Role of N6‑methyladenosine in the pathogenesis, diagnosis and treatment of pancreatic cancer (Review)
- Authors:
- Tong Ye
- Jiaxin Wang
- Haiying Zhao
- Guiping Zhao
- Peng Li
-
Affiliations: Department of Gastroenterology, Beijing Friendship Hospital, Capital Medical University, Beijing 100050, P.R. China - Published online on: November 10, 2022 https://doi.org/10.3892/ijo.2022.5452
- Article Number: 4
-
Copyright: © Ye et al. This is an open access article distributed under the terms of Creative Commons Attribution License.
This article is mentioned in:
Abstract
 |
 |
|
Mizrahi JD, Surana R, Valle JW and Shroff RT: Pancreatic cancer. Lancet. 395:2008–2020. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Martin-Perez E, Domínguez-Muñoz JE, Botella-Romero F, Cerezo L, Matute Teresa F, Serrano T and Vera R: Multidisciplinary consensus statement on the clinical management of patients with pancreatic cancer. Clin Transl Oncol. 22:1963–1975. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
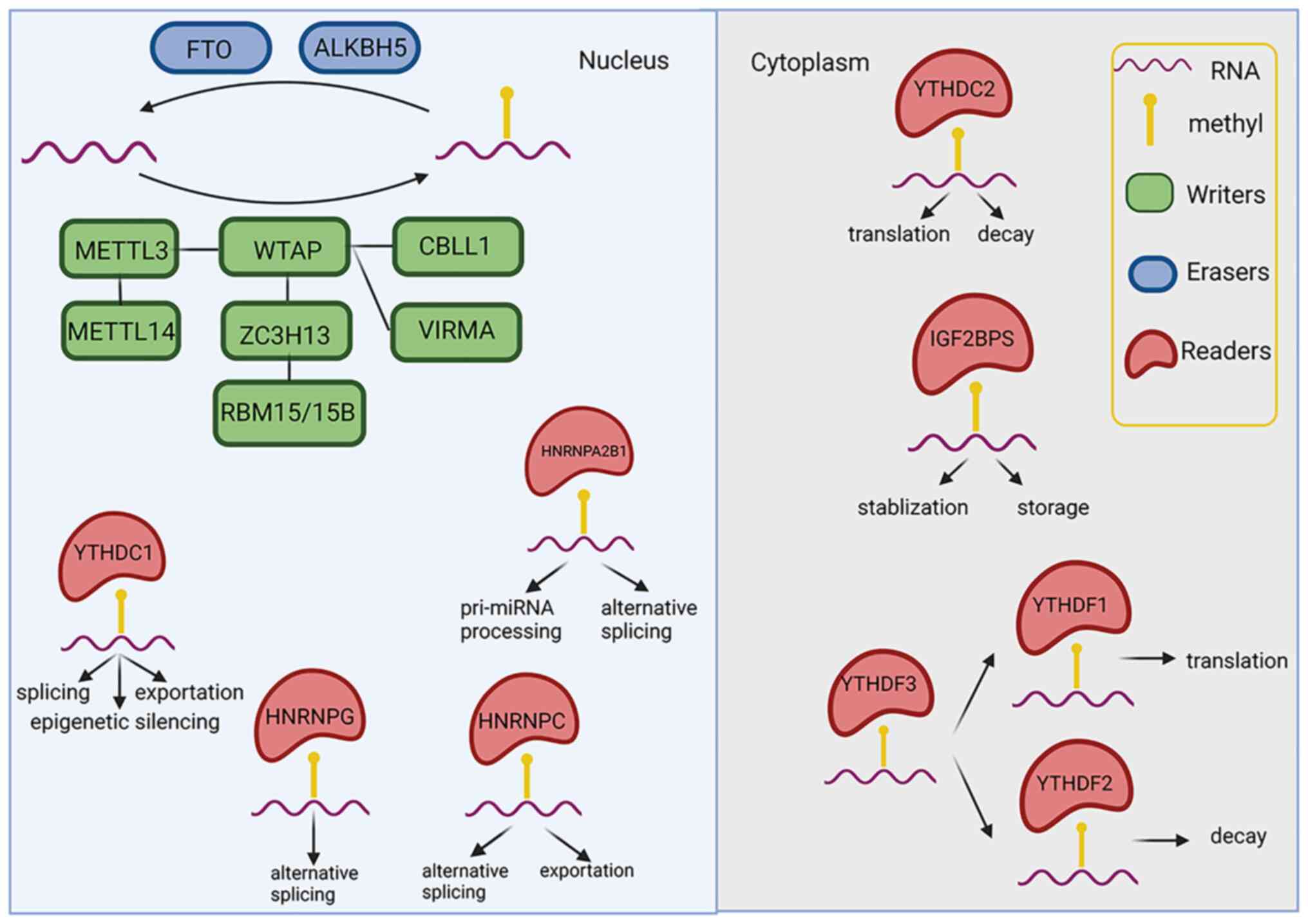
Zaccara S, Ries RJ and Jaffrey SR: Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 20:608–624. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Fu Y, Dominissini D, Rechavi G and He C: Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 15:293–306. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
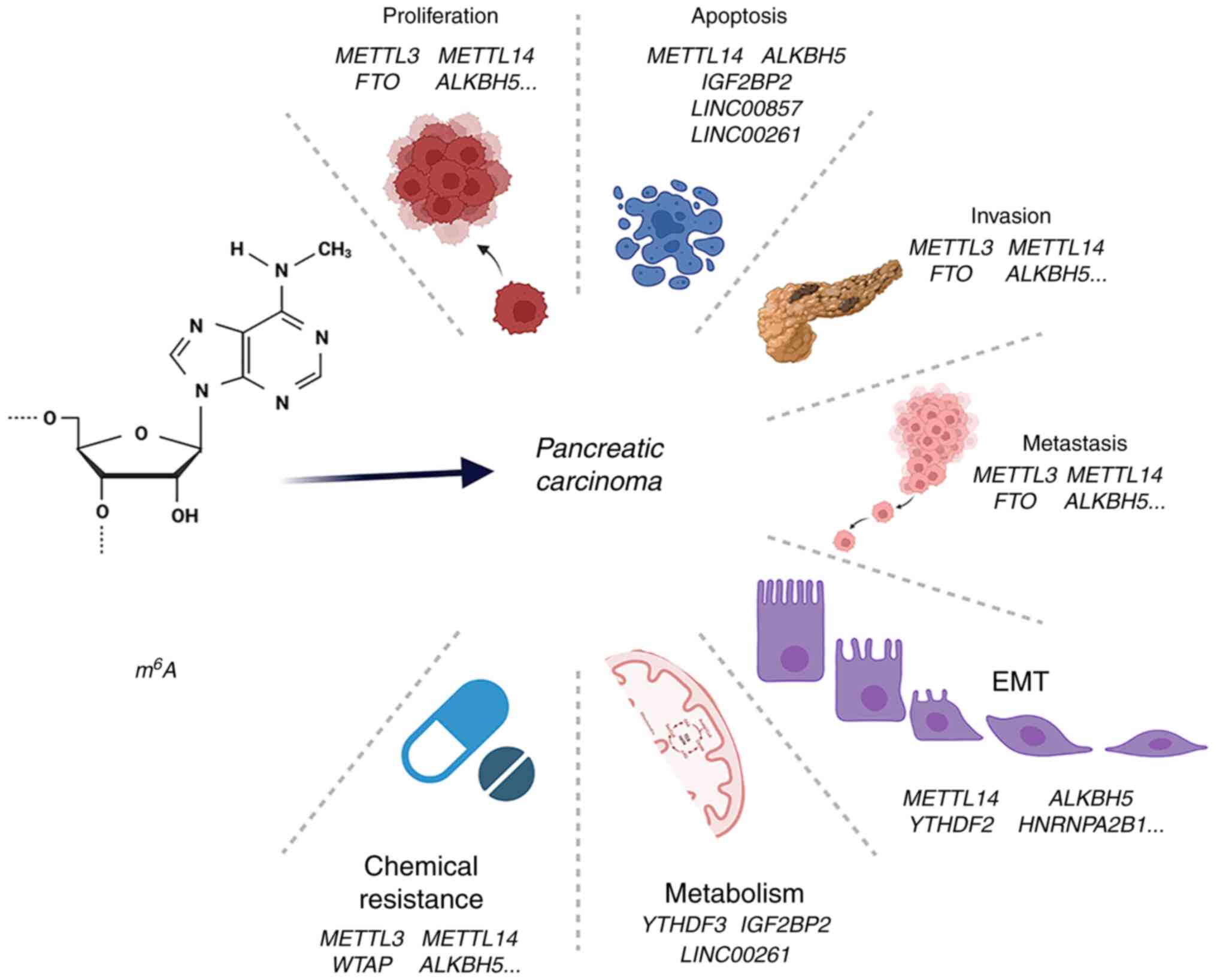
Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D, Wu Q, Yuan B, Lu Q and Yang H: Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 19:1042020. View Article : Google Scholar : PubMed/NCBI | |
|
He L, Li H, Wu A, Peng Y, Shu G and Yin G: Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 18:1762019. View Article : Google Scholar : PubMed/NCBI | |
|
Frye M, Harada BT, Behm M and He C: RNA modifications modulate gene expression during development. Science. 361:1346–1349. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Sun T, Wu R and Ming L: The role of m6A RNA methylation in cancer. Biomed Pharmacother. 112:1086132019. View Article : Google Scholar : PubMed/NCBI | |
|
Chen XY, Zhang J and Zhu JS: The role of m6A RNA methylation in human cancer. Mol Cancer. 18:1032019. View Article : Google Scholar | |
|
Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S and Liu T: The critical role of RNA m6A methylation in cancer. Cancer Res. 79:1285–1292. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Wei CM, Gershowitz A and Moss B: Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell. 4:379–386. 1975. View Article : Google Scholar : PubMed/NCBI | |
|
Rottman F, Shatkin AJ and Perry RP: Sequences containing methylated nucleotides at the 5′ termini of messenger RNAs: Possible implications for processing. Cell. 3:197–199. 1974. View Article : Google Scholar : PubMed/NCBI | |
|
Desrosiers R, Friderici K and Rottman F: Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975. 1974. View Article : Google Scholar : PubMed/NCBI | |
|
Adams JM and Cory S: Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature. 255:28–33. 1975. View Article : Google Scholar : PubMed/NCBI | |
|
Schäfer KP: RNA synthesis and processing reactions in a subcellular system from mouse L cells. Hoppe Seylers Z Physiol Chem. 363:33–43. 1982. View Article : Google Scholar : PubMed/NCBI | |
|
Bokar JA, Shambaugh ME, Polayes D, Matera AG and Rottman FM: Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 3:1233–1247. 1997.PubMed/NCBI | |
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI | |
|
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Schibler U, Kelley DE and Perry RP: Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 115:695–714. 1977. View Article : Google Scholar : PubMed/NCBI | |
|
Rottman FM, Desrosiers RC and Friderici K: Nucleotide methylation patterns in eukaryotic mRNA. Prog Nucleic Acid Res Mol Biol. 19:21–38. 1976. View Article : Google Scholar : PubMed/NCBI | |
|
Wei CM and Moss B: Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 16:1672–1676. 1977. View Article : Google Scholar : PubMed/NCBI | |
|
Krug RM: Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methyl-guanosine in cap structures. J Virol. 20:45–53. 1976. View Article : Google Scholar : PubMed/NCBI | |
|
Beemon K and Keith J: Localization of N6-methyladenosine in the rous sarcoma viru genome. J Mol Bid. 113:165–179. 1977. View Article : Google Scholar | |
|
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al: Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 485:201–206. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N, Cacchiarelli D, et al: Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al: Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 347:1002–1006. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, Chen K, Lu Z, Chen H, Shi YG, et al: N6-methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 15:88–94. 2019. View Article : Google Scholar | |
|
van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, Bohnsack KE, Bohnsack MT, Jaffrey SR, Graille M and Lafontaine DLJ: The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47:7719–7733. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, Hase H, Harada K, Hirata K and Tsujikawa K: AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 7:422712017. View Article : Google Scholar : PubMed/NCBI | |
|
Luo H, Zhu G, Xu J, Lai Q, Yan B, Guo Y, Fung TK, Zeisig BB, Cui Y, Zha J, et al: HOTTIP lncRNA promotes hematopoietic stem cell self-renewal leading to AML-like disease in mice. Cancer Cell. 36:645–659.e8. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, Huang XD, Liu RQ, Chen F, He XJ, et al: Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 79:4612–4626. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Cesarini V, Silvestris DA, Tassinari V, Tomaselli S, Alon S, Eisenberg E, Locatelli F and Gallo A: ADAR2/miR-589-3p axis controls glioblastoma cell migration/invasion. Nucleic Acids Res. 46:2045–2059. 2018. View Article : Google Scholar : | |
|
Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, Yuan WB, Lu JC, Zhou ZJ, Lu Q, et al: METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 18:1102019. View Article : Google Scholar : PubMed/NCBI | |
|
Li J, Huang C, Zou Y, Ye J, Yu J and Gui Y: CircTLK1 promotes the proliferation and metastasis of renal cell carcinoma by sponging miR-136-5p. Mol Cancer. 19:1032020. View Article : Google Scholar : PubMed/NCBI | |
|
Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP and Conrad NK: The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 169:824–835.e14. 2017. View Article : Google Scholar | |
|
Roundtree IA, Evans ME, Pan T and He C: Dynamic RNA modifications in gene expression regulation. Cell. 169:1187–1200. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE and Jaffrey SR: Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 12:767–772. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T and Hamakubo T: Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 288:33292–33302. 2013. View Article : Google Scholar : PubMed/NCBI | |
|
Narayan P and Rottman FM: An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science. 242:1159–1162. 1988. View Article : Google Scholar : PubMed/NCBI | |
|
Schumann U, Shafik A and Preiss T: METTL3 gains R/W access to the epitranscriptome. Mol Cell. 62:323–324. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Śledź P and Jinek M: Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 5:e184342016. View Article : Google Scholar : PubMed/NCBI | |
|
Wang P, Doxtader KA and Nam Y: Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 63:306–317. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 10:93–95. 2014. View Article : Google Scholar : | |
|
Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, et al: Corrigendum: Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. 542:2602017. View Article : Google Scholar | |
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24:177–189. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Schöller E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, Feederle R, Bruckmann A and Meister G: Interactions, localization, and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex. RNA. 24:499–512. 2018. View Article : Google Scholar | |
|
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, Cheng T, Gao M, Shu X, Ma H, et al: VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:102018. View Article : Google Scholar | |
|
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M and Jaffrey SR: m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 537:369–373. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villaseñor R, Hess D, et al: Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 32:415–429. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Wen J, Lv R, Ma H, Shen H, He C, Wang J, Jiao F, Liu H, Yang P, Tan L, et al: Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 69:1028–1038.e6. 2018. View Article : Google Scholar | |
|
Fujita Y, Krause G, Scheffner M, Zechner D, Leddy HE, Behrens J, Sommer T and Birchmeier W: Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat Cell Biol. 4:222–231. 2002. View Article : Google Scholar : PubMed/NCBI | |
|
Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D, Heidelberger JB, Richter FM, Nallasivan MP, Morin V, et al: Hakai is required for stabilization of core components of the m6A mRNA methylation machinery. Nat Commun. 12:37782021. View Article : Google Scholar | |
|
Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al: Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 71:973–985.e5. 2018. View Article : Google Scholar | |
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar : | |
|
Aik W, Scotti JS, Choi H, Gong L, Demetriades M, Schofield CJ and McDonough MA: Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 42:4741–4754. 2014. View Article : Google Scholar : PubMed/NCBI | |
|
Wang T, Kong S, Tao M and Ju S: The potential role of RNA N6-methyladenosine in cancer progression. Mol Cancer. 19:882020. View Article : Google Scholar : PubMed/NCBI | |
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, et al: The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 48:3816–3831. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang C, Chen L, Peng D, Jiang A, He Y, Zeng Y, Xie C, Zhou H, Luo X, Liu H, et al: METTL3 and N6-methyladenosine promote homologous recombination-mediated repair of DSBs by modulating DNA-RNA hybrid accumulation. Mol Cell. 79:425–442.e7. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Liu SY, Feng Y, Wu JJ, Zou ML, Sun ZL, Li X and Yuan FL: m6A facilitates YTHDF-independent phase separation. J Cell Mol Med. 24:2070–2072. 2020. View Article : Google Scholar | |
|
Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH and Jaffrey SR: m6A enhances the phase separation potential of mRNA. Nature. 571:424–428. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC and Li P: Multivalent m6A motifs promote phase separation of YTHDF proteins. Cell Res. 29:767–769. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Wang J, Wang L, Diao J, Shi YG, Shi Y, Ma H and Shen H: Binding to m6A RNA promotes YTHDF2-mediated phase separation. Protein Cell. 11:304–307. 2020. View Article : Google Scholar | |
|
Sheth U and Parker R: Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 300:805–808. 2003. View Article : Google Scholar : PubMed/NCBI | |
|
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J and Wu L: YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 7:126262016. View Article : Google Scholar : PubMed/NCBI | |
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C and He C: YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27:315–328. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al: Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 61:507–519. 2016. View Article : Google Scholar : PubMed/NCBI | |
|
Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, Schultz RM and Wang PJ: Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. 14. pp. e10074122018, View Article : Google Scholar | |
|
Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al: YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 6:e313112017. View Article : Google Scholar | |
|
Lesbirel S, Viphakone N, Parker M, Parker J, Heath C, Sudbery I and Wilson SA: The m6A-methylase complex recruits TREX and regulates mRNA export. Sci Rep. 8:138272018. View Article : Google Scholar | |
|
Brockdorff N, Bowness JS and Wei G: Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 34:733–744. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Liang D, Lin WJ, Ren M, Qiu J, Yang C, Wang X, Li N, Zeng T, Sun K, You L, et al: m6A reader YTHDC1 modulates autophagy by targeting SQSTM1 in diabetic skin. Autophagy. 18:1318–1337. 2022. View Article : Google Scholar | |
|
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al: Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27:1115–1127. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM and Ule J: iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 17:909–915. 2010. View Article : Google Scholar : PubMed/NCBI | |
|
McCloskey A, Taniguchi I, Shinmyozu K and Ohno M: hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science. 335:1643–1646. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Liu N, Dai Q, Zheng G, He C, Parisien M and Pan T: N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 518:560–564. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Ż, Pan JN, He C, Parisien M and Pan T: Regulation of co-transcriptional pre-mRNA splicing by m6A through the low-complexity protein hnRNPG. Mol Cell. 76:70–81.e9. 2019. View Article : Google Scholar | |
|
Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L and Pan T: N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45:6051–6063. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S and Tavazoie SF: HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 162:1299–1308. 2015. View Article : Google Scholar : PubMed/NCBI | |
|
Fabbiano F, Corsi J, Gurrieri E, Trevisan C, Notarangelo M and D'Agostino VG: RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J Extracell Vesicles. 10:e120432020. View Article : Google Scholar | |
|
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al: Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 20:285–295. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Wu Z, Shi Y, Lu M, Song M, Yu Z, Wang J, Wang S, Ren J, Yang YG, Liu GH, et al: METTL3 counteracts premature aging via m6A-dependent stabilization of MIS12 mRNA. Nucleic Acids Res. 48:11083–11096. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, Wang SY, Baltissen MPA, Jansen PWTC, Rossa M, et al: N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 24:870–878. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, Li S, Tan L, Mai D, Li G, et al: Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 10:18582019. View Article : Google Scholar : | |
|
Hua YQ, Zhang K, Sheng J, Ning ZY, Li Y, Shi WD and Liu LM: NUCB1 suppresses growth and shows additive effects with gemcitabine in pancreatic ductal adenocarcinoma via the unfolded protein response. Front Cell Dev Biol. 9:6418362021. View Article : Google Scholar : PubMed/NCBI | |
|
Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, Li X, Xu S, Miao J, Guo J, et al: Upregulation of METTL14 mediates the elevation of PERP mRNA N6 adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 19:1302020. View Article : Google Scholar | |
|
Xie F, Liu S and Wang H: M6A methyltransferase METTL16 suppresses pancreatic cancer proliferation through p21 pathways. Pancreas. 49:14372020. | |
|
Huang H, Li H, Pan R, Wang S, Khan AA, Zhao Y, Zhu H and Liu X: Ribosome 18S m6A methyltransferase METTL5 promotes pancreatic cancer progression by modulating c-Myc translation. Int J Oncol. 60:92022. View Article : Google Scholar | |
|
Zeng J, Zhang H, Tan Y, Wang Z, Li Y and Yang X: m6A demethylase FTO suppresses pancreatic cancer tumorigenesis by demethylating PJA2 and inhibiting Wnt signaling. Mol Ther Nucleic Acids. 25:277–292. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Tang X, Liu S, Chen D, Zhao Z and Zhou J: The role of the fat mass and obesity-associated protein in the proliferation of pancreatic cancer cells. Oncol Lett. 17:2473–2478. 2019.PubMed/NCBI | |
|
Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, Feng Y, Pan Q and Wan R: RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol Cancer. 19:912020. View Article : Google Scholar : PubMed/NCBI | |
|
He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, Liu D, Tian L, Yin J, Jiang K and Miao Y: ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. 48:838–846. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
He Y, Yue H, Cheng Y, Ding Z, Xu Z, Lv C, Wang Z, Wang J, Yin C, Hao H and Chen C: ALKBH5-mediated m6A demethylation of KCNK15-AS1 inhibits pancreatic cancer progression via regulating KCNK15 and PTEN/AKT signaling. Cell Death Dis. 12:11212021. View Article : Google Scholar | |
|
Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, He S and Shimamoto F: m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 19:32020. View Article : Google Scholar | |
|
Huang R, Yang L, Zhang Z, Liu X, Fei Y, Tong WM, Niu Y and Liang Z: RNA m6A demethylase ALKBH5 protects against pancreatic ductal adenocarcinoma via targeting regulators of iron metabolism. Front Cell Dev Biol. 9:7242822021. View Article : Google Scholar | |
|
Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, Mo YY and Yang L: IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 27:1782–1794. 2020. View Article : Google Scholar : | |
|
Meng X, Deng Y, He S, Niu L and Zhu H: m6A-mediated upregulation of LINC00857 promotes pancreatic cancer tumorigenesis by regulating the miR-150-5p/E2F3 axis. Front Oncol. 11:6299472021. View Article : Google Scholar | |
|
Zhai S, Xu Z, Xie J, Zhang J, Wang X, Peng C, Li H, Chen H, Shen B and Deng X: Epigenetic silencing of LncRNA LINC00261 promotes c-myc-mediated aerobic glycolysis by regulating miR-222-3p/HIPK2/ERK axis and sequestering IGF2BP1. Oncogene. 40:277–291. 2021. View Article : Google Scholar : | |
|
Geng Y, Guan R, Hong W, Huang B, Liu P, Guo X, Hu S, Yu M and Hou B: Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann Transl Med. 8:3872020. View Article : Google Scholar : PubMed/NCBI | |
|
Ying P, Li Y, Yang N, Wang X, Wang H, He H, Li B, Peng X, Zou D, Zhu Y, et al: Identification of genetic variants in m6A modification genes associated with pancreatic cancer risk in the Chinese population. Arch Toxicol. 95:1117–1128. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Tian J, Zhu Y, Rao M, Cai Y, Lu Z, Zou D, Peng X, Ying P, Zhang M, Niu S, et al: N6-methyladenosine mRNA methylation of PIK3CB regulates AKT signalling to promote PTEN-deficient pancreatic cancer progression. Gut. 69:2180–2192. 2020. View Article : Google Scholar : PubMed/NCBI | |
|
Brabletz T, Kalluri R, Nieto MA and Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018. View Article : Google Scholar : PubMed/NCBI | |
|
Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang J, Lu Z, Wu P, Cai B, Miao Y and Jiang K: The RNA m6A methyltransferase METTL3 promotes pancreatic cancer cell proliferation and invasion. Pathol Res Pract. 215:1526662019. View Article : Google Scholar : PubMed/NCBI | |
|
Li BQ, Huang S, Shao QQ, Sun J, Zhou L, You L, Zhang TP, Liao Q, Guo JC and Zhao YP: WT1-associated protein is a novel prognostic factor in pancreatic ductal adenocarcinoma. Oncol Lett. 13:2531–2538. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, Lu Y, Zeng J, Du F, Gong A and Xu M: YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 16:2259–2271. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Dai S, Zhang J, Huang S, Lou B, Fang B, Ye T, Huang X, Chen B and Zhou M: HNRNPA2B1 regulates the epithelial-mesenchymal transition in pancreatic cancer cells through the ERK/snail signalling pathway. Cancer Cell Int. 17:122017. View Article : Google Scholar : PubMed/NCBI | |
|
Dahlem C, Barghash A, Puchas P, Haybaeck J and Kessler SM: The insulin-like growth factor 2 mRNA binding protein IMP2/IGF2BP2 is overexpressed and correlates with poor survival in pancreatic cancer. Int J Mol Sci. 20:32042019. View Article : Google Scholar : PubMed/NCBI | |
|
Chen S, Yang C, Wang ZW, Hu JF, Pan JJ, Liao CY, Zhang JQ, Chen JZ, Huang Y, Huang L, et al: CLK1/SRSF5 pathway induces aberrant exon skipping of METTL14 and Cyclin L2 and promotes growth and metastasis of pancreatic cancer. J Hematol Oncol. 14:602021. View Article : Google Scholar : PubMed/NCBI | |
|
Huang XT, Li JH, Zhu XX, Huang CS, Gao ZX, Xu QC, Zhao W and Yin XY: HNRNPC impedes m6A-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett. 518:196–206. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Hu Y, Tang J, Xu F, Chen J, Zeng Z, Han S, Wang F, Wang D, Huang M, Zhao Y, et al: A reciprocal feedback between N6-methyladenosine reader YTHDF3 and lncRNA DICER1-AS1 promotes glycolysis of pancreatic cancer through inhibiting maturation of miR-5586-5p. J Exp Clin Cancer Res. 41:692022. View Article : Google Scholar : PubMed/NCBI | |
|
Huang S, Wu Z, Cheng Y, Wei W and Hao L: Insulin-like growth factor 2 mRNA binding protein 2 promotes aerobic glycolysis and cell proliferation in pancreatic ductal adenocarcinoma via stabilizing GLUT1 mRNA. Acta Biochim Biophys Sin (Shanghai). 51:743–752. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T, Doki Y, Mori M, Ishii H and Ogawa K: The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 52:621–629. 2018.PubMed/NCBI | |
|
Zhang C, Ou S, Zhou Y, Liu P, Zhang P, Li Z, Xu R and Li Y: m6A methyltransferase METTL14-mediated upregulation of cytidine deaminase promoting gemcitabine resistance in pancreatic cancer. Front Oncol. 11:6963712021. View Article : Google Scholar | |
|
Kong F, Liu X, Zhou Y, Hou X, He J, Li Q, Miao X and Yang L: Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int J Biochem Cell Biol. 122:1057312020. View Article : Google Scholar : PubMed/NCBI | |
|
Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, Peng S, Chen K, Wang M, Gong S, et al: Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 134:17963–17971. 2012. View Article : Google Scholar : PubMed/NCBI | |
|
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, Wang Y, Li X, Xiong XF, Wei B, et al: RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 18:462019. View Article : Google Scholar : PubMed/NCBI | |
|
Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, Gan J, Jiang H, Jia GF, Luo C and Yang CG: Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43:373–384. 2015. View Article : Google Scholar : | |
|
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al: m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 18:2622–2634. 2017. View Article : Google Scholar : PubMed/NCBI | |
|
Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, Ni T, Zhang ZS, Zhang T, Li C, et al: Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 35:677–691.e10. 2019. View Article : Google Scholar : PubMed/NCBI | |
|
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al: R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 172:90–105.e23. 2018. View Article : Google Scholar | |
|
Xu P and Ge R: Roles and drug development of METTL3 (methyltransferase-like 3) in anti-tumor therapy. Eur J Med Chem. 230:1141182022. View Article : Google Scholar : PubMed/NCBI | |
|
Forino NM, Hentschel J and Stone MD: Cryo-EM structures tell a tale of two telomerases. Nat Struct Mol Biol. 28:457–459. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D, Hendrick AG, et al: Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 593:597–601. 2021. View Article : Google Scholar : PubMed/NCBI | |
|
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al: METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 22:191–205.e9. 2018. View Article : Google Scholar | |
|
Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang K, Go MY, Ng SC, Chan FK, Sung JJ and Yu J: Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut. 65:1482–1493. 2016. View Article : Google Scholar | |
|
Huang C, Zhou S, Zhang C, Jin Y, Xu G, Zhou L, Ding G, Pang T, Jia S and Cao L: ZC3H13-mediated N6-methyladenosine modification of PHF10 is impaired by fisetin which inhibits the DNA damage response in pancreatic cancer. Cancer Lett. 530:16–28. 2022. View Article : Google Scholar : PubMed/NCBI |









