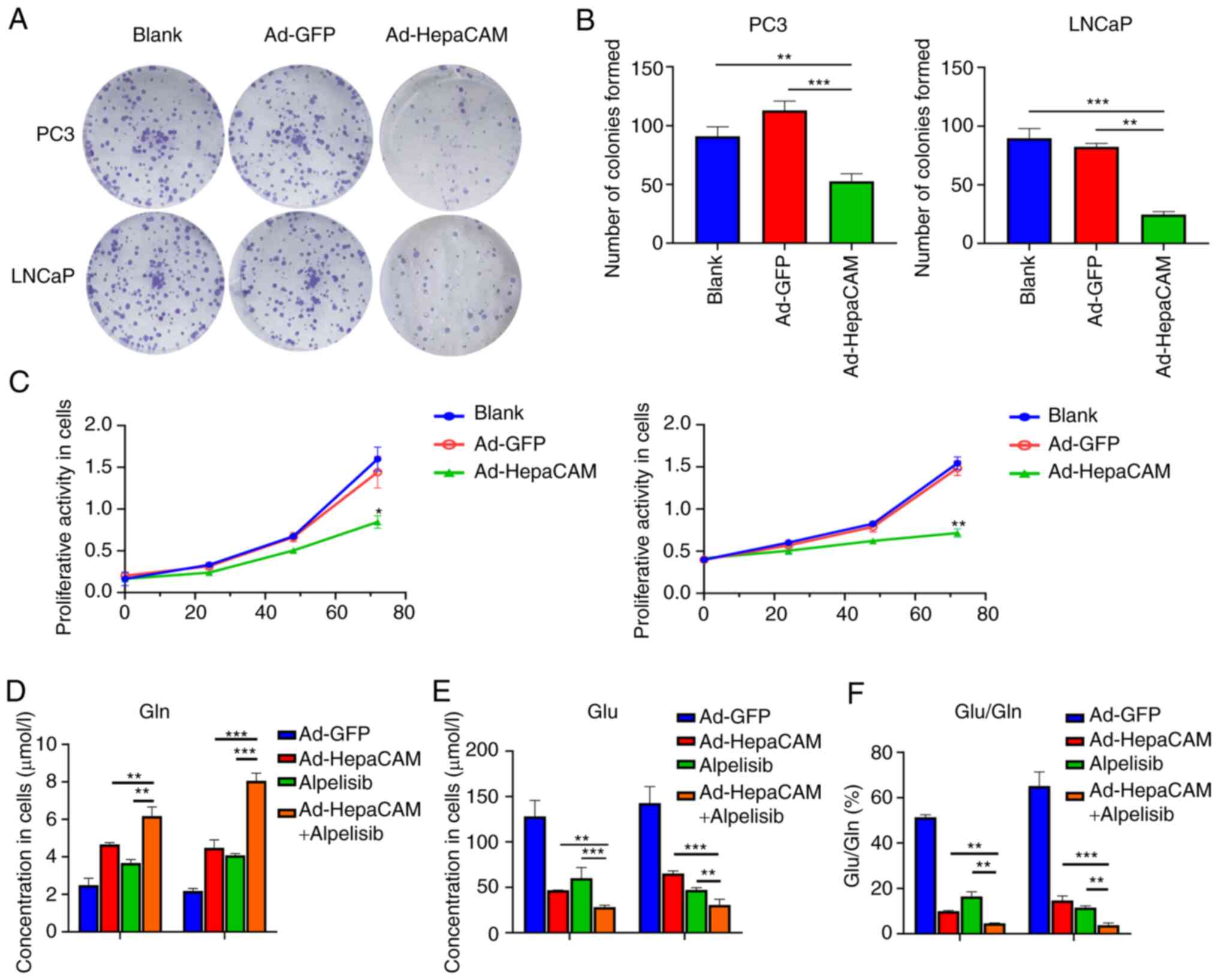
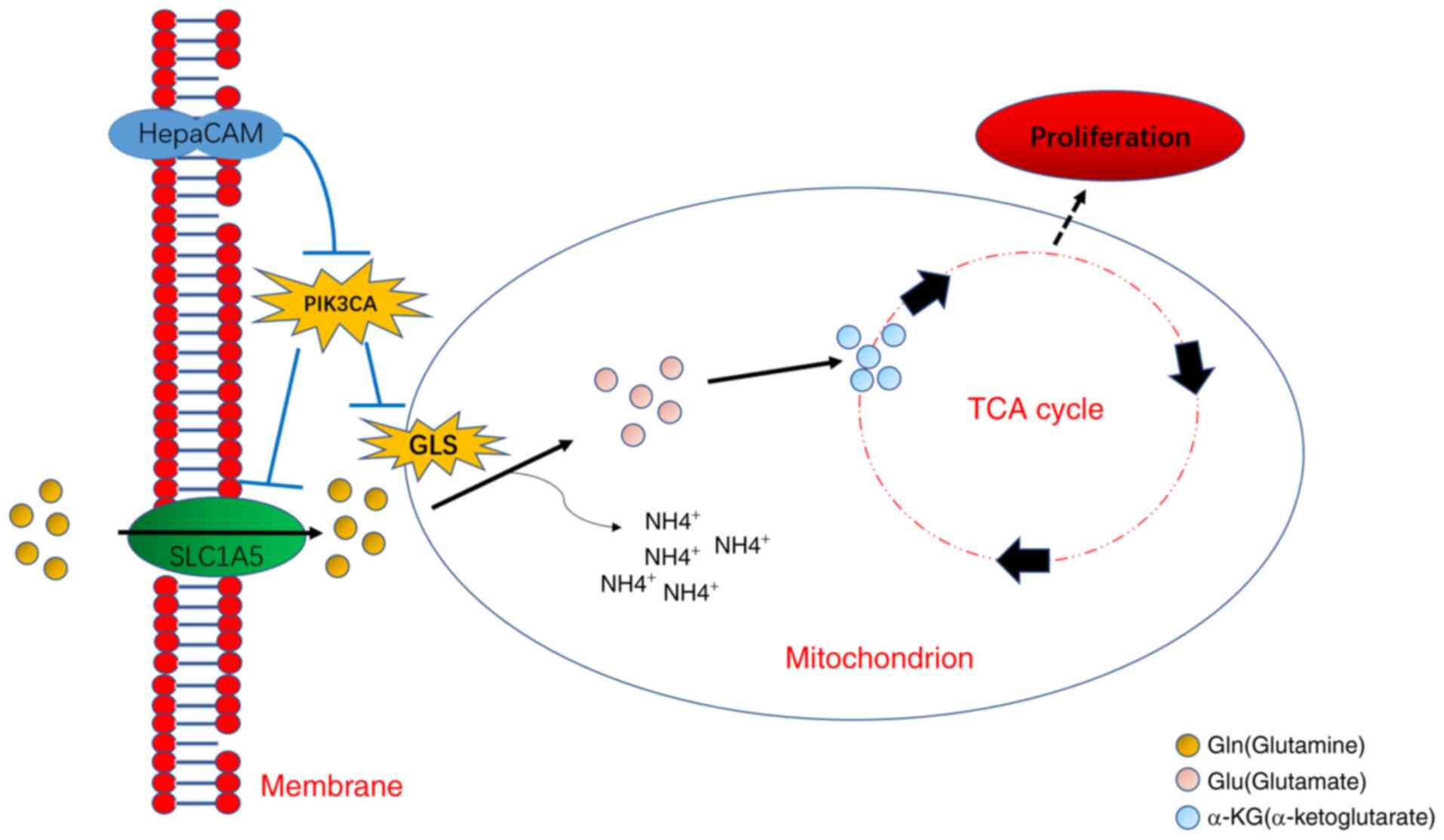
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fu ZT, Guo XL, Zhang SW, Zheng RS, Zeng
HM, Chen R, Wang SM, Sun KX, Wei WW and He J: Statistical analysis
of incidence and mortality of prostate cancer in China, 2015.
Zhonghua Zhong Liu Za Zhi. 42:718–722. 2020.In Chinese. PubMed/NCBI
|
|
3
|
Li Z and Zhang H: Reprogramming of
glucose, fatty acid and amino acid metabolism for cancer
progression. Cell Mol Life Sci. 73:377–392. 2016. View Article : Google Scholar
|
|
4
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Altman B, Stine Z and Dang C: From Krebs
to clinic: Glutamine metabolism to cancer therapy. Nat Rev Cancer.
16:619–634. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang J, Pavlova NN and Thompson CB:
Cancer cell metabolism: The essential role of the nonessential
amino acid, glutamine. EMBO J. 36:1302–1315. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mates JM, Segura JA, Martin-Rufian M,
Campos-Sandoval JA, Alonso FJ and Marquez J: Glutaminase isoenzymes
as key regulators in metabolic and oxidative stress against cancer.
Curr Mol Med. 13:514–534. 2013. View Article : Google Scholar
|
|
8
|
Hassanein M, Hoeksema MD, Shiota M, Qian
J, Harris BK, Chen H, Clark JE, Alborn WE, Eisenberg R and Massion
PP: SLC1A5 mediates glutamine transport required for lung cancer
cell growth and survival. Clin Cancer Res. 19:560–570. 2013.
View Article : Google Scholar :
|
|
9
|
Strmiska V, Michalek P, Eckschlager T,
Stiborova M, Adam V, Krizkova S and Heger Z: Prostate
cancer-specific hallmarks of amino acids metabolism: Towards a
paradigm of precision medicine. Biochim Biophys Acta Rev Cancer.
1871:248–258. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martinez-Outschoorn U, Lisanti M and
Sotgia F: Catabolic cancer-associated fibroblasts transfer energy
and biomass to anabolic cancer cells, fueling tumor growth. Semin
Cancer Biol. 25:47–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang C and McConathy J: Radiolabeled
amino acids for oncologic imaging. J Nucl Med. 54:1007–1010. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Twum-Ampofo J, Fu D, Passaniti A, Hussain
A and Siddiqui M: Metabolic targets for potential prostate cancer
therapeutics. Curr Opin Oncol. 28:241–247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chung Moh M, Hoon Lee L and Shen S:
Cloning and characterization of hepaCAM, a novel Ig-like cell
adhesion molecule suppressed in human hepatocellular carcinoma. J
Hepatol. 42:833–841. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moh MC, Zhang C, Luo C, Lee LH and Shen S:
Structural and functional analyses of a novel ig-like cell adhesion
molecule, hepaCAM, in the human breast carcinoma MCF7 cells. J Biol
Chem. 280:27366–27374. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song X, Wang Y, Du H, Fan Y, Yang X, Wang
X, Wu X and Luo C: Overexpression of HepaCAM inhibits cell
viability and motility through suppressing nucleus translocation of
androgen receptor and ERK signaling in prostate cancer. Prostate.
74:1023–1033. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang M, Zhao Y, Liu N, Chen E, Quan Z, Wu
X and Luo C: Overexpression of HepaCAM inhibits bladder cancer cell
proliferation and viability through the AKT/FoxO pathway. J Cancer
Res Clin Oncol. 143:793–805. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Chen E, Yang X, Wang Y, Quan Z, Wu
X and Luo C: 5-azacytidine inhibits the proliferation of bladder
cancer cells via reversal of the aberrant hypermethylation of the
hepaCAM gene. Oncol Rep. 35:1375–1384. 2016. View Article : Google Scholar
|
|
18
|
Quan Z, He Y, Luo C, Xia Y, Zhao Y, Liu N
and Wu X: Interleukin 6 induces cell proliferation of clear cell
renal cell carcinoma by suppressing hepaCAM via the STAT3-dependent
up-regulation of DNMT1 or DNMT3b. Cell Signal. 32:48–58. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan Y, Ou L, Fan J, Li L, Wu X, Luo C, Gao
Y and Niu L: HepaCAM regulates warburg effect of renal cell
carcinoma via HIF-1α/NF-κB signaling pathway. Urology. 127:61–67.
2019. View Article : Google Scholar
|
|
20
|
Deng Q, Luo L, Quan Z, Liu N, Du Z, Sun W,
Luo C and Wu X: HepaCAM inhibits cell proliferation and invasion in
prostate cancer by suppressing nuclear translocation of the
androgen receptor via its cytoplasmic domain. Mol Med Rep.
19:2115–2124. 2019.PubMed/NCBI
|
|
21
|
Yuan TL and Cantley LC: PI3K pathway
alterations in cancer: Variations on a theme. Oncogene.
27:5497–5510. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu W, Yuan Y, Sun C, Balasubramanian B,
Zhao Z and An L: Effects of dietary betaine on growth performance,
digestive function, carcass traits, and meat quality in indigenous
yellow-feathered broilers under long-term heat stress. Animals
(Basel). 9:5062019. View Article : Google Scholar
|
|
23
|
Liu WC, Guo Y, Zhao ZH, Jha R and
Balasubramanian B: Algae-derived polysaccharides promote growth
performance by improving antioxidant capacity and intestinal
barrier function in broiler chickens. Front Vet Sci. 7:6013362020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo Y, Zhao ZH, Pan ZY, An LL,
Balasubramanian B and Liu WC: New insights into the role of dietary
marine-derived polysaccharides on productive performance, egg
quality, antioxidant capacity, and jejunal morphology in late-phase
laying hens. Poult Sci. 99:2100–2107. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hiles ID, Otsu M, Volinia S, Fry MJ, Gout
I, Dhand R, Panayotou G, Ruiz-Larrea F, Thompson A, Totty NF, et
al: Phosphatidylinositol 3-kinase: Structure and expression of the
110 kd catalytic subunit. Cell. 70:419–429. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Holst F, Werner HM, Mjøs S, Hoivik EA,
Kusonmano K, Wik E, Berg A, Birkeland E, Gibson WJ, Halle MK, et
al: PIK3CA amplification associates with aggressive phenotype but
not markers of AKT-MTOR signaling in endometrial carcinoma. Clin
Cancer Res. 25:334–345. 2019. View Article : Google Scholar :
|
|
27
|
Han C, Yang L, Choi HH, Baddour J, Achreja
A, Liu Y, Li Y, Li J, Wan G, Huang C, et al: Amplification of USP13
drives ovarian cancer metabolism. Nat Commun. 7:135252016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hao Y, Samuels Y, Li Q, Krokowski D, Guan
BJ, Wang C, Jin Z, Dong B, Cao B, Feng X, et al: Oncogenic PIK3CA
mutations reprogram glutamine metabolism in colorectal cancer. Nat
Commun. 7:119712016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boku S, Watanabe M, Sukeno M, Yaoi T,
Hirota K, Iizuka-Ohashi M, Itoh K and Sakai T: Deactivation of
glutaminolysis sensitizes PIK3CA-mutated colorectal cancer cells to
aspirin-induced growth inhibition. Cancers (Basel). 12:10972020.
View Article : Google Scholar
|
|
30
|
Lau CE, Tredwell GD, Ellis JK, Lam EW and
Keun H: Metabolomic characterisation of the effects of oncogenic
PIK3CA transformation in a breast epithelial cell line. Sci Rep.
7:460792017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Du Z, Li L, Sun W, Wang X, Zhang Y, Chen
Z, Yuan M, Quan Z, Liu N, Hao Y, et al: HepaCAM inhibits the
malignant behavior of castration-resistant prostate cancer cells by
downregulating Notch signaling and PF-3084014 (a γ-secretase
inhibitor) partly reverses the resistance of refractory prostate
cancer to docetaxel and enzalutamide in vitro. Int J Oncol.
53:99–112. 2018.PubMed/NCBI
|
|
32
|
Zhao Y, Balasubramanian B, Guo Y, Qiu SJ,
Jha R and Liu WC: Dietary enteromorpha polysaccharides
supplementation improves breast muscle yield and is associated with
modification of mrna transcriptome in broiler chickens. Front Vet
Sci. 8:6639882021. View Article : Google Scholar :
|
|
33
|
Guo Y, Balasubramanian B, Zhao ZH and Liu
WC: Marine algal polysaccharides alleviate aflatoxin B1-induced
bursa of Fabricius injury by regulating redox and apoptotic
signaling pathway in broilers. Poult Sci. 100:844–857. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu WC, Ou BH, Liang ZL, Zhang R and Zhao
ZH: Algae-derived polysaccharides supplementation ameliorates heat
stress-induced impairment of bursa of Fabricius via modulating
NF-κB signaling pathway in broilers. Poult Sci. 100:1011392021.
View Article : Google Scholar
|
|
35
|
Liu WC, Zhu YR, Zhao ZH, Jiang P and Yin
FQ: Effects of dietary supplementation of algae-derived
polysaccharides on morphology, tight junctions, antioxidant
capacity and immune response of duodenum in broilers under heat
stress. Animals (Basel). 11:22792021. View Article : Google Scholar
|
|
36
|
Yu C, Huang S, Wang M, Zhang J, Liu H,
Yuan Z, Wang X, He X, Wang J and Zou L: A novel tandem mass
spectrometry method for first-line screening of mainly
beta-thalassemia from dried blood spots. J Proteomics. 154:78–84.
2017. View Article : Google Scholar
|
|
37
|
Geng HT, Cao RJ, Cheng L and Liu CY:
Overexpression of hepatocyte cell adhesion molecule (hepaCAM)
inhibits the proliferation, migration, and invasion in colorectal
cancer cells. Oncol Res. 25:1039–1046. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang QL, Luo CL, Wu XH, Wang CY, Xu X,
Zhang YY, Liu Q and Shen SL: HepaCAM induces G1 phase arrest and
promotes c-Myc degradation in human renal cell carcinoma. J Cell
Biochem. 112:2910–2919. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wise DR, DeBerardinis RJ, Mancuso A, Sayed
N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon
SB and Thompson CB: Myc regulates a transcriptional program that
stimulates mitochondrial glutaminolysis and leads to glutamine
addiction. Proc Natl Acad Sci USA. 105:18782–18787. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013. View Article : Google Scholar
|
|
41
|
Lawrence MS, Stojanov P, Mermel CH,
Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander
ES and Getz G: Discovery and saturation analysis of cancer genes
across 21 tumour types. Nature. 505:495–501. 2014. View Article : Google Scholar
|
|
42
|
Torre LA, Sauer AM, Chen MS Jr,
Kagawa-Singer M, Jemal A and Siegel R: Cancer statistics for Asian
Americans, Native Hawaiians, and Pacific Islanders, 2016:
Converging incidence in males and females. CA Cancer J Clin.
66:182–202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mayers JR, Torrence ME, Danai LV,
Papagiannakopoulos T, Davidson SM, Bauer MR, Lau AN, Ji BW, Dixit
PD and Hosios AM: Tissue of origin dictates branched-chain amino
acid metabolism in mutant Kras-driven cancers. Science.
353:1161–1165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yuneva MO, Fan TW, Allen TD, Higashi RM,
Ferraris DV, Tsukamoto T, Matés JM, Alonso FJ, Wang C, Seo Y, et
al: The metabolic profile of tumors depends on both the responsible
genetic lesion and tissue type. 15:157–170. 2012.PubMed/NCBI
|
|
45
|
Li T, Liu N, Gao Y, Quan Z, Hao Y, Yu C,
Li L, Yuan M, Niu L, Luo C and Wu X: Long noncoding RNA HOTAIR
regulates the invasion and metastasis of prostate cancer by
targeting hepaCAM. Br J Cancer. 124:247–258. 2020. View Article : Google Scholar
|
|
46
|
McGuirk S, Gravel S, Deblois G, Papadopoli
DJ, Faubert B, Wegner A, Hiller K, Avizonis D, Akavia UD, Jones RG,
et al: PGC-1α supports glutamine metabolism in breast cancer.
Cancer Metab. 1:222013. View Article : Google Scholar
|
|
47
|
DeBerardinis RJ and Cheng T: Q's next: The
diverse functions of glutamine in metabolism, cell biology and
cancer. Oncogene. 29:313–324. 2010. View Article : Google Scholar :
|
|
48
|
Biancur DE, Paulo JA, Małachowska B,
Quiles Del Rey M, Sousa CM, Wang X, Sohn AS, Chu GC, Gygi SP,
Harper JW, et al: Compensatory metabolic networks in pancreatic
cancers upon perturbation of glutamine metabolism. Nat Commun.
8:159652017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lowman XH, Hanse EA, Yang Y, Ishak Gabra
MB, Tran TQ, Li H and Kong M: p53 promotes cancer cell adaptation
to glutamine deprivation by upregulating Slc7a3 to increase
arginine uptake. Cell Rep. 26:3051–3060.e3054. 2019. View Article : Google Scholar :
|
|
50
|
Jiang W, He T, Liu S, Zheng Y, Xiang L,
Pei X, Wang Z and Yang H: The PIK3CA E542K and E545K mutations
promote glycolysis and proliferation via induction of the
β-catenin/SIRT3 signaling pathway in cervical cancer. J Hematol
Oncol. 11:1392018. View Article : Google Scholar
|
|
51
|
André F, Ciruelos E, Rubovszky G, Campone
M, Loibl S, Rugo HS, Iwata H, Conte P, Mayer IA, Kaufman B, et al:
Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced
breast cancer. N Engl J Med. 380:1929–1940. 2019. View Article : Google Scholar
|
|
52
|
Juric D, Janku F, Rodón J, Burris HA,
Mayer IA, Schuler M, Seggewiss-Bernhardt R, Gil-Martin M, Middleton
MR, Baselga J, et al: Alpelisib plus fulvestrant in PIK3CA-altered
and PIK3CA-wild-type estrogen receptor-positive advanced breast
cancer: A phase 1b clinical trial. JAMA Oncol. 5:e1844752019.
View Article : Google Scholar
|
|
53
|
Konstantinopoulos PA, Barry WT, Birrer M,
Westin SN, Cadoo KA, Shapiro GI, Mayer EL, O'Cearbhaill RE, Coleman
RL, Kochupurakkal B, et al: Olaparib and α-specific PI3K inhibitor
alpelisib for patients with epithelial ovarian cancer: A
dose-escalation and dose-expansion phase 1b trial. Lancet Oncol.
20:570–580. 2019. View Article : Google Scholar : PubMed/NCBI
|