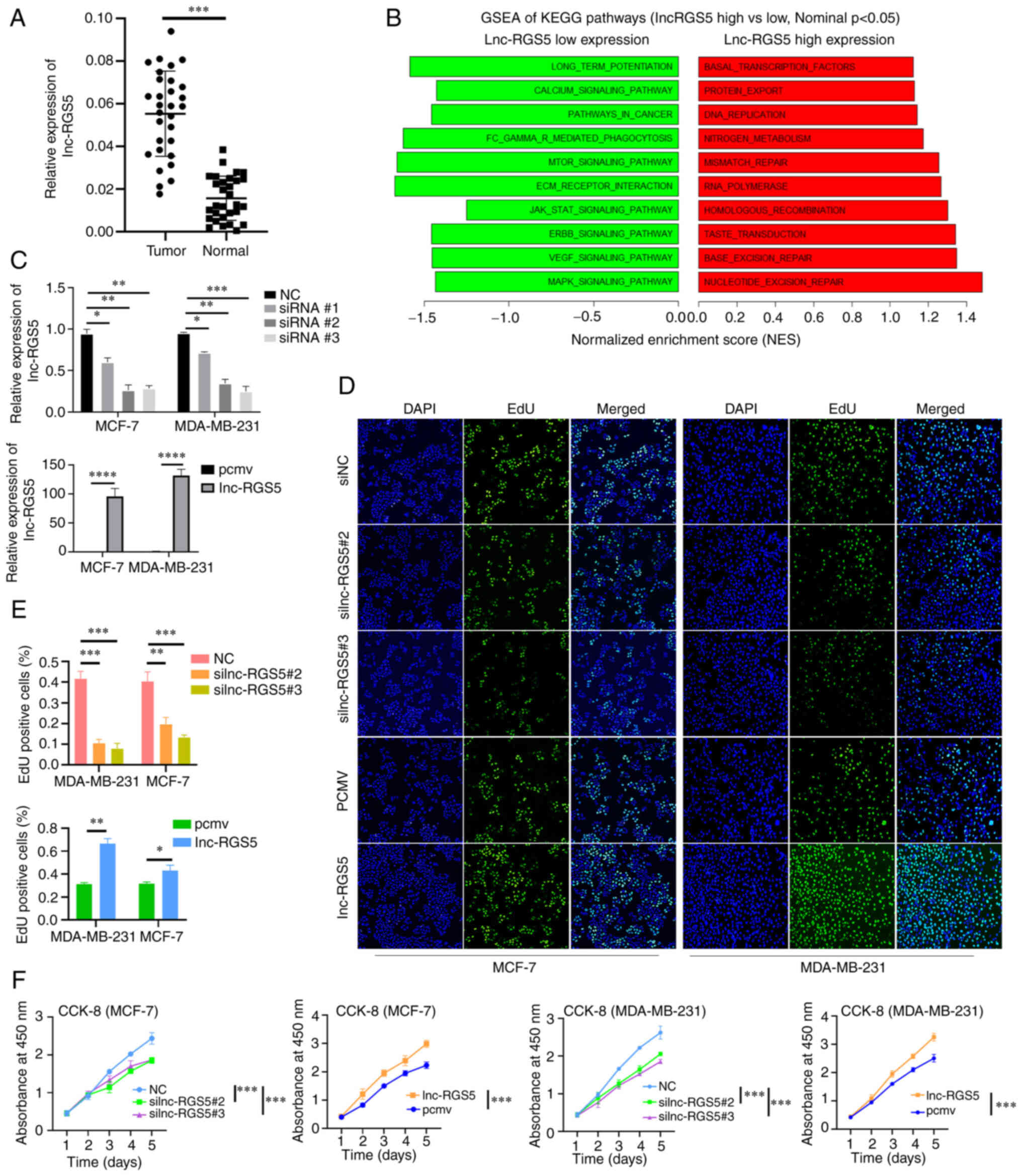
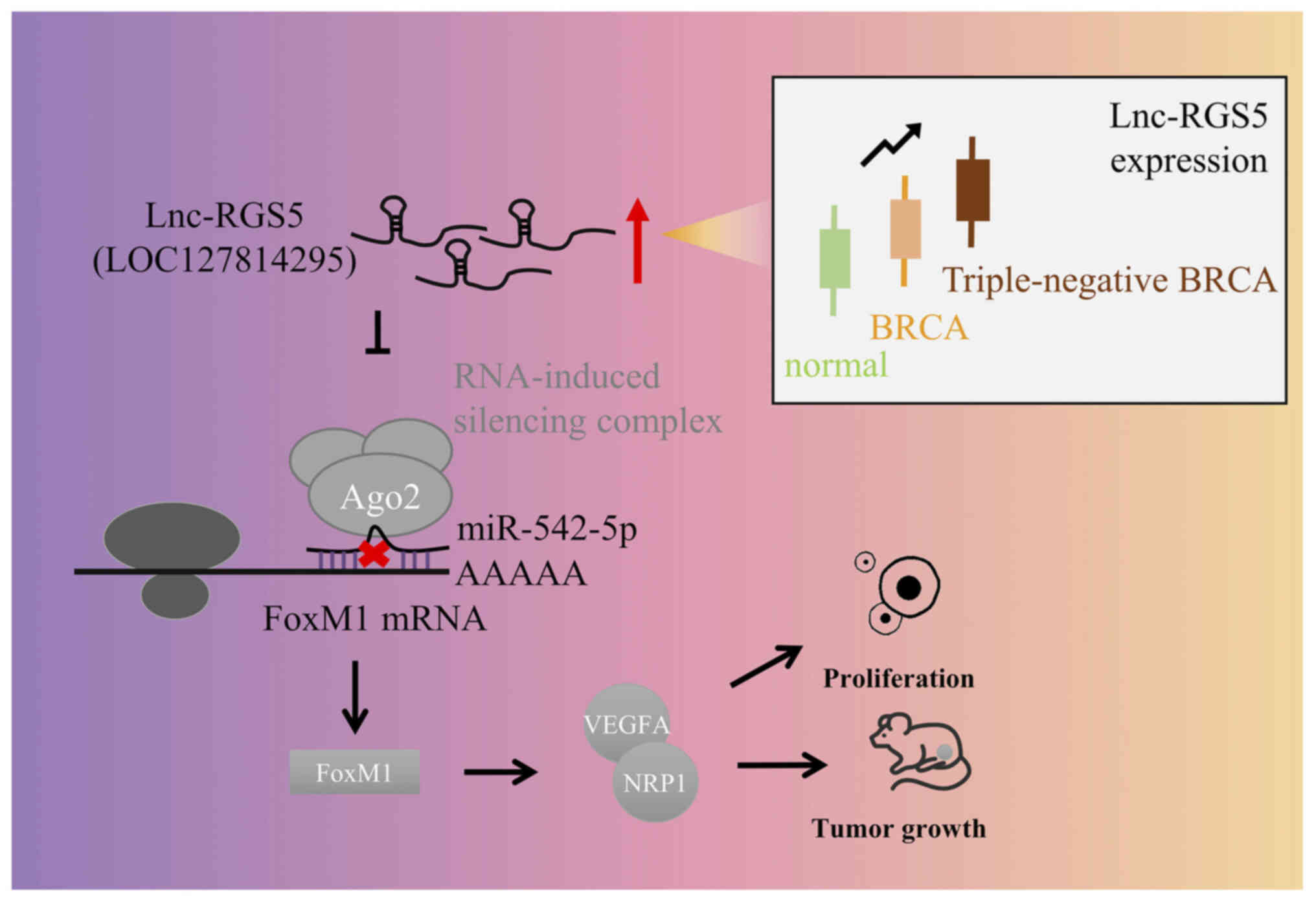
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Denkert C, Liedtke C, Tutt A and von
Minckwitz G: Molecular alterations in triple-negative breast
cancer-the road to new treatment strategies. Lancet. 389:2430–2442.
2017. View Article : Google Scholar
|
|
3
|
Li Y, Zhang H, Merkher Y, Chen L, Liu N,
Leonov S and Chen Y: Recent advances in therapeutic strategies for
triple-negative breast cancer. J Hematol Oncol. 15:1212022.
View Article : Google Scholar :
|
|
4
|
Murphy CG: The Role of CDK4/6 inhibitors
in breast cancer. Curr Treat Options Oncol. 20:522019. View Article : Google Scholar
|
|
5
|
Kim C, Gao R, Sei E, Brandt R, Hartman J,
Hatschek T, Crosetto N, Foukakis T and Navin NE: Chemoresistance
Evolution in triple-negative breast cancer delineated by
single-cell sequencing. Cell. 173:879–893.e13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta GK, Collier AL, Lee D, Hoefer RA,
Zheleva V, Siewertsz van Reesema LL, Tang-Tan AM, Guye ML, Chang
DZ, Winston JS, et al: Perspectives on triple-negative breast
cancer: Current treatment strategies, unmet needs, and potential
targets for future therapies. Cancers (Basel). 12:23922020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mahdi A, Darvishi B, Majidzadeh AK, Salehi
M and Farahmand L: Challenges facing antiangiogenesis therapy: The
significant role of hypoxia-inducible factor and MET in development
of resistance to anti-vascular endothelial growth factor-targeted
therapies. J Cell Physiol. 234:5655–5663. 2019. View Article : Google Scholar
|
|
8
|
Wang J, Xie S, Yang J, Xiong H, Jia Y,
Zhou Y, Chen Y, Ying X, Chen C, Ye C, et al: The long noncoding RNA
H19 promotes tamoxifen resistance in breast cancer via autophagy. J
Hematol Oncol. 12:812019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q,
Qin Q, Zhao L, Huang Q, Luo Z, et al: Long noncoding RNA LINC00511
contributes to breast cancer tumourigenesis and stemness by
inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res.
37:2892018. View Article : Google Scholar :
|
|
10
|
Shahi P, Wang CY, Chou J, Hagerling C,
Gonzalez Velozo H, Ruderisch A, Yu Y, Lai MD and Werb Z: GATA3
targets semaphorin 3B in mammary epithelial cells to suppress
breast cancer progression and metastasis. Oncogene. 36:5567–5575.
2017. View Article : Google Scholar
|
|
11
|
Zhang M, Wang N, Song P, Fu Y, Ren Y, Li Z
and Wang J: LncRNA GATA3-AS1 facilitates tumour progression and
immune escape in triple-negative breast cancer through
destabilization of GATA3 but stabilization of PD-L1. Cell Prolif.
53:e128552020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dasgupta S, Ghosh T, Dhar J, Bhuniya A,
Nandi P, Das A, Saha A, Das J, Guha I, Banerjee S, et al:
RGS5-TGFβ-Smad2/3 axis switches proto anti-apoptotic signaling in
tumor-residing pericytes, assisting tumor growth. Cell Death
Differ. 28:3052–3076. 2021. View Article : Google Scholar :
|
|
13
|
Silini A, Ghilardi C, Figini S, Sangalli
F, Fruscio R, Dahse R, Pedley RB, Giavazzi R and Bani M: Regulator
of G-protein signaling 5 (RGS5) protein: A novel marker of cancer
vasculature elicited and sustained by the tumor's proangiogenic
microenvironment. Cell Mol Life Sci. 69:1167–1178. 2012. View Article : Google Scholar
|
|
14
|
Boss CN, Grünebach F, Brauer K, Häntschel
M, Mirakaj V, Weinschenk T, Stevanovic S, Rammensee HG and Brossart
P: Identification and characterization of T-cell epitopes deduced
from RGS5, a novel broadly expressed tumor antigen. Clin Cancer
Res. 13:3347–3355. 2007. View Article : Google Scholar
|
|
15
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krüger J and Rehmsmeier M: RNAhybrid:
MicroRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34(Web Server Issue): W451–W454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar
|
|
19
|
Yevshin I, Sharipov R, Valeev T, Kel A and
Kolpakov F: GTRD: A database of transcription factor binding sites
identified by ChIP-seq experiments. Nucleic Acids Res. 45(D1):
D61–D67. 2017. View Article : Google Scholar :
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Wang S, Zhen L, Liu Z, Ai Q, Ji Y, Du G,
Wang Y and Bu Y: Identification and analysis of the promoter region
of the human HAS3 gene. Biochem Biophys Res Commun. 460:1008–1014.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tian Y, Ma R, Sun Y, Liu H, Zhang H, Sun
Y, Liu L, Li Y, Song L and Gao P: SP1-activated long noncoding RNA
lncRNA GCMA functions as a competing endogenous RNA to promote
tumor metastasis by sponging miR-124 and miR-34a in gastric cancer.
Oncogene. 39:4854–4868. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang C, Chen H, Yu L, Shan L, Xie L, Hu J,
Chen T and Tan Y: Inhibition of FOXM1 transcription factor
suppresses cell proliferation and tumor growth of breast cancer.
Cancer Gene Ther. 20:117–124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Luo M, Hou L, Li J, Shao S, Huang S, Meng
D, Liu L, Feng L, Xia P, Qin T and Zhao X: VEGF/NRP-1axis promotes
progression of breast cancer via enhancement of
epithelial-mesenchymal transition and activation of NF-κB and
β-catenin. Cancer Lett. 373:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yao S, Fan LYN and Lam EWF: The
FOXO3-FOXM1 axis: A key cancer drug target and a modulator of
cancer drug resistance. Semin Cancer Biol. 50:77–89. 2018.
View Article : Google Scholar
|
|
26
|
Kalathil D, John S and Nair AS: FOXM1 and
cancer: Faulty cellular signaling derails homeostasis. Front Oncol.
10:6268362021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peng WX, Han X, Zhang CL, Ge L, Du FY, Jin
J and Gong AH: FoxM1-mediated RFC5 expression promotes temozolomide
resistance. Cell Biol Toxicol. 33:527–537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bushweller JH: Targeting transcription
factors in cancer-from undruggable to reality. Nat Rev Cancer.
19:611–624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang SH, Ma F, Tang ZH, Wu XC, Cai Q,
Zhang MD, Weng MZ, Zhou D, Wang JD and Quan ZW: Long non-coding RNA
H19 regulates FOXM1 expression by competitively binding endogenous
miR-342-3p in gallbladder cancer. J Exp Clin Cancer Res.
35:1602016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao Y, E G, Wang E, Pal K, Dutta SK,
Bar-Sagi D and Mukhopadhyay D: VEGF exerts an
angiogenesis-independent function in cancer cells to promote their
malignant progression. Cancer Res. 72:3912–3918. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Simons M, Gordon E and Claesson-Welsh L:
Mechanisms and regulation of endothelial VEGF receptor signalling.
Nat Rev Mol Cell Biol. 17:611–625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng S, Zhang Z, Hu C, Xing N, Xia Y and
Pang B: Pristimerin suppressed breast cancer progression via
miR-542-5p/DUB3 axis. Onco Targets Ther. 13:6651–6660. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He RQ, Li XJ, Liang L, Xie Y, Luo DZ, Ma
J, Peng ZG, Hu XH and Chen G: The suppressive role of miR-542-5p in
NSCLC: The evidence from clinical data and in vivo validation using
a chick chorioallantoic membrane model. BMC Cancer. 17:6552017.
View Article : Google Scholar : PubMed/NCBI
|