradova M, Gehling VS, Gustafson A,
Arora S, Tindell CA, Wilson C, Williamson KE, Guler GD, Gangurde P,
Manieri W, et al: An inhibitor of KDM5 demethylases reduces
survival of drug-tolerant cancer cells. Nat Chem Biol. 12:531–538.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xue XJ, Li FR and Yu J: Mitochondrial
pathway of the lysine demethylase 5C inhibitor CPI-455 in the
Eca-109 esophageal squamous cell carcinoma cell line. World J
Gastroenterol. 27:1805–1815. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pang SW, Awi NJ, Armon S, Lim WW, Low JS,
Peh KB, Peh SC and Teow SY: Current update of laboratory molecular
diagnostics advancement in management of colorectal cancer (CRC).
Diagnostics (Basel). 10:92019. View Article : Google Scholar
|
|
22
|
Mattiuzzi C, Sanchis-Gomar F and Lippi G:
Concise update on colorectal cancer epidemiology. Ann Transl Med.
7:6092019. View Article : Google Scholar
|
|
23
|
He Q, Long J, Yin Y, Li Y, Lei X, Li Z and
Zhu W: Emerging roles of lncRNAs in the formation and progression
of colorectal cancer. Front Oncol. 9:15422020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mas-Ponte D, Carlevaro-Fita J, Palumbo E,
Hermoso Pulido T, Guigo R and Johnson R: LncATLAS database for
subcellular localization of long noncoding RNAs. RNA. 23:1080–1087.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu H, Luo J, Luan S, He C and Li Z: Long
non-coding RNAs involved in cancer metabolic reprogramming. Cell
Mol Life Sci. 76:495–504. 2019. View Article : Google Scholar
|
|
26
|
Li MY, Tang XH, Fu Y, Wang TJ and Zhu JM:
Regulatory mechanisms and clinical applications of the long
non-coding RNA PVT1 in cancer treatment. Front Oncol. 9:7872019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Petronikolou N, Longbotham JE and Fujimori
DG: Extended recognition of the histone H3 tail by histone
demethylase KDM5A. Biochemistry. 59:647–651. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen X, Hu K, Cheng G, Xu L, Chen Z, Du P
and Zhuang Z: KDM5D inhibit epithelial-mesenchymal transition of
gastric cancer through demethylation in the promoter of Cul4A in
male. J Cell Biochem. 120:12247–12258. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Patel S, Alam A, Pant R and Chattopadhyay
S: Wnt signaling and its significance within the tumor
microenvironment: Novel therapeutic insights. Front Immunol.
10:28722019. View Article : Google Scholar
|
|
30
|
Martin-Orozco E, Sanchez-Fernandez A,
Ortiz-Parra I and Ayala-San Nicolas M: WNT signaling in tumors: The
way to evade drugs and immunity. Front Immunol. 10:28542019.
View Article : Google Scholar
|
|
31
|
Yu R, Cai L, Chi Y, Ding X and Wu X:
miR-377 targets CUL4A and regulates metastatic capability in
ovarian cancer. Int J Mol Med. 41:3147–3156. 2018.PubMed/NCBI
|
|
32
|
Goodlad RA: Quantification of epithelial
cell proliferation, cell dynamics, and cell kinetics in vivo. Wiley
Interdiscip Rev Dev Biol. 6:2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gellad ZF and Provenzale D: Colorectal
cancer: National and international perspective on the burden of
disease and public health impact. Gastroenterology. 138:2177–2190.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar :
|
|
35
|
Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong
H, Zhou JY and Yang SM: Long non-coding RNAs in colorectal cancer.
Oncotarget. 7:5226–5239. 2016. View Article : Google Scholar :
|
|
36
|
Liu H, Li A, Sun Z, Zhang J and Xu H: Long
non-coding RNA NEAT1 promotes colorectal cancer progression by
regulating miR-205-5p/VEGFA axis. Hum Cell. 33:386–396. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu Z, Du S, Yin K, Ai S, Yu M, Liu Y,
Shen Y, Liu M, Jiao R, Chen X and Guan W: Knockdown long noncoding
RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal
cancer through modulating miR-193a-3p/KRAS. Cancer Med. 8:261–275.
2019. View Article : Google Scholar
|
|
38
|
Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan
C, Xu M, Sun H, Liu C, Wei P and Du X: The lncRNA NEAT1 activates
Wnt/β-catenin signaling and promotes colorectal cancer progression
via interacting with DDX5. J Hematol Oncol. 11:1132018. View Article : Google Scholar
|
|
39
|
Zhao JP and Chen LL: Circular RNA MAT2B
induces colorectal cancer proliferation via sponging miR-610,
resulting in an increased E2F1 expression. Cancer Manag Res.
12:7107–7116. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Denechaud PD, Fajas L and Giralt A: E2F1,
a novel regulator of metabolism. Front Endocrinol (Lausanne).
8:3112017. View Article : Google Scholar
|
|
41
|
Chen J, Gong C, Mao H, Li Z, Fang Z, Chen
Q, Lin M, Jiang X, Hu Y, Wang W, et al: E2F1/SP3/STAT6 axis is
required for IL-4-induced epithelial-mesenchymal transition of
colorectal cancer cells. Int J Oncol. 53:567–578. 2018.PubMed/NCBI
|
|
42
|
Fang Z, Gong C, Liu H, Zhang X, Mei L,
Song M, Qiu L, Luo S, Zhu Z, Zhang R, et al: E2F1 promote the
aggressiveness of human colorectal cancer by activating the
ribonucleotide reductase small subunit M2. Biochem Biophys Res
Commun. 464:407–415. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Torres IO, Kuchenbecker KM, Nnadi CI,
Fletterick RJ, Kelly MJ and Fujimori DG: Histone demethylase KDM5A
is regulated by its reader domain through a positive-feedback
mechanism. Nat Commun. 6:62042015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hung MS, Chen YC, Lin P, Li YC, Hsu CC,
Lung JH, You L, Xu Z, Mao JH, Jablons DM and Yang CT: Cul4A
modulates invasion and metastasis of lung cancer through regulation
of ANXA10. Cancers (Basel). 11:6182019. View Article : Google Scholar :
|
|
45
|
Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang
P, He X, Wang Q, Huang Y, Jen KY, et al: CUL4A induces
epithelial-mesenchymal transition and promotes cancer metastasis by
regulating ZEB1 expression. Cancer Res. 74:520–531. 2014.
View Article : Google Scholar :
|
|
46
|
Ren S, Xu C, Cui Z, Yu Y, Xu W, Wang F, Lu
J, Wei M, Lu X, Gao X, et al: Oncogenic CUL4A determines the
response to thalidomide treatment in prostate cancer. J Mol Med
(Berl). 90:1121–1132. 2012. View Article : Google Scholar
|
|
47
|
Jia L, Yan F, Cao W, Chen Z, Zheng H, Li
H, Pan Y, Narula N, Ren X, Li H and Zhou P: Dysregulation of CUL4A
and CUL4B ubiquitin ligases in lung cancer. J Biol Chem.
292:2966–2978. 2017. View Article : Google Scholar :
|
|
48
|
Sui X, Zhou H, Zhu L, Wang D, Fan S and
Zhao W: CUL4A promotes proliferation and metastasis of colorectal
cancer cells by regulating H3K4 trimethylation in
epithelial-mesenchymal transition. Onco Targets Ther. 10:735–743.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li C, Bu J, Liao Y, Zhang J, Han J, Zhang
H, Xing H, Li Z, Wu H, Liang L, et al: High expressions of CUL4A
and TP53 in colorectal cancer predict poor survival. Cell Physiol
Biochem. 51:2829–2842. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar :
|
|
51
|
Novellasdemunt L, Antas P and Li VS:
Targeting Wnt signaling in colorectal cancer. A review in the
theme: Cell signaling: proteins, pathways and mechanisms. Am J
Physiol Cell Physiol. 309:C511–C521. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Miranda-Carboni GA, Krum SA, Yee K, Nava
M, Deng QE, Pervin S, Collado-Hidalgo A, Galic Z, Zack JA, Nakayama
K, et al: A functional link between Wnt signaling and
SKP2-independent p27 turnover in mammary tumors. Genes Dev.
22:3121–3134. 2008. View Article : Google Scholar : PubMed/NCBI
|