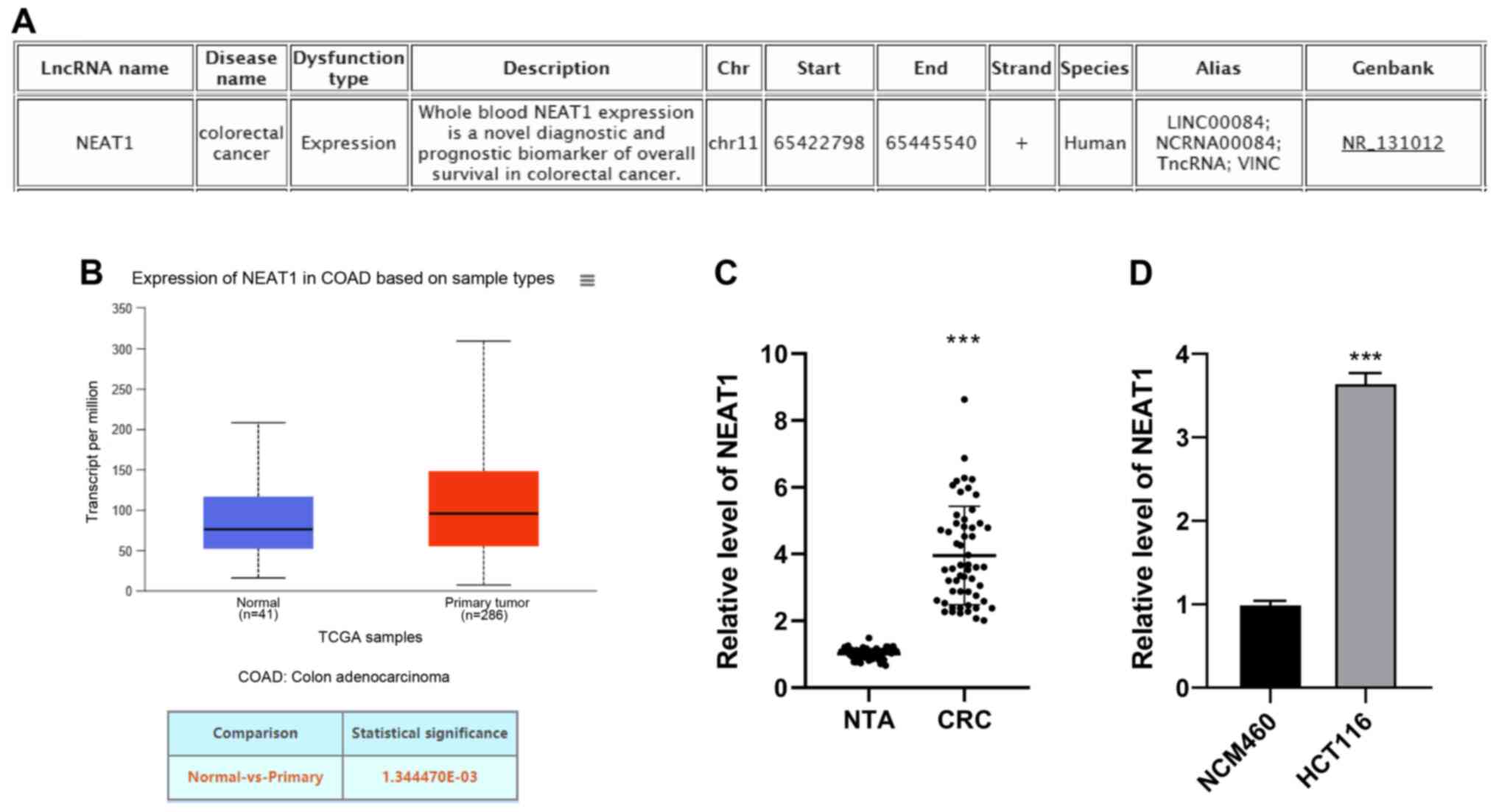
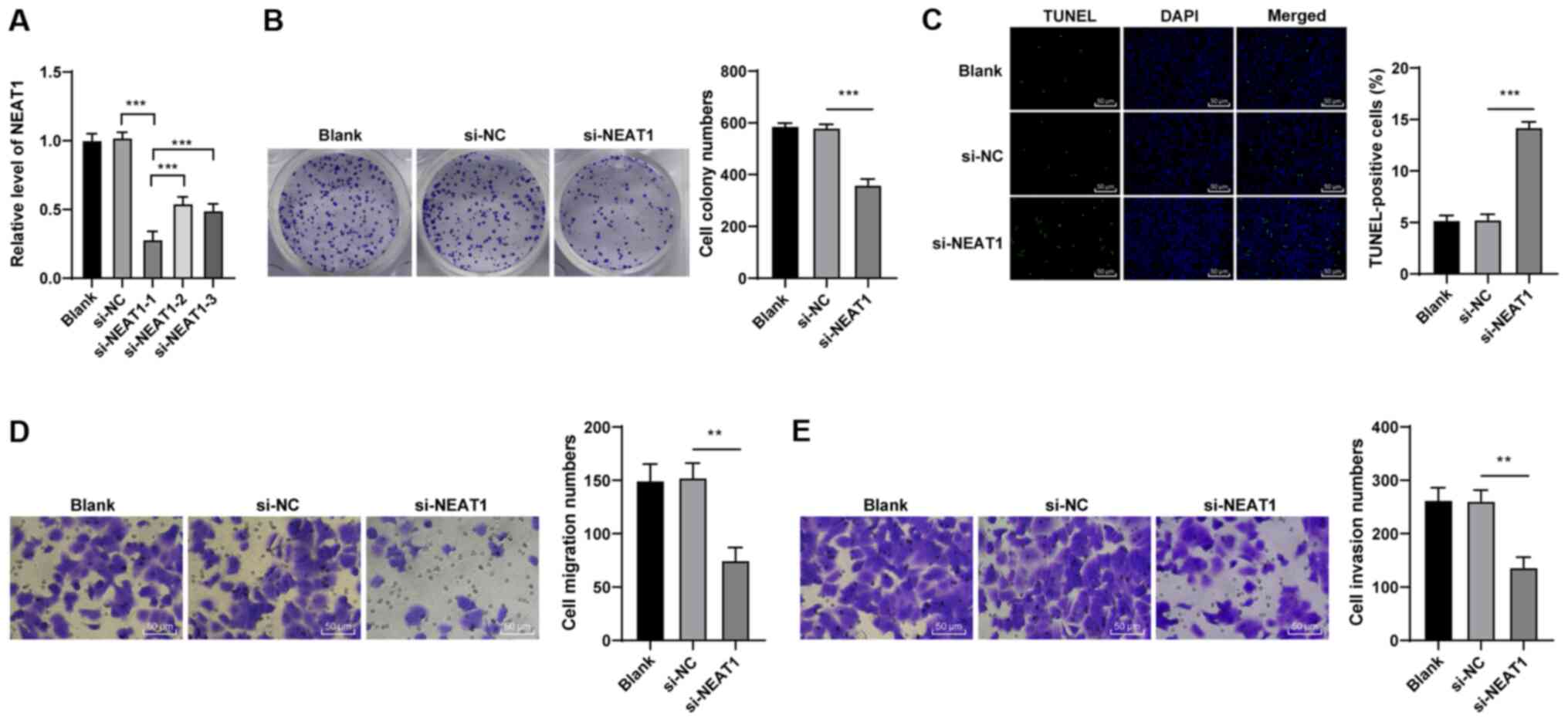
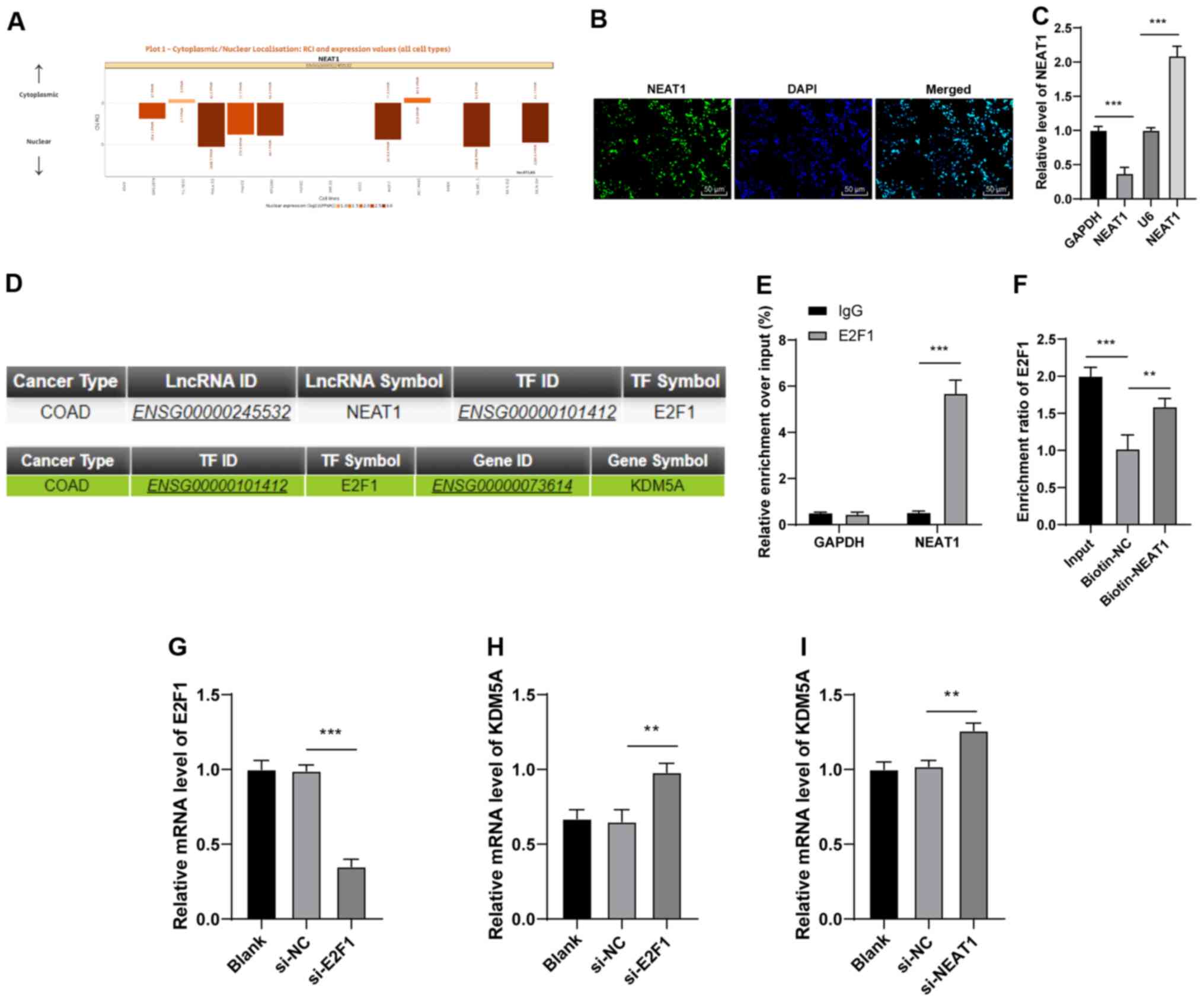
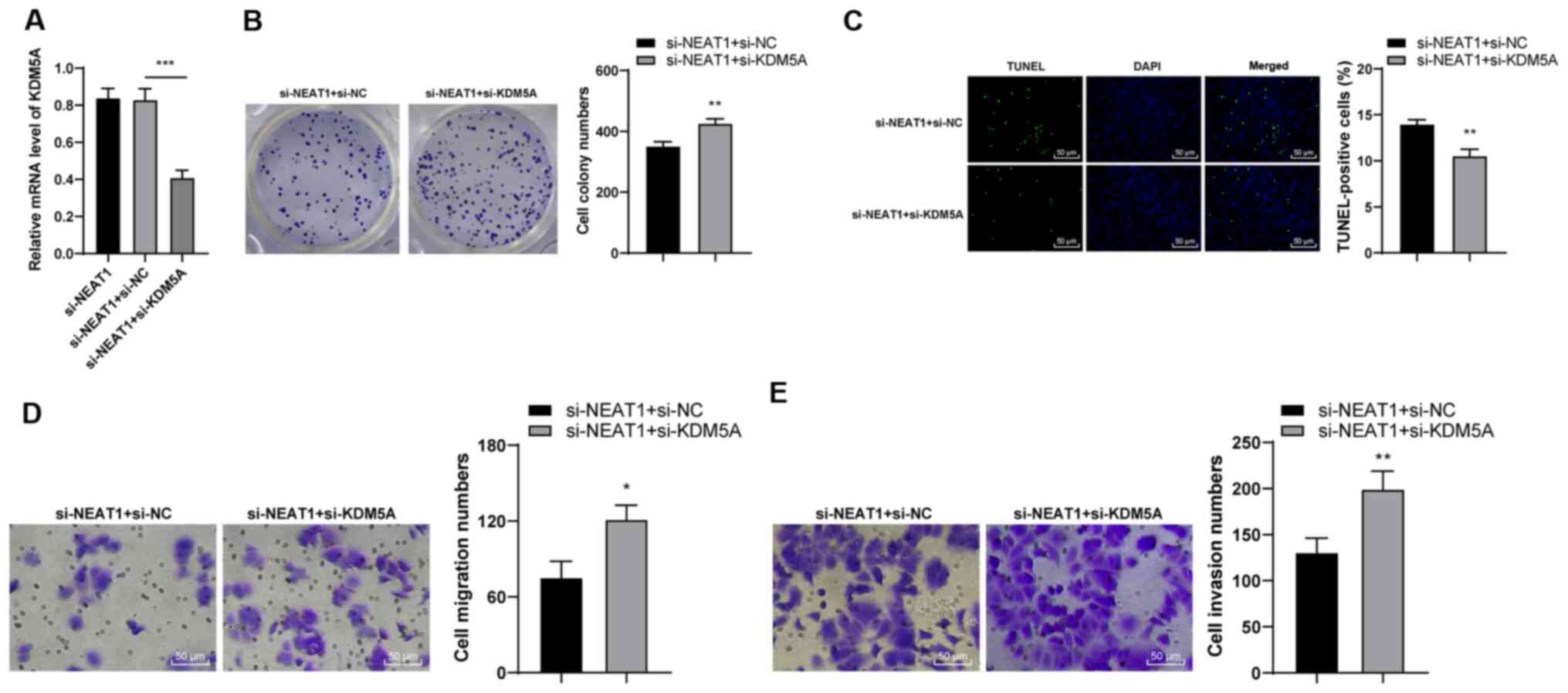
|
1
|
Mármol I, Sánchez-de-Diego C, Pradilla
Dieste A, Cerrada E and Rodriguez Yoldi MJ: Colorectal carcinoma: A
general overview and future perspectives in colorectal cancer. Int
J Mol Sci. 18:1972017. View Article : Google Scholar :
|
|
2
|
Abbaszadegan MR and Moghbeli M: Genetic
and molecular origins of colorectal cancer among the Iranians: An
update. Diagn Pathol. 13:972018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cunningham D, Atkin W, Lenz HJ, Lynch HT,
Minsky B, Nordlinger B and Starling N: Colorectal cancer. Lancet.
375:1030–1047. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Okugawa Y, Grady WM and Goel A: Epigenetic
alterations in colorectal cancer: Emerging biomarkers.
Gastroenterology. 149:1204–1225.e12. 2015. View Article : Google Scholar :
|
|
5
|
Cheng J, Chen J, Zhang X, Mei H, Wang F
and Cai Z: Overexpression of CRNDE promotes the progression of
bladder cancer. Biomed Pharmacother. 99:638–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar :
|
|
7
|
Wei L, Wang X, Lv L, Zheng Y, Zhang N and
Yang M: The emerging role of noncoding RNAs in colorectal cancer
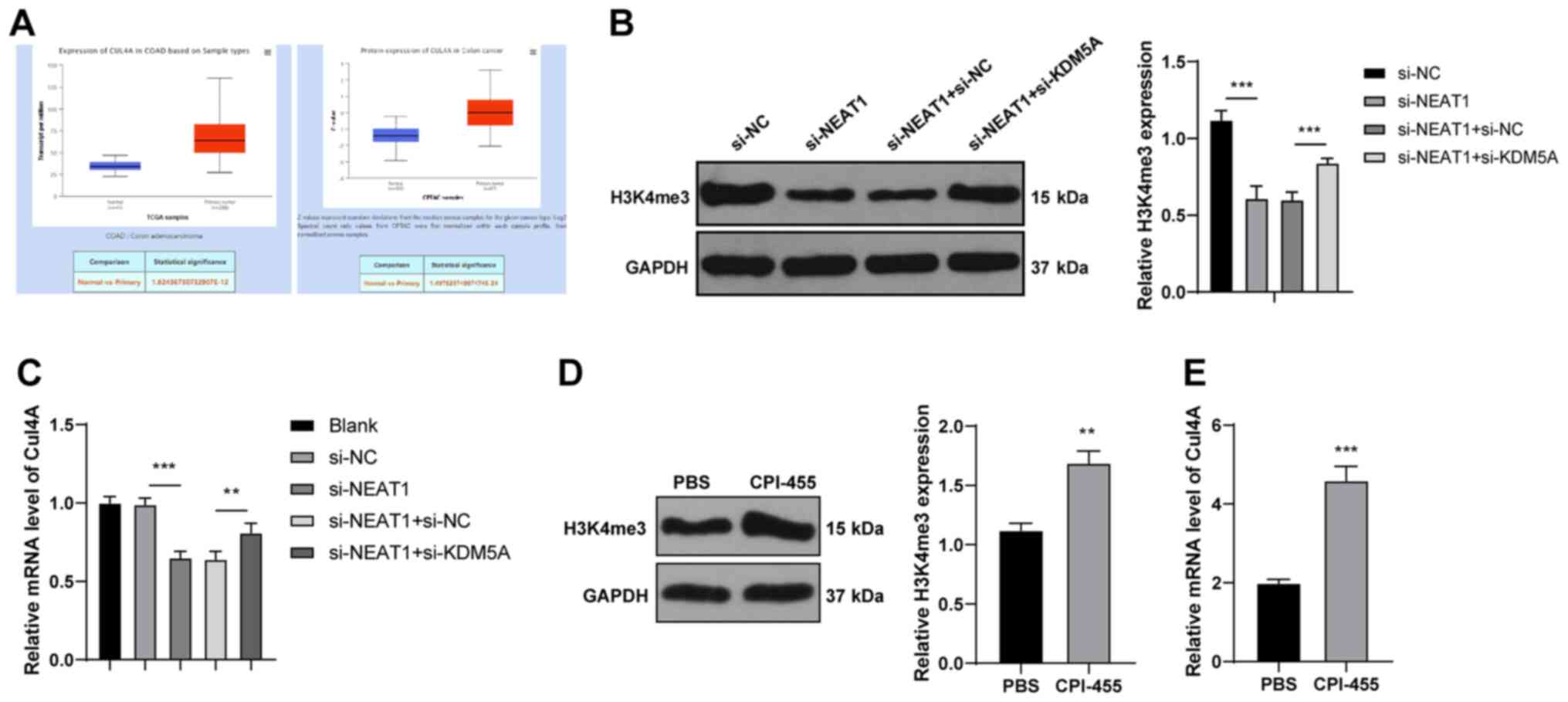
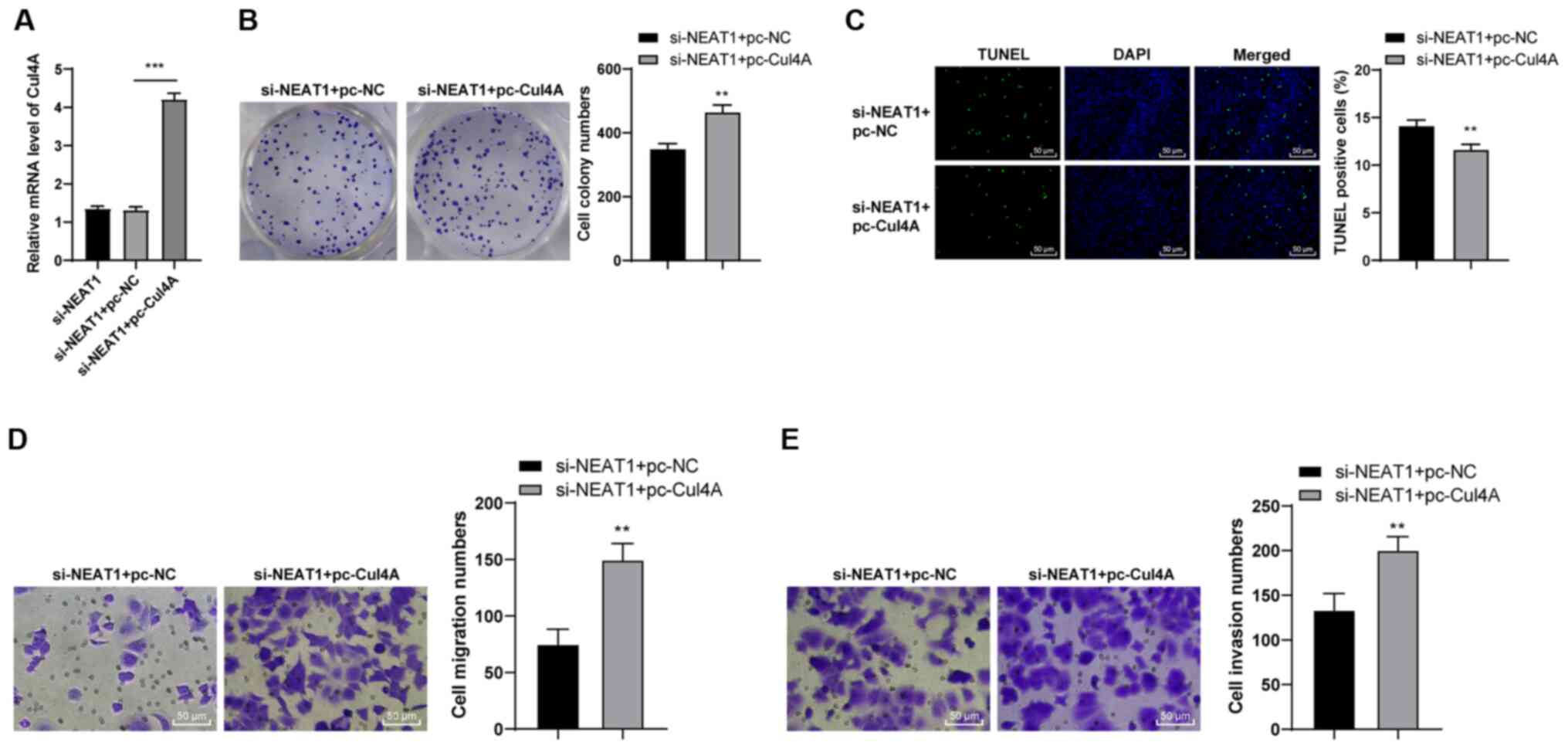
chemoresistance. Cell Oncol (Dordr). 42:757–768. 2019. View Article : Google Scholar
|
|
8
|
Xu MD, Qi P and Du X: Long non-coding RNAs
in colorectal cancer: Implications for pathogenesis and clinical
application. Mod Pathol. 27:1310–1320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu X, Li Z, Zheng H, Chan MT and Wu WK:
NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif.
50:e123292017. View Article : Google Scholar
|
|
11
|
Dong P, Xiong Y, Yue J, Hanley SJB,
Kobayashi N, Todo Y and Watari H: Long non-coding RNA NEAT1: A
novel target for diagnosis and therapy in human tumors. Front
Genet. 9:4712018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu F, Ai FY, Zhang DC, Tian L, Yang ZY
and Liu SJ: LncRNA NEAT1 knockdown attenuates autophagy to elevate
5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer
Med. 9:1079–1091. 2020. View Article : Google Scholar
|
|
13
|
Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F,
Zhuo C, Zheng Y, Li B, Wang Z and Xu Y: Nuclear-enriched abundant
transcript 1 as a diagnostic and prognostic biomarker in colorectal
cancer. Mol Cancer. 14:1912015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41(Database Issue): D983–D986. 2013. View Article : Google Scholar :
|
|
15
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Li L, Wang Z, Pan T, Sahni N, Jin X,
Wang G, Li J, Zheng X, Zhang Y, et al: LncMAP: Pan-cancer atlas of
long noncoding RNA-mediated transcriptional network perturbations.
Nucleic Acids Res. 46:1113–1123. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vinogradova M, Gehling VS, Gustafson A,
Arora S, Tindell CA, Wilson C, Williamson KE, Guler GD, Gangurde P,
Manieri W, et al: An inhibitor of KDM5 demethylases reduces
survival of drug-tolerant cancer cells. Nat Chem Biol. 12:531–538.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xue XJ, Li FR and Yu J: Mitochondrial
pathway of the lysine demethylase 5C inhibitor CPI-455 in the
Eca-109 esophageal squamous cell carcinoma cell line. World J
Gastroenterol. 27:1805–1815. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
20
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pang SW, Awi NJ, Armon S, Lim WW, Low JS,
Peh KB, Peh SC and Teow SY: Current update of laboratory molecular
diagnostics advancement in management of colorectal cancer (CRC).
Diagnostics (Basel). 10:92019. View Article : Google Scholar
|
|
22
|
Mattiuzzi C, Sanchis-Gomar F and Lippi G:
Concise update on colorectal cancer epidemiology. Ann Transl Med.
7:6092019. View Article : Google Scholar
|
|
23
|
He Q, Long J, Yin Y, Li Y, Lei X, Li Z and
Zhu W: Emerging roles of lncRNAs in the formation and progression
of colorectal cancer. Front Oncol. 9:15422020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mas-Ponte D, Carlevaro-Fita J, Palumbo E,
Hermoso Pulido T, Guigo R and Johnson R: LncATLAS database for
subcellular localization of long noncoding RNAs. RNA. 23:1080–1087.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu H, Luo J, Luan S, He C and Li Z: Long
non-coding RNAs involved in cancer metabolic reprogramming. Cell
Mol Life Sci. 76:495–504. 2019. View Article : Google Scholar
|
|
26
|
Li MY, Tang XH, Fu Y, Wang TJ and Zhu JM:
Regulatory mechanisms and clinical applications of the long
non-coding RNA PVT1 in cancer treatment. Front Oncol. 9:7872019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Petronikolou N, Longbotham JE and Fujimori
DG: Extended recognition of the histone H3 tail by histone
demethylase KDM5A. Biochemistry. 59:647–651. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen X, Hu K, Cheng G, Xu L, Chen Z, Du P
and Zhuang Z: KDM5D inhibit epithelial-mesenchymal transition of
gastric cancer through demethylation in the promoter of Cul4A in
male. J Cell Biochem. 120:12247–12258. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Patel S, Alam A, Pant R and Chattopadhyay
S: Wnt signaling and its significance within the tumor
microenvironment: Novel therapeutic insights. Front Immunol.
10:28722019. View Article : Google Scholar
|
|
30
|
Martin-Orozco E, Sanchez-Fernandez A,
Ortiz-Parra I and Ayala-San Nicolas M: WNT signaling in tumors: The
way to evade drugs and immunity. Front Immunol. 10:28542019.
View Article : Google Scholar
|
|
31
|
Yu R, Cai L, Chi Y, Ding X and Wu X:
miR-377 targets CUL4A and regulates metastatic capability in
ovarian cancer. Int J Mol Med. 41:3147–3156. 2018.PubMed/NCBI
|
|
32
|
Goodlad RA: Quantification of epithelial
cell proliferation, cell dynamics, and cell kinetics in vivo. Wiley
Interdiscip Rev Dev Biol. 6:2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gellad ZF and Provenzale D: Colorectal
cancer: National and international perspective on the burden of
disease and public health impact. Gastroenterology. 138:2177–2190.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang L, Cho KB, Li Y, Tao G, Xie Z and Guo
B: Long noncoding RNA (lncRNA)-mediated competing endogenous RNA
networks provide novel potential biomarkers and therapeutic targets
for colorectal cancer. Int J Mol Sci. 20:57582019. View Article : Google Scholar :
|
|
35
|
Xie X, Tang B, Xiao YF, Xie R, Li BS, Dong
H, Zhou JY and Yang SM: Long non-coding RNAs in colorectal cancer.
Oncotarget. 7:5226–5239. 2016. View Article : Google Scholar :
|
|
36
|
Liu H, Li A, Sun Z, Zhang J and Xu H: Long
non-coding RNA NEAT1 promotes colorectal cancer progression by
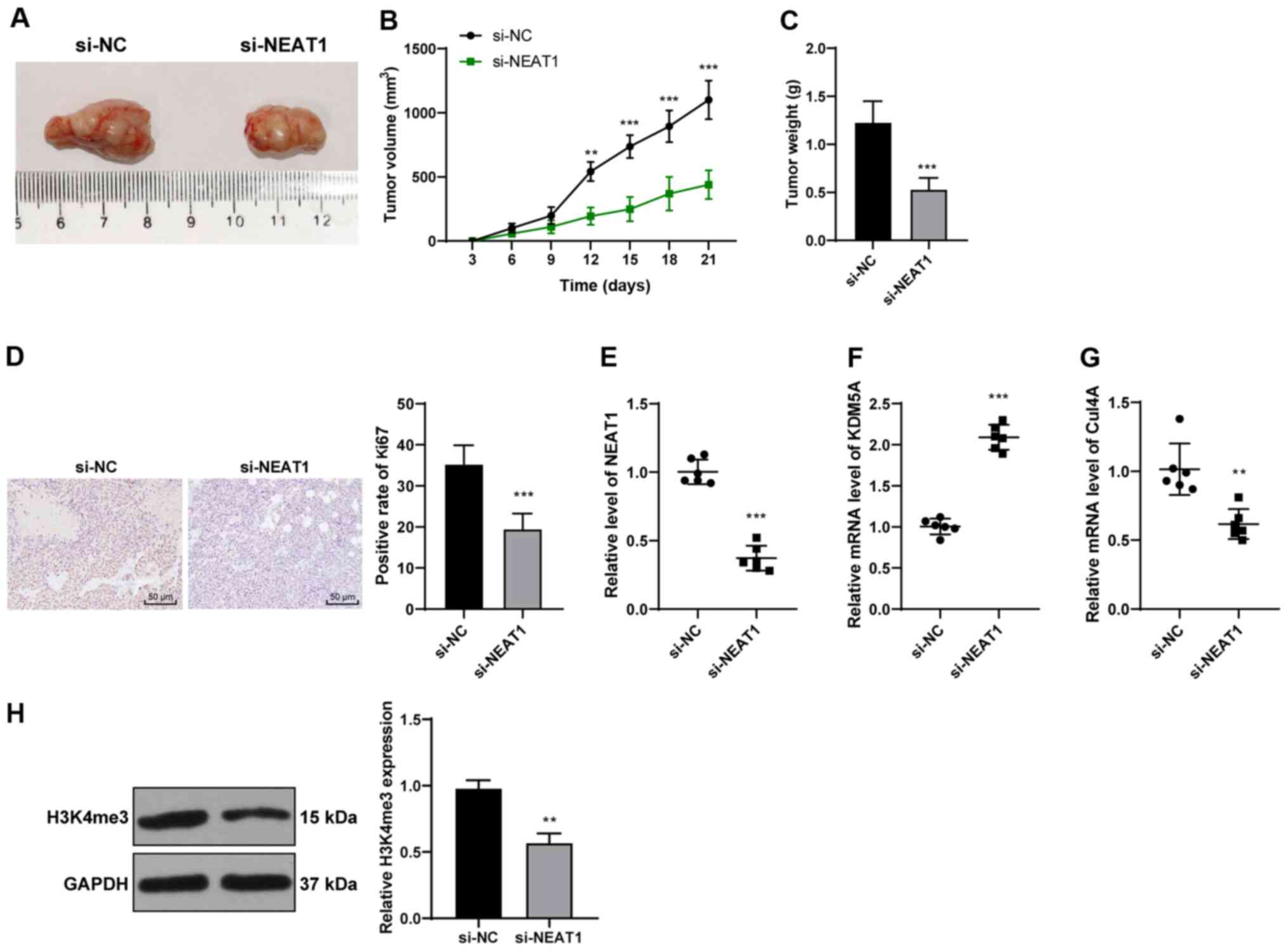
regulating miR-205-5p/VEGFA axis. Hum Cell. 33:386–396. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu Z, Du S, Yin K, Ai S, Yu M, Liu Y,
Shen Y, Liu M, Jiao R, Chen X and Guan W: Knockdown long noncoding
RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal
cancer through modulating miR-193a-3p/KRAS. Cancer Med. 8:261–275.
2019. View Article : Google Scholar
|
|
38
|
Zhang M, Weng W, Zhang Q, Wu Y, Ni S, Tan
C, Xu M, Sun H, Liu C, Wei P and Du X: The lncRNA NEAT1 activates
Wnt/β-catenin signaling and promotes colorectal cancer progression
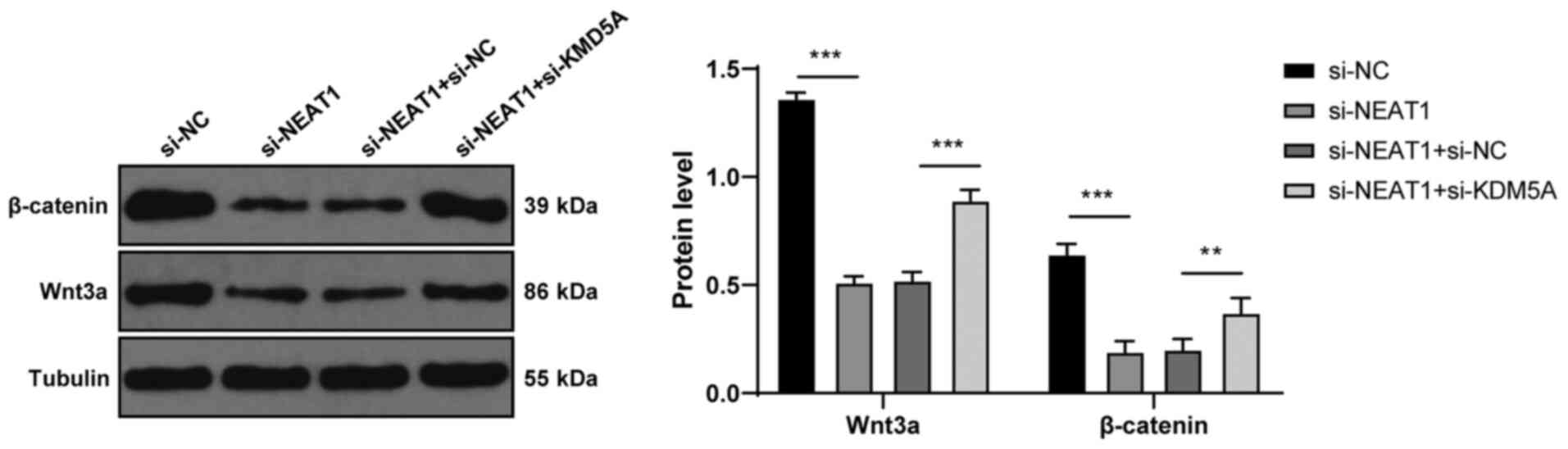
via interacting with DDX5. J Hematol Oncol. 11:1132018. View Article : Google Scholar
|
|
39
|
Zhao JP and Chen LL: Circular RNA MAT2B
induces colorectal cancer proliferation via sponging miR-610,
resulting in an increased E2F1 expression. Cancer Manag Res.
12:7107–7116. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Denechaud PD, Fajas L and Giralt A: E2F1,
a novel regulator of metabolism. Front Endocrinol (Lausanne).
8:3112017. View Article : Google Scholar
|
|
41
|
Chen J, Gong C, Mao H, Li Z, Fang Z, Chen
Q, Lin M, Jiang X, Hu Y, Wang W, et al: E2F1/SP3/STAT6 axis is
required for IL-4-induced epithelial-mesenchymal transition of
colorectal cancer cells. Int J Oncol. 53:567–578. 2018.PubMed/NCBI
|
|
42
|
Fang Z, Gong C, Liu H, Zhang X, Mei L,
Song M, Qiu L, Luo S, Zhu Z, Zhang R, et al: E2F1 promote the
aggressiveness of human colorectal cancer by activating the
ribonucleotide reductase small subunit M2. Biochem Biophys Res
Commun. 464:407–415. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Torres IO, Kuchenbecker KM, Nnadi CI,
Fletterick RJ, Kelly MJ and Fujimori DG: Histone demethylase KDM5A
is regulated by its reader domain through a positive-feedback
mechanism. Nat Commun. 6:62042015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hung MS, Chen YC, Lin P, Li YC, Hsu CC,
Lung JH, You L, Xu Z, Mao JH, Jablons DM and Yang CT: Cul4A
modulates invasion and metastasis of lung cancer through regulation
of ANXA10. Cancers (Basel). 11:6182019. View Article : Google Scholar :
|
|
45
|
Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang
P, He X, Wang Q, Huang Y, Jen KY, et al: CUL4A induces
epithelial-mesenchymal transition and promotes cancer metastasis by
regulating ZEB1 expression. Cancer Res. 74:520–531. 2014.
View Article : Google Scholar :
|
|
46
|
Ren S, Xu C, Cui Z, Yu Y, Xu W, Wang F, Lu
J, Wei M, Lu X, Gao X, et al: Oncogenic CUL4A determines the
response to thalidomide treatment in prostate cancer. J Mol Med
(Berl). 90:1121–1132. 2012. View Article : Google Scholar
|
|
47
|
Jia L, Yan F, Cao W, Chen Z, Zheng H, Li
H, Pan Y, Narula N, Ren X, Li H and Zhou P: Dysregulation of CUL4A
and CUL4B ubiquitin ligases in lung cancer. J Biol Chem.
292:2966–2978. 2017. View Article : Google Scholar :
|
|
48
|
Sui X, Zhou H, Zhu L, Wang D, Fan S and
Zhao W: CUL4A promotes proliferation and metastasis of colorectal
cancer cells by regulating H3K4 trimethylation in
epithelial-mesenchymal transition. Onco Targets Ther. 10:735–743.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li C, Bu J, Liao Y, Zhang J, Han J, Zhang
H, Xing H, Li Z, Wu H, Liang L, et al: High expressions of CUL4A
and TP53 in colorectal cancer predict poor survival. Cell Physiol
Biochem. 51:2829–2842. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017. View Article : Google Scholar :
|
|
51
|
Novellasdemunt L, Antas P and Li VS:
Targeting Wnt signaling in colorectal cancer. A review in the
theme: Cell signaling: proteins, pathways and mechanisms. Am J
Physiol Cell Physiol. 309:C511–C521. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Miranda-Carboni GA, Krum SA, Yee K, Nava
M, Deng QE, Pervin S, Collado-Hidalgo A, Galic Z, Zack JA, Nakayama
K, et al: A functional link between Wnt signaling and
SKP2-independent p27 turnover in mammary tumors. Genes Dev.
22:3121–3134. 2008. View Article : Google Scholar : PubMed/NCBI
|