Introduction
Breast cancer (BRCA) is the most common cancer in
women worldwide. In total, >43,000 BRCA-related deaths are
estimated in 2023 (1), and this
may be due to a high degree of tumor heterogeneity. The
histological subtypes of BRCA observed in clinical practice are
Her2 (ER−, PR−, HER2+), LumA
(ER+/PR+, HER2−), LumB
(ER+/PR+, HER2+) and
triple-negative BRCA (ER−, PR−,
HER2−). Although a further understanding of BRCA
subtypes has led to improved outcomes using targeted therapies,
patients with triple-negative BRCA exhibit a poor prognosis due to
augmented proliferative activity and acquired treatment resistance
(2). Numerous drugs targeting the
cell cycle have been developed to inhibit augmented proliferative
activity, including gemcitabine, a chemotherapeutic agent for G1/S
phase, and tyrosine kinase inhibitors, such as CDK4/6 (3). However, acquired treatment resistance
to the aforementioned drugs was observed in patients following
therapy (4,5). Notably, triple-negative BRCA may also
be treated using anti-vascular endothelial growth factor (VEGF)
therapy, including Avastin and Lucentis, which inhibit
proliferative activity and angiogenesis. However, decreased tumor
vessel and drug penetration, and increased hypoxia stimulated
increased VEGF expression, resulting in resistance (6,7).
Thus, the development of novel therapeutic targets is required to
overcome the excessive proliferation and drug resistance of
BRCA.
Long non-coding RNAs (lncRNAs), exhibit no coding
potential and are >200 nucleotides in length. Notably, lncRNAs
play a critical role in BRCA. For example, the upregulation of H19
inhibited the binding of DNA methyltransferase 3 β (DNMT3B) to the
Beclin1 promoter region, resulting in tamoxifen resistance in BRCA
cells. Moreover, H19 knockdown reversed this effect (8). In addition, LINC00511 promoted the
proliferation of BRCA cells via sponging miR-185-3p to activate E2F
transcription factor 1/Nanog signaling (9). Notably, lncRNA may be divided into
numerous subgroups according to the location on the genome,
including intronic, intergenic, divergent and antisense lncRNA.
Thus, lncRNA and protein-coding transcripts may overlap in the
genome but exhibit different functions. GATA binding protein 3
(GATA3) transcription activates Semaphorin 3B to inhibit BRCA
development (10). By contrast,
GATA3-AS1 destabilized the GATA3 protein, and enhanced the
progression and immune escape of triple-negative BRCA through
promoting GATA3 ubiquitination (11).
LOC127814295 [known as lnc-regulator of G protein
signaling 5 (RGS5) or ENSG00000232995] is a novel lncRNA with a
genomic region overlapping with protein-coding gene RGS5. As a
protein-coding gene, RGS5 is involved in tumor development and
tumor microenvironment remodeling. Results of a previous study
demonstrated that the RGS5-TGFβ-PSmad2 axis reduces RGS5and
TGFβ-dependent cell apoptosis through promoting PI3K-AKT signaling,
and preventing mitochondrial damage and activation of caspases.
This process leads to sustained pericyte survival and expansion in
the tumor microenvironment (12).
Notably, RGS5 promotes the occurrence of tumor angiogenesis in the
tumor microenvironment of human melanoma and renal cancer
xenografts (13). In addition,
RGS5 exhibits potential as a widely expressed tumor antigen for
identifying and characterizing T cell epitopes (14). However, studies focusing on the
specific role of lnc-RGS5 in cancer are lacking, despite the
deregulation of lnc-RGS5 in numerous cancer types. Therefore, the
present study aimed to investigate the functional role of lnc-RGS5
and determine the clinical implications in BRCA. The present study
also aimed to further elucidate the mechanistic role of lnc-RGS5 in
regulating BRCA proliferation.
Materials and methods
Patient tissues and ethics approval
A total of 30 pairs of tissues from patients with
BRCA were collected from The First Affiliated Hospital of Chongqing
Medical University (Chongqing, China). Written informed consent was
obtained from all patients. Women patients who were diagnosed with
BRCA by two pathologists were included in the study (October 1,
2020 to December 30, 2020). All experimental procedures were
approved by Ethics Committee of Chongqing Medical University
(Chongqing, China) (approval date: September 20, 2020). Informed
consent was obtained from all patients.
Analysis of gene expression and survival
in public databases
Gene expression was analyzed in diverse cancer types
detailed in The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). A comparison of
gene expression between tumors and healthy samples was performed
using the DESeq2 package (15) in
R (version 4.2.3; https://www.r-project.org/). The expression
correlation between gene pairs was performed using Pearson's
correlation coefficient based on TCGA-BRCA dataset (https://portal.gdc.cancer.gov/projects/TCGA-BRCA).
Benjamini-Hochberg adjusted P<0.05 was used as a threshold.
Kaplan-Meier survival analysis was performed for triple-negative
BRCA based on TCGA-BRCA dataset using R software (v4.2.3). Log-rank
test was used for survival analysis and P<0.05 was used as a
threshold. Median as cut-off was used.
Gene set enrichment analysis (GSEA)
The Gene Ontology (GO) functions and Kyoto
Encyclopedia of Genes and Genomes pathways of lnc-RGS5 were
analyzed using GSEA software (16). Patients were divided into high and
low lnc-RGS5 expression groups according to the median expression
of lnc-RGS5. Nominal P<0.05 was used as a threshold.
Regulation network analysis
A regulation network was constructed based on the
putative interactions of genes. lnc-RGS5 and miRNA interactions,
and miRNA-coding gene interactions were determined based on
RNAhybrid (17) and miRanda
(18) software. Notably, genes
that interacted with miRNAs are often negatively correlated with
miRNAs. Only genes differentially expressed in BRCA were
considered. For transcription factors, putative targets were
obtained from the Gene Transcription Regulation Database (GTRD)
database (19).
Cell lines and culture
MCF-7 and MDA-MB-231 cell lines were purchased from
the American Type Culture Collection (ATCC), and cultured in
complete DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing
10% fetal bovine serum (FBS; Shanghai ExCell Biology, Inc.), 100
U/ml penicillin and 0.1 mg/ml streptomycin (Beyotime Institute of
Biotechnology) at 37°C in a humidified atmosphere containing 5%
CO2.
Cell transfection
Small interfering RNA (siRNA) targeting lnc-RGS5 and
the negative control (NC) were purchased from Shanghai GenePharma
Co., Ltd. Micro (mi)RNA inhibitor and mimics were purchased from
Tsingke Biological Technology. Following the manufacturer's
instructions, cells were transfected using siRNA-Mate (Shanghai
GenePharma Co., Ltd.). The pcDNA3.1/forkhead box M1 (FoxM1) and
pcDNA3.1/lnc-RGS5 plasmids were purchased from Tsingke Biological
Technology and cells were transfected using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were transfected with siRNAs (1 μg)
or plasmids (2 μg) and cultured at 37°C for 48 h to perform
subsequent experiments. Sequences were as follows: silnc-RGS5-1,
5′-UUU AAA GUG CAG UCU CUG UAC-3′; silnc-RGS5-2, 5′-UUU AAU GCC AUC
CUG GCC AGA-3′; silnc-RGS5-3, 5′-UUU AAA CAG GUG AUC CCU AGA-3′;
siFoxM1, 5′-GGA CCA CUU UCC CUA CUU U-3′; siNC, 5′-UUC UCC GAA CGU
GUC ACG UTT-3′; miR-542-5p inhibitor, 5′-UCU CGU GAC AUG AUG AUC
CCC GA-3′; NC inhibitor, 5′-UCU ACU CUU UCU AGG AGG UUG UGA-3′;
miR-542-5p mimics, 5′-UCG UGA CAU GAU GAU CCC CGA UU-3′; and NC
mimics, 5′-UCA CAA CCU CCU AGA AAG AGU AGA-3′.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA was extracted from MCF-7 and MDA-MB-231
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. RNA
(500 mg) was reverse transcribed into a 10 μl final volume
of cDNA using Reverse Transcription kit (Takara Bio, Inc.),
following the manufacturer's instructions. lnc-RGS5 and miR-542-5p
RNA expression were measured using qPCR on the Step One Plus
Real-Time PCR system. qPCR was conducted in three independent
experiments using SYBR® Premix Ex Taq™ II (Takara Bio,
Inc.) and analyzed using the 2−ΔΔCq method (20). qPCR reaction conditions were as
follows: Initial denaturation at 95°C for 30 sec,
followed by 39 cycles at 95°C for 5 sec and
60°C for 30 sec. GAPDH and U6 were used as endogenous
controls. The primers were designed by Tsingke Biological
Technology. qPCR primer sequences were as follows: GAPDH forward,
5′-CCA TGG GGA AGG TGA AGG TC-3′ and reverse, 5′-AGT GAT GGC ATG
GAC TGT GG-3′; U6 forward, 5′-GCT TCG GCA GCA CAT ATA CTA AAA T-3′
and reverse, 5′-CGC TTC ACG AAT TTG CGT GTC AT-3′; lnc-RGS5
forward, 5′-AGT GAC AAG ATG GGG GTG TTC-3′ and reverse, 5′-CTG GTG
GCT TCT GTT GGT TTG-3′; miR-542-5p forward, 5′-TCG GGG ATC ATC ATG
T-3′ and reverse, 5′-GTG CAG GGT CCG AGG T-3′; and miR-542-5p,
5′-GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG ACC TGC
GGT CTC GTG-3′.
Western blot analysis
MCF-7 and MDA-MB-231 cells were transfected with
miRNA and siRNA and cultured for 48 h. Total protein was extracted
from cells using lysis buffer (cat. no. KGP250; Keygene Biotech,
Inc.; http://www.keygentec.com.cn/), and
the protein concentration was measured using Enhanced BCA Protein
Assay Kit (cat. no. P0010; Beyotime Institute of Biotechnology). A
total of 40 μg proteins were loaded per lane in SDS-PAGE.
Proteins were separated via SDS PAGE on a 10% gel. The separated
proteins were subsequently transferred onto PVDF membranes. The
membranes were then blocked with 5% non-fat milk in TBST (1 ml/l
Tween-20; cat. no. ST825; Beyotime Institute of Biotechnology) at
room temperature (RT) for 1 h, and incubated with the following
primary antibodies: Anti-β-actin (1:1,000; cat. no. 20536-1-AP;
ProteinTech Group, Inc.), anti-FoxM1 (1:1,000; cat. no. ab207298;
Abcam), anti-VEGFA (1:1,000; cat. no. ab214424; Abcam) and
anti-Neuropilin 1 (NRP1; 1:1,000; cat. no. ab81321; Abcam)
overnight at 4°C. Following primary incubation, membranes were
incubated with the HRP-conjugated secondary antibody (1:5,000; cat.
no. SA00001-2; ProteinTech Group, Inc.) at RT for 1 h. The protein
ladder was purchased from Shanghai Epizyme Biomedical Technology
Co., Ltd. (cat. no. WJ103; 10~250 kDa). Protein bands were
visualized using the Pierce ECL Plus Western Blotting Substrate
(Bio-Rad Laboratories, Inc.). Protein expression was quantified
using ImageJ 5.2.1 software (National Institutes of Health) with
β-actin as the loading control.
RNA immunoprecipitation assay
RNA immunoprecipitation assays were performed using
a BersinBio™ RIP kit according to the manufacturer's instructions.
The MCF-7 and MDA-MB-231 cells (density: ~90%) were lysed using
lysis buffer containing a protease inhibitor cocktail and RNase
inhibitor, and the lysates were immunoprecipitated with anti-Ago2
(5 μl; cat. no. 67934-1-Ig; ProteinTech Group, Inc.) and IgG
(5 μl; cat. no. 30000-0-AP; ProteinTech Group, Inc.)
antibodies at 4°C overnight. A total of 30 μl Protein
A-Agarose beads (cat. no. sc-2001; Santa Cruz Biotechnology, Inc.)
were added and incubated at 4°C for 2 h. After centrifugation at
200 x g for 30 sec, beads were 3 times washed with wash buffer (20
mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 0.4
U/μl RNase inhibitor, and 0.4 U/μl Protease Inhibitor
Cocktail). The retrieved RNAs were quantified using RT-qPCR.
Dual-luciferase reporter assay
Transfection and luciferase reporter assays were
performed as previously described (21). lnc-RGS5 cDNA containing the
predictive binding site of miR-542-5p was cloned into the pmirGLO
Dual-Luciferase miRNA Target Expression Vector (Promega
Corporation) to form the wild-type vector (lnc-RGS5-WT). Mutant
(Mut) lnc-RGS5 containing mutations of the miR-542-5p binding site
was specifically synthesized and inserted into the aforementioned
vector (lnc-RGS5-Mut). BRCA cells were cultured and co-transfected
with pmirGLO-lnc-RGS5-3′-untranslated region (UTR) vectors,
including WT or Mut fragments and miR-542-5p mimics. The pmirGLO
vector was used as the NC. FoxM1-WT and FoxM1-Mut were cloned into
the pmirGLO vector (Promega Corporation) using the one-step
directed cloning kit (Novoprotein Scientific, Inc.). miR-542-5p
mimics were co-transfected with FoxM1-WT or FoxM1-Mut vector into
BRCA cells using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Luciferase activity was evaluated using
Dual-Luciferase® Reporter Assay System (Promega Corp.)
after 48-h transfection. Data were presented as a ratio of Firefly
to Renilla luciferase activity.
Short hairpin (sh)RNA transfection
shRNA targeting lnc-RGS5 (shlnc-RGS5 sequence,
5′-GCA TGG TTG GAG ACA ATA AGT CTC GAG ACT TAT TGT CTC CAA CCA
TGC-3′; target sequence, 5′-GCA TGG TTG GAG ACA ATA AGT-3′; 1
μg) was expressed using the pLKO.1-TRC-copGFP-T2A-Puro
vector (TsingKe Biological Technology). A scrambled shRNA (5′-TTC
TCC GAA CGT GTC ACG T-3′; 1 μg) was used as a negative
control (shNC). MDA-MB-231 cells expressing green fluorescent
protein were screened after 72 h transfection at 37°C. HiTransG P
transfection agent (Shanghai Genechem Co., Ltd.) was used. To
generate stable lnc-RGS5-knockdown cells, 2 μg/ml puromycin
was used for induction, and 1 μg/ml puromycin was used for
maintenance.
Cell viability and proliferation
assay
A Cell Counting Kit-8 (CCK-8) assay (cat. no.
40203ES60; Shanghai Yeasen Biotechnology Co., Ltd.) was used to
measure cell proliferation. In total, 2,000 cells were seeded in
96-well plates, and 10 μl CCK-8 solution was diluted and
added. After incubation with CCK-8 reagent at 37°C for 1 h in dark,
absorbance was measured at 450 nm. Cell proliferation was also
detected using a BeyoClickTM EdU Cell proliferation kit according
to the manufacturer's instructions (Beyotime Institute of
Biotechnology). Cells were stained with Alexa Fluor 488 at RT for
30 min in dark, and observed using a fluorescence microscope using
a 10X objective lens (magnification, ×100; Nikon Eclipse Ts2R;
Nikon Corporation).
RNA expression in the nucleus and
cytoplasm
RNA was extracted from the nuclei and cytoplasm
using the PARIS kit (Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. An RNase inhibitor was used in RNA
extraction. The purity of extracted RNA was evaluated, and
high-quality RNA (260/280 nm ratio >1.8) was used for subsequent
RT-qPCR experiments. U2 and β-actin were used as endogenous
controls, and the primer sequences were as follows: U2 forward,
5′-CCT TTT GGC TAA GAT CAA GTG TAG TAT CTG TT-3′ and reverse,
5′-AGC AAG CTC CTA TTC CAT CTC CCT G-3′; and β-actin forward,
5′-CCT TCC TGG GCA TGG AGT C-3′ and reverse, 5′-TGA TCT TCA TTG TGC
TGG GTG-3′.
In vivo tumor formation assay
Nude BALB/c-nu mice (age, 4 weeks; sex, female;
weight, 16-18 g; n=16) were purchased from Huafukang Biotechnology
Company. The in vivo tumor formation assay was performed as
previously described (22).
Briefly, 6×105 MDA-MB-231 cells resuspended using
phosphate buffered saline (PBS) stably transfected with
LV-shlnc-RGS5 or LV-NC were inoculated subcutaneously into the
axillary fossa. Antagomir-542-5p and antagomir-NC (5 nmol/mouse
each time), shFoxM1 (target sequence, 5′-CTCTTCTCCCTCAGATATA-3′),
or shNC plasmids (10 μg/mouse each time) were injected into
the tumors every three days following tumor formation (n=4 in each
group, randomly allocated). The diameter of the largest tumor
observed did not exceed 1.5 cm. At the end of the experiment, all
mice were sacrificed by cervical dislocation. All animal
experiments were blinded and carried out in the IVC Laboratory
under specific pathogen-free (SPF) conditions (4 nude mice per
cage) of Barrier Facilities of Chongqing Medical University. All
mice were group housed on a 12-h dark/12-h light cycle at
temperatures of 18-23°C with 40-60% humidity, and were provided
with sufficient food and water. All animal experiments were
reviewed and approved (approval no. IACUC-CQMU-2022-0016) by the
Experimental Animal Management and Use Committee of Chongqing
Medical University (Chongqing, China).
Statistical analysis
Bioinformatics analysis was conducted using R
software (version 4.2.3; https://www.r-project.org/). Statistical analysis for
experiment results was conducted based on three replications using
Prism 8 (Dotmatics). Unpaired t-tests were used for two-group
comparisons. A P<0.05 was considered to indicate a statistically
significant difference. Error bars (mean ± standard deviation) were
shown.
Results
Expression and clinical analyses of
lnc-RGS5 in BRCA
In Fig. 1A, the
lnc-RGS5 (LOC127814295) gene located on 1q23.3 is
demonstrated. As a lncRNA, the transcripts of lnc-RGS5 were >200
nucleotides in length, with very low protein-coding potential (The
Coding Potential Assessment Tool; CPAT software; 1.97%), no open
reading frames and no translation initiation site (Fig. 1A). Results of TCGA data analysis
demonstrated that lnc-RGS5 was upregulated in diverse cancer types,
such as bladder cancer, esophagus cancer and BRCA (Fig. 1B). Moreover, lnc-RGS5
expression was significantly higher in basal-like or
triple-negative BRCA than in Her2 (P=0.0006), LumA (P=0.028) or
LumB (P=0.048) subtypes (Fig. 1C).
However, lnc-RGS5 demonstrated no significant association with
lymph node or distant metastasis (P>0.05; Fig. 1D). Results of the survival analysis
demonstrated that high lnc-RGS5 expression was associated with poor
overall survival of patients with triple-negative BRCA (cut-off,
median lnc-RGS5 expression; P=0.045; Fig. 1E) and progression-free survival
(cut-off, median lnc-RGS5 expression; P=0.018; Fig. 1F) of patients with BRCA. Results of
the multivariate analysis demonstrated that lnc-RGS5 may act as an
independent factor for progression-free survival (Fig. 1G). These results implied that
lnc-RGS5 may play a critical role in the tumorigenesis of
BRCA and may exhibit potential as a prognostic biomarker in
triple-negative BRCA.
 | Figure 1Expression and clinical analysis of
lnc-RGS5 in BRCA. (A) Genomic location and protein-coding potential
of lnc-RGS5. (B) Expression of lnc-RGS5 in TCGA cancer types. (C)
Expression of lnc-RGS5 in BRCA subtypes. (D) Expression of lnc-RGS5
in patients with lymph node (N) and distant metastasis (M) compared
with patients without metastasis. (E) Overall survival analysis of
lnc-RGS5 in patients with BRCA and triple-negative BRCA. Median
lnc-RGS5 expression was used as the cut-off value. (F and G)
Univariate and multivariate progression-free survival analyses of
lnc-RGS5 in patients with BRCA. *P<0.05,
**P<0.01 and ***P<0.001. N1, 1-3 lymph
node metastasis; N2, 4-9 lymph node metastasis; N3, >9 lymph
node metastasis; M0, without distant metastasis; M1, with distant
metastasis; lncRNA, long non-coding RNA; BRCA, breast cancer; TCGA,
The Cancer Genome Atlas; RGS5, regulator of G protein signaling
5. |
lnc-RGS5 is upregulated and increases
cell proliferation in BRCA
Results of the RT-qPCR analyses indicated that
lnc-RGS5 was upregulated in BRCA compared with healthy
samples (Fig. 2A, Table I). GSEA demonstrated that high
lnc-RGS5 expression was associated with increased activities of DNA
repair, protein export and DNA replication. By contrast, low
lnc-RGS5 expression was associated with decreased activities of
mTOR, MAPK, Erb-B2 Receptor Tyrosine Kinase (ERBB) and VEGF
signaling pathways (Fig. 2B).
Collectively, these results suggested that lnc-RGS5 may be involved
in sustaining the proliferative signaling of BRCA. Thus, the role
of lnc-RGS5 was further validated in BRCA cell proliferation.
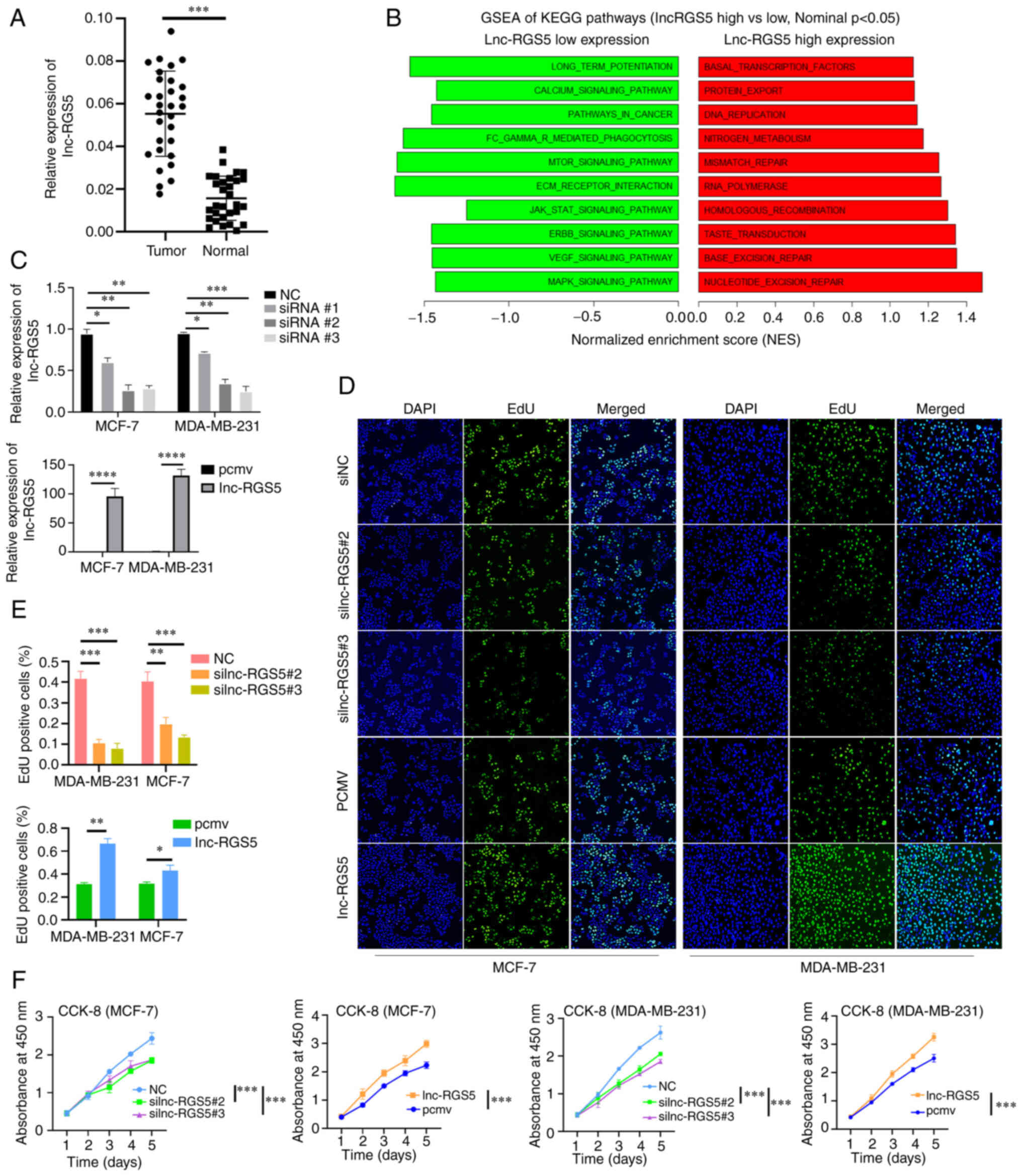 | Figure 2Upregulation of lnc-RGS5 promotes
BRCA cell proliferation. (A) Expression of lnc-RGS5 in BRCA tissues
and healthy tissues (n=30). (B) GSEA of KEGG pathways in high and
low lnc-RGS5 expression groups. (C) Transfection efficiency of RGS5
knockdown and RGS5 overexpression. (D-F) EdU and CCK-8 assays
following siRGS5 and RGS5 overexpression in MCF7 and MDA-MB-231
cells, compared with the negative control. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001. PCMV, pcDNA3,1 empty vector; lncRNA,
long non-coding RNA; BRCA, breast cancer; GSEA, gene set enrichment
analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; CCK-8,
Cell Counting Kit-8; RGS5, regulator of G protein signaling 5;
siRNA, small-interfering RNA. |
 | Table IThe clinical information of patients
with breast cancer. |
Table I
The clinical information of patients
with breast cancer.
| Variable | Number of patients
(%) |
|---|
| Age (median, 58
years old) | |
| <60 | 7 (23.3) |
| ≥60 | 23 (76.7) |
| Sex | |
| Female | 30 (100) |
| Tumor size (T) | |
| T1 | 10 (33.3) |
| T2 | 8 (26.7) |
| T3 | 8 (26.7) |
| T4 | 4 (13.3) |
| Lymph node (N) | |
| N0 | 22 (73.3) |
| N1 | 8 (26.7) |
| Distant metastasis
(M) | |
| M0 | 16 (86.7) |
| M1 | 4 (13.3) |
| PAM50 Subtype | |
| Luminal A | 10 (33.3) |
| Luminal B | 6 (20) |
| Her2-enriched | 5 (16.7) |
| Basal-like | 5 (16.7) |
| Normal-like | 4 (13.3) |
In the present study, lnc-RGS5 was overexpressed and
silenced following transfection with lnc-RGS5 overexpression
plasmid and lnc-RGS5 siRNAs, respectively (Fig. 2C). Results of the CCK-8 and EdU
assays indicated that overexpression of lnc-RGS5 promoted
proliferation, while si-lnc-RGS5 transfection inhibited the growth
of BRCA cells (Fig. 2D-F). These
findings confirmed that lnc-RGS5 significantly enhances the growth
of BRCA cells in vitro.
Lnc-RGS5 functions as a competing
endogenous RNA (ceRNA) for miR-542-5p in BRCA
To investigate the mechanism of lnc-RGS5 in BRCA, GO
enrichment analysis was performed. Results of the present study
demonstrated that lnc-RGS5 was associated with RNA binding involved
in post-transcriptional gene silencing (Fig. 3A), such as ceRNA mechanisms.
Results of the RT-qPCR analysis revealed that lnc-RGS5 was mainly
expressed in the cytoplasm (Fig.
3B). Subsequently, an RNA immunoprecipitation assay was
performed to determine whether Ago2, a key component of the
RNA-induced silencing complex, may combine with lnc-RGS5 and
further mediate the binding of miRNAs with lnc-RGS5. Results of the
present study demonstrated the significant enrichment of lnc-RGS5
immunoprecipitated by the anti-Ago2 antibody, compared with
anti-IgG (Fig. 3C). Collectively,
results of the present study indicated that lnc-RGS5 may function
as a ceRNA.
A ceRNA regulation network of lnc-RGS5 was
subsequently constructed based on an integrative analysis (Fig. 3D). Results of the present study
demonstrated that miR-542-5p (degree; 26) was the potential target
of lnc-RGS5. VEGFA was the hub gene (degree; 15) of the
downstream network regulated by miR-542-5p/FoxM1 signaling.
NRP1 was one of the receptors of ligand VEGFA.
Moreover, expression correlation analysis revealed that miR-542-5p
was negatively correlated with lnc-RGS5 and FoxM1, while
lnc-RGS5 was positively correlated with FoxM1 in BRCA
(P<0.05; Fig. 3E). In addition,
miR-542-5p was downregulated and FoxM1 was upregulated in
BRCA, compared with healthy samples (Fig. 3F). These results were consistent
with those demonstrating the ceRNA mechanism.
Subsequently, the binding sites of miR-542-5p on
lnc-RGS5 and FoxM1 were mutated (Fig. 4A), and miR-542-5p was overexpressed
following transfection with the miRNA mimics (Fig. 4B). Results of the dual-luciferase
assay indicated that miR-542-5p significantly reduced the
luciferase activity of lnc-RGS5 and FoxM1 in BRCA cells.
However, no significant differences in the luciferase activity of
lnc-RGS5-Mut and FoxM1-Mut were observed following miR-542-5p
overexpression (Fig. 4C). Notably,
transfection with the lnc-RGS5 overexpression vector reversed the
decreased pmirGLO-FoxM1 3′UTR luciferase activity induced by
miR-542-5p; however, this was not observed with the
lnc-RGS5-Mut in which the miR-542-5p binding site was mutated
(Fig. 4D). Collectively, these
results demonstrated that miR-542-5p directly binds to lnc-RGS5 and
FoxM1.
 | Figure 4lnc-RGS5 acts as a ceRNA to promote
BRCA cell proliferation in vitro. (A) The mutation sites of
lnc-RGS5 and FoxM1 that bind to miR-542-5p. (B) Transfection
efficiency of miR-542 mimics in MCF-7 and MDA-MB-231 cells. (C)
Relative luciferase activity of lnc-RGS5, lnc-RGS5-Mut, FoxM1 and
FoxM1-Mut in BRCA cells co-transfected with miR-542-5p. (D)
Dual-luciferase assays indicated that lnc-RGS5 overexpression
plasmid, but not lnc-RGS5-Mut, reversed the miR-542-5p
overexpression-mediated decreased luciferase activity of FoxM1
3′UTR. (E) Expression of miR-542-5p following lnc-RGS5
transfection. (F-H) Expression of FoxM1 and VEGFA/NRP1 following
lnc-RGS5, miR-542-5p or FoxM1 transfection in BRCA cells.
*P<0.05, **P<0.01 and
***P<0.001. PCMV, pcDNA3,1 empty vector; lncRNA, long
non-coding RNA; ceRNA, competing endogenous RNA; BRCA, breast
cancer; FoxM1, forkhead box M1; miRNA, microRNA; Mut, mutant; RGS5,
regulator of G protein signaling 5; UTR, untranslated region; NRP1,
Neuropilin 1; NC, negative control. |
lnc-RGS5 promotes BRCA cell proliferation
through the miR-542-5p/FoxM1 axis in vitro
Results of the RT-q-PCR analysis revealed that
miR-542-5p was upregulated in silnc-RGS5 cells compared with siNC
cells, while miR-542-5p was downregulated in cells overexpressing
lnc-RGS5, compared with those transfected with the empty vector
(Fig. 4E). Transfection with the
lnc-RGS5 overexpression vector or miR-542-5p inhibitor upregulated
the expression of FoxM1 and VEGFA/NRP1, while
transfection with si-lnc-RGS5 or miR-542-5p mimics downregulated
the corresponding expression (Fig. 4F
and G). Moreover, FoxM1 overexpression promoted the
expression of VEGFA/NRP1, while transfection with
siFoxM1 inhibited the corresponding expression (Fig. 4H). Subsequently, the efficiency of
shlnc-RGS5 and shFoxM1 transfection was determined (Fig. 5A and B). The decreased
proliferative ability and FoxM1/VEGFA/NRP1 expression
induced by shlnc-RGS5 were regained following
co-transfection with the miR-542-5p inhibitor. Notably, these
results were inhibited following transfection with shFoxM1
in BRCA cells (Fig. 5C-E). The
decreased proliferative ability and protein expression levels of
FoxM1/VEGFA/NRP1 induced by miR-542-5p mimics were regained
following co-transfection with lnc-RGS5. Notably, these results
were inhibited following transfection with shFoxM1 in BRCA
cells (Fig. 6A-C). Thus, lnc-RGS5
competitively sponges miR-542-5p to prevent miR-542-5p binding to
FoxM1 3′UTRs, resulting in FoxM1 upregulation and
increased proliferation of BRCA cells.
 | Figure 5lnc-RGS5 promotes BRCA cell growth
through miR-542-5p/FoxM1 in vitro. (A and B) Transfection
efficiency of shlnc-RGS5 and shFoxM1. (C) Western blot analysis
indicated that inhibitor-542-5p rescued shlnc-RGS5-mediated
inhibition of FoxM1 and VEGFA/NRP1 expression in BRCA cells, and
this was reversed following FoxM1 knockdown. (D and E) EdU and
CCK-8 assays indicated that inhibitor-542-5p rescued the
shlnc-RGS5-mediated inhibition of BRCA cell proliferation, and this
was reversed following FoxM1 knockdown. *P<0.05,
**P<0.01 and ***P<0.001. lncRNA, long
non-coding RNA; BRCA, breast cancer; miRNA, microRNA; FoxM1,
forkhead box M1; shRNA, short hairpin RNA; RGS5, regulator of G
protein signaling 5; NRP1, Neuropilin 1; CCK-8, Cell Counting
Kit-8; NC, negative control. |
 | Figure 6lnc-RGS5 promotes BRCA cell growth
through miR-542-5p/FoxM1 in vitro and in vivo. (A and
B) EdU and CCK-8 assays indicated that lnc-RGS5 rescued the
miR-542-5p mimics-mediated inhibition of BRCA cell proliferation,
and this was reversed following FoxM1 knockdown. (C) Western blot
assays indicated that lnc-RGS5 rescued the miR-542-5p
mimics-mediated inhibition of FoxM1 and VEGFA/NRP1 expression in
BRCA cells, and this was reversed following FoxM1 knockdown. (D)
Relative expression of lnc-RGS5 and miR-542-5p between T3/T4 and
T1/T2 BRCA tissues. (E) Treatment with antagomir-542-5p rescued the
shlnc-RGS5-mediated inhibition of BRCA cell growth, and this was
reversed following FoxM1 knockdown. (F) Western blot assays of
xenograft-derived tissues indicated that antagomir-542-5p treatment
rescued the shlnc-RGS5-mediated inhibition of FoxM1 and VEGFA/NRP1
expression in BRCA cells, and this was reversed following FoxM1
knockdown. MDA-MB-231 cells stably expressing LV-shlnc-RGS5 or
LV-NC were used for in vivo experiments. T1/T2, maximum
tumor diameter was ≤5 cm; T3/T4, maximum tumor diameter was >5
cm. *P<0.05, **P<0.01 and
***P<0.001. PCMV, pcDNA3.1 empty vector; lncRNA, long
non-coding RNA; BRCA, breast cancer; miRNA, microRNA; FoxM1,
forkhead box M1; CCK-8, Cell Counting Kit-8; NRP1, Neuropilin 1;
RGS5, regulator of G protein signaling 5; shRNA, short hairpin RNA;
NC, negative control; LV, lentiviral vector; NC, negative
control. |
lnc-RGS5 promotes BRCA cell proliferation
through a ceRNA pattern in vivo
Analysis of BRCA tissues revealed that the
expression of lnc-RGS5 was higher in patients with T3/T4 tumors
compared with patients with T1/T2 tumors, while the expression of
miR-542-5p was lower in patients with T3/T4 tumors compared with
patients with T1/T2 tumors. Rescue experiments explored whether
lnc-RGS5 exerts biological functions via a ceRNA pattern in
vivo. Treatment with antagomir-542-5p significantly abolished
the decreased tumor growth in LV-shlnc-RGS5 tumors, which
was reversed following FoxM1 knockdown (Fig. 6E). Results of the western blot
analysis of xenograft tumors demonstrated that treatment with
antagomir-542-5p recovered the decreased protein expression of
FoxM1/VEGFA/NRP1 in LV-shlnc-RGS5 tumors, and this was
reduced following FoxM1 knockdown (Fig. 6F). Collectively, these data
demonstrated that lnc-RGS5 acts as a ceRNA for miR-542-5p to
promote BRCA cell proliferation in vitro and in vivo
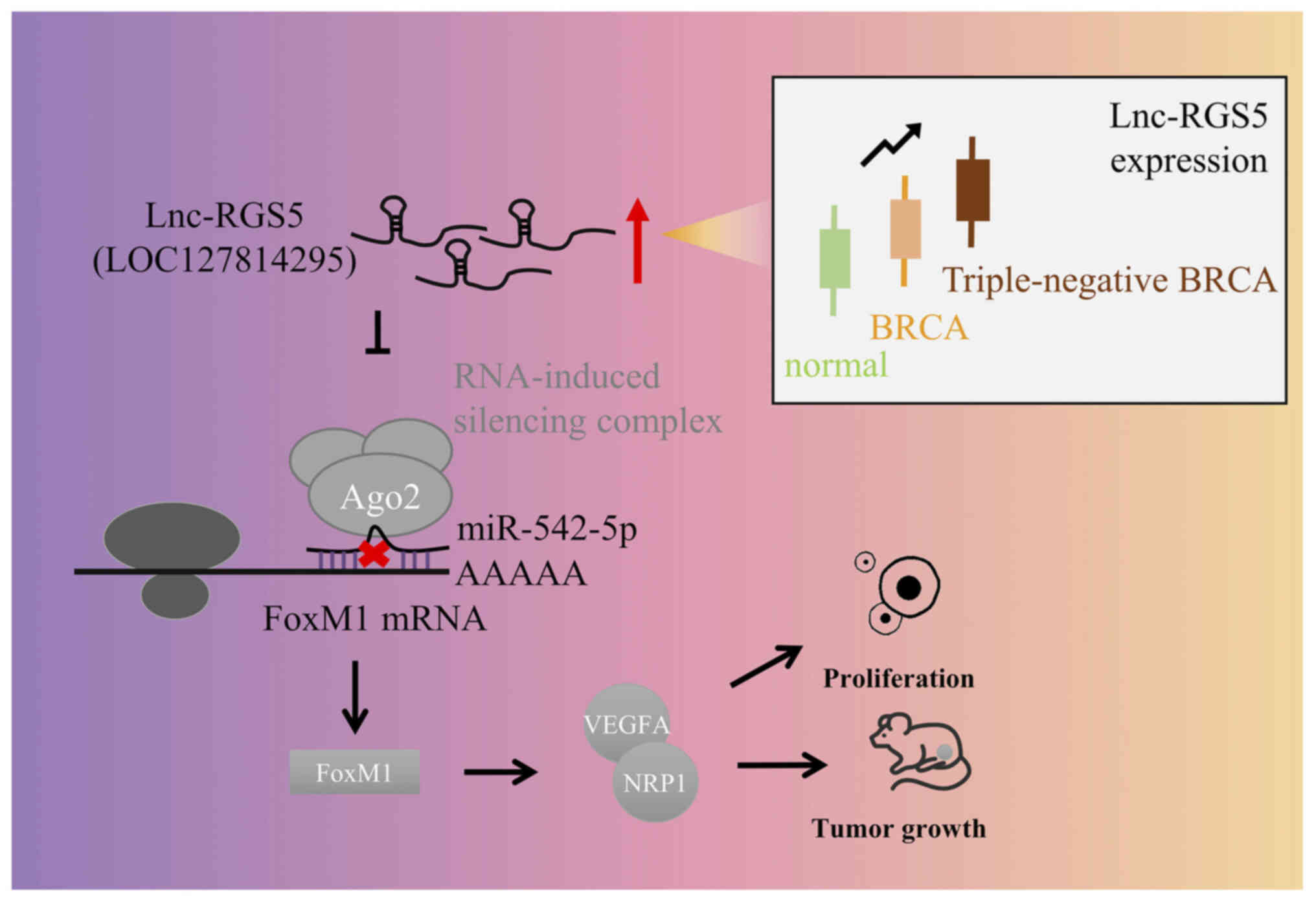
(Fig. 7).
Discussion
Results of the present study demonstrated that
lnc-RGS5 was upregulated in BRCA tissues compared with healthy
samples and associated with the overall survival of patients with
triple-negative BRCA. Functionally, lnc-RGS5 promoted the
proliferation of BRCA cells in vitro and in vivo.
Mechanistically, lnc-RGS5 functions by competitively sponging
miR-542-5p to promote FoxM1/VEGFA signaling. Thus, lnc-RGS5
may exhibit potential as a novel target for the treatment of
BRCA.
Results of the present study revealed that lnc-RGS5
may act as a cancer-associated lncRNA. Pathway and functional
analyses demonstrated that lnc-RGS5 was involved in DNA replication
and signaling pathways associated with cell proliferation,
differentiation and metastasis, such as mTOR, MAPK and VEGF
signaling pathways. However, clinical data analysis revealed that
the expression of lnc-RGS5 was not significantly associated with
lymph node or distant metastasis. Thus, the present study focused
on the regulatory role of lnc-RGS5 in the proliferation of BRCA
cells. Notably, results of the GSEA revealed that RNA binding
involved in post-transcriptional gene silencing was the most
significantly enriched molecular function. Thus, a ceRNA network
was constructed to investigate the potential mechanistic role of
lnc-RGS5.
Results of the present study demonstrated that
lnc-RGS5 knockdown inhibited FoxM1/VEGFA signaling via ceRNA
mechanisms, implying that lnc-RGS5 may act as an alternative target
for combined therapy with anti-VEGFA therapies, as anti-VEGFA
monotherapy stimulated higher VEGF expression in BRCA
(6,7). lnc-RGS5 may also be involved in DNA
repair pathways, such as mismatch repair, base/nucleotide excision
repair and homologous recombination. DNA damage is a hallmark of
cancer as it may lead to tumor evolution and microenvironment
remodeling, which is consistent with the functional results of the
present study, in which lnc-RGS5 was associated with
immune-associated pathways, such as FcγR-mediated phagocytosis.
Further experiments are required to elucidate the molecular
mechanisms underlying lnc-RGS5 in the tumor microenvironment.
FoxM1 transcription factor is a member of the
forkhead box family. Results of a previous study revealed that
FoxM1 upregulation promoted BRCA tumorigenesis (23), and VEGFA and NRP1
were also upregulated (24).
However, the corresponding expression levels and roles in cell
proliferation in BRCA were not further validated. FoxM1 is a
critical downstream effector of PI3K-AKT and JNK/MAPK signaling
pathways for cell proliferation, cell cycle control and DNA damage
repair (25). Results of the
present study revealed that FoxM1 may also be regulated by
lnc-RGS5/miR-542-5p signaling to promote cell proliferation.
FOXM1 is involved in three main cellular mechanisms of
single-strand break repair; namely, nucleotide excision repair,
base excision repair and mismatch repair (26). For example, FoxM1
transcriptionally promotes the expression of replication factor c
subunit 5 to participate in nucleotide excision repair (27). Notably, results of the present
study revealed the potential function of lnc-RGS5 in the regulation
of DNA damage repair through regulating FoxM1. In addition,
results of a previous study indicated that FoxM1 may exhibit
potential as a target in the treatment of tumors (25). However, transcription factors are
complex drug targets due to disordered structures and a lack of
significant small molecule binding pockets (28). A previous study revealed an
alternative approach to targeting transcription factors; for
example, targeting the upstream non-coding RNA. Specifically,
targeting lncRNA H19 inhibited FoxM1 in gallbladder cancer
(29). Thus, lnc-RGS5 may act as a
novel upstream target to suppress FoxM1 and
VEGFA.
Results of a previous study revealed that the
VEGFA/NRP1 axis may regulate cell proliferation in BRCA
(30). Notably, autocrine
VEGFA produced by tumor cells promoted tumor-forming
capacity in vivo, independent of effects on angiogenesis
through interaction with NRP1 (30). Considering the effect of VEGFA in
angiogenesis with VEGF receptor-1/2 (31), VEGFA may promote cell growth
through activating both VEGFR-1/2 and NRP1/Ras signaling pathways
in BRCA. Future studies are required to determine whether lnc-RGS5
promotes BRCA cell growth via angiogenesis in vivo.
Moreover, in future studies, more animals will be used for in
vivo experiments.
As a tumor suppressor gene, miR-542-5p plays an
important role in various tumors (32). miR-542-5p promotes the progression
of BRCA through inhibiting Ubiquitin Specific Peptidase 17 Like
Family Member 2 (USP17L2, also known as DUB3), and treatment with
pristimerin reversed this process at the cellular level (32). Results of a previous study also
demonstrated that miR-542-5p inhibits tumor progression in lung
cancer through inhibiting EGFR (33). Thus, lnc-RGS5 may exhibit potential
as a targeted therapeutic drug to supplement pristimerin, or as a
target of combined therapy.
In conclusion, results of the present study revealed
that lnc-RGS5 may act as a novel oncogenic lncRNA in BRCA, and may
exhibit potential in the treatment of BRCA.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from TCGA (https://portal.gdc.cancer.gov/).
Authors' contributions
JS, YT and FS were responsible for
conceptualization. JS and YT were responsible for the methodology.
JS was responsible for software and validation. JS and YT were
responsible for normal analysis, and JS was responsible for
investigation. JS was responsible for resources, data curation and
writing the original draft. JS and YT were responsible for
reviewing and editing. JS was responsible for visualization. FS was
responsible for supervision. FS was responsible for project
administration and funding acquisition. JS and YT confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
All animal experiments were reviewed and approved
(approval no. IACUC-CQMU-2022-0016) by the Experimental Animal
Management and Use Committee of Chongqing Medical University
(Chongqing, China). Informed consent was obtained from all
patients. All experimental procedures were approved by Chongqing
Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by Key Research and Development
of Social and People's Livelihood (grant nos.
cstc2018jscx-mszdX0031 and CSTC510215195605120418).
References
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Denkert C, Liedtke C, Tutt A and von
Minckwitz G: Molecular alterations in triple-negative breast
cancer-the road to new treatment strategies. Lancet. 389:2430–2442.
2017. View Article : Google Scholar
|
|
3
|
Li Y, Zhang H, Merkher Y, Chen L, Liu N,
Leonov S and Chen Y: Recent advances in therapeutic strategies for
triple-negative breast cancer. J Hematol Oncol. 15:1212022.
View Article : Google Scholar :
|
|
4
|
Murphy CG: The Role of CDK4/6 inhibitors
in breast cancer. Curr Treat Options Oncol. 20:522019. View Article : Google Scholar
|
|
5
|
Kim C, Gao R, Sei E, Brandt R, Hartman J,
Hatschek T, Crosetto N, Foukakis T and Navin NE: Chemoresistance
Evolution in triple-negative breast cancer delineated by
single-cell sequencing. Cell. 173:879–893.e13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta GK, Collier AL, Lee D, Hoefer RA,
Zheleva V, Siewertsz van Reesema LL, Tang-Tan AM, Guye ML, Chang
DZ, Winston JS, et al: Perspectives on triple-negative breast
cancer: Current treatment strategies, unmet needs, and potential
targets for future therapies. Cancers (Basel). 12:23922020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mahdi A, Darvishi B, Majidzadeh AK, Salehi
M and Farahmand L: Challenges facing antiangiogenesis therapy: The
significant role of hypoxia-inducible factor and MET in development
of resistance to anti-vascular endothelial growth factor-targeted
therapies. J Cell Physiol. 234:5655–5663. 2019. View Article : Google Scholar
|
|
8
|
Wang J, Xie S, Yang J, Xiong H, Jia Y,
Zhou Y, Chen Y, Ying X, Chen C, Ye C, et al: The long noncoding RNA
H19 promotes tamoxifen resistance in breast cancer via autophagy. J
Hematol Oncol. 12:812019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q,
Qin Q, Zhao L, Huang Q, Luo Z, et al: Long noncoding RNA LINC00511
contributes to breast cancer tumourigenesis and stemness by
inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res.
37:2892018. View Article : Google Scholar :
|
|
10
|
Shahi P, Wang CY, Chou J, Hagerling C,
Gonzalez Velozo H, Ruderisch A, Yu Y, Lai MD and Werb Z: GATA3
targets semaphorin 3B in mammary epithelial cells to suppress
breast cancer progression and metastasis. Oncogene. 36:5567–5575.
2017. View Article : Google Scholar
|
|
11
|
Zhang M, Wang N, Song P, Fu Y, Ren Y, Li Z
and Wang J: LncRNA GATA3-AS1 facilitates tumour progression and
immune escape in triple-negative breast cancer through
destabilization of GATA3 but stabilization of PD-L1. Cell Prolif.
53:e128552020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dasgupta S, Ghosh T, Dhar J, Bhuniya A,
Nandi P, Das A, Saha A, Das J, Guha I, Banerjee S, et al:
RGS5-TGFβ-Smad2/3 axis switches proto anti-apoptotic signaling in
tumor-residing pericytes, assisting tumor growth. Cell Death
Differ. 28:3052–3076. 2021. View Article : Google Scholar :
|
|
13
|
Silini A, Ghilardi C, Figini S, Sangalli
F, Fruscio R, Dahse R, Pedley RB, Giavazzi R and Bani M: Regulator
of G-protein signaling 5 (RGS5) protein: A novel marker of cancer
vasculature elicited and sustained by the tumor's proangiogenic
microenvironment. Cell Mol Life Sci. 69:1167–1178. 2012. View Article : Google Scholar
|
|
14
|
Boss CN, Grünebach F, Brauer K, Häntschel
M, Mirakaj V, Weinschenk T, Stevanovic S, Rammensee HG and Brossart
P: Identification and characterization of T-cell epitopes deduced
from RGS5, a novel broadly expressed tumor antigen. Clin Cancer
Res. 13:3347–3355. 2007. View Article : Google Scholar
|
|
15
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krüger J and Rehmsmeier M: RNAhybrid:
MicroRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34(Web Server Issue): W451–W454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar
|
|
19
|
Yevshin I, Sharipov R, Valeev T, Kel A and
Kolpakov F: GTRD: A database of transcription factor binding sites
identified by ChIP-seq experiments. Nucleic Acids Res. 45(D1):
D61–D67. 2017. View Article : Google Scholar :
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Wang S, Zhen L, Liu Z, Ai Q, Ji Y, Du G,
Wang Y and Bu Y: Identification and analysis of the promoter region
of the human HAS3 gene. Biochem Biophys Res Commun. 460:1008–1014.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tian Y, Ma R, Sun Y, Liu H, Zhang H, Sun
Y, Liu L, Li Y, Song L and Gao P: SP1-activated long noncoding RNA
lncRNA GCMA functions as a competing endogenous RNA to promote
tumor metastasis by sponging miR-124 and miR-34a in gastric cancer.
Oncogene. 39:4854–4868. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang C, Chen H, Yu L, Shan L, Xie L, Hu J,
Chen T and Tan Y: Inhibition of FOXM1 transcription factor
suppresses cell proliferation and tumor growth of breast cancer.
Cancer Gene Ther. 20:117–124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo M, Hou L, Li J, Shao S, Huang S, Meng
D, Liu L, Feng L, Xia P, Qin T and Zhao X: VEGF/NRP-1axis promotes
progression of breast cancer via enhancement of
epithelial-mesenchymal transition and activation of NF-κB and
β-catenin. Cancer Lett. 373:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao S, Fan LYN and Lam EWF: The
FOXO3-FOXM1 axis: A key cancer drug target and a modulator of
cancer drug resistance. Semin Cancer Biol. 50:77–89. 2018.
View Article : Google Scholar
|
|
26
|
Kalathil D, John S and Nair AS: FOXM1 and
cancer: Faulty cellular signaling derails homeostasis. Front Oncol.
10:6268362021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peng WX, Han X, Zhang CL, Ge L, Du FY, Jin
J and Gong AH: FoxM1-mediated RFC5 expression promotes temozolomide
resistance. Cell Biol Toxicol. 33:527–537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bushweller JH: Targeting transcription
factors in cancer-from undruggable to reality. Nat Rev Cancer.
19:611–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang SH, Ma F, Tang ZH, Wu XC, Cai Q,
Zhang MD, Weng MZ, Zhou D, Wang JD and Quan ZW: Long non-coding RNA
H19 regulates FOXM1 expression by competitively binding endogenous
miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res.
35:1602016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao Y, E G, Wang E, Pal K, Dutta SK,
Bar-Sagi D and Mukhopadhyay D: VEGF exerts an
angiogenesis-independent function in cancer cells to promote their
malignant progression. Cancer Res. 72:3912–3918. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Simons M, Gordon E and Claesson-Welsh L:
Mechanisms and regulation of endothelial VEGF receptor signalling.
Nat Rev Mol Cell Biol. 17:611–625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng S, Zhang Z, Hu C, Xing N, Xia Y and
Pang B: Pristimerin suppressed breast cancer progression via
miR-542-5p/DUB3 axis. Onco Targets Ther. 13:6651–6660. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He RQ, Li XJ, Liang L, Xie Y, Luo DZ, Ma
J, Peng ZG, Hu XH and Chen G: The suppressive role of miR-542-5p in
NSCLC: The evidence from clinical data and in vivo validation using
a chick chorioallantoic membrane model. BMC Cancer. 17:6552017.
View Article : Google Scholar : PubMed/NCBI
|