Introduction
Hepatocellular carcinoma (HCC) is the 6th ubiquitous
tumor worldwide and the main cause of cancer-related deaths
(1). Previous evidence revealed
that after decades of dramatic increasing, the incidence of primary
liver cancer in men has stabilized, but the incidence in women
continues to rise by >2% per year (2). Recently, there have been novel
advances in the therapy of precision management of HCC, such as
accurate surgical resection, immunotherapy and targeted molecular
therapies (3). Patients with
multinodular HCC are preferred for hepatectomy and have a
satisfactory long-term survival (4). For advanced-stage HCC, the sequence
of first-line immunotherapy helps to distinguish relevant clinical
and molecular markers for treatment selection (5). Although a large number of newly
approved therapies have emerged, the treatment for HCC remains
limited and the prognosis is poor (6). Thus, it is considered that the
identification of potential therapeutic targets for HCC is an
overwhelming and urgent requirement.
Pyroptosis is an inflammatory form of programmed
cell death that was first identified in 1992 (7,8).
Pyroptosis is caused by caspase-1 activation triggered by
inflammasomes, which then cleave interleukin (IL)-1β and IL-18
precursors and mediate the cleavage of gasdermin D (GSDMD) into an
N-terminal pore-forming domain, which translocates into the
membrane and releases matured IL-1β and IL-18 (9,10).
The non-classical pathway does not depend on caspase-1 for
activation of the inflammasomes, but rather on
lipopolysaccharide-activated caspase-11 or caspase-4/5 (11). Intracellular LPS-activated
caspase-11 directly caused GSDMD cleavage to undergo pyroptosis,
whereas caspase-11 activation-induced secretion of IL-1β and IL-18
was indirectly mediated through caspase-1 (12,13). Estrogen suppresses malignant
behaviors of HCC cells by targeting the NOD-like receptor thermal
protein domain associated protein 3 (NLRP3) inflammasomes (14), suggesting that there is a
necessity to understand the mechanisms of pyroptosis-related
factors in the proliferation, migration and invasion of HCC.
The Endosomal Sorting Complexes Required for
Transport system ((ESCRTs) consists of ESCRT-0, ESCRT-I, ESCRT-II,
ESCRT-III and Vps4-VTA1, as well as some accessory proteins (for
example ALIX homodimers) which are responsible for catalyzing the
sorting of receptors into vesicles on the endosomal membrane to
produce multivesicular bodies (15). ESCRT-III complexes, also known as
charged multivesicular body proteins (CHMPs), are considered to
copolymerize on endosomal membranes (16) and are targets of Vps4, an ATPase
associated with various cellular activities that provides energy
for ESCRTs to disassemble from the membrane (17,18). CHMP2A and CHMP3 participate in the
later stages of ESCRT-III assembly, recruiting VPS4 and preventing
Snf7 (CHMP4) aggregation (19,20). A study has found that higher
levels of CHMP3 in triple-negative breast cancer (TNBC) predict
longer 3- and 5-year outcomes. Overexpression of CHMP3 in TNBC
cells suppressed cell growth and invasion through inhibiting EMT
process and MAPL (mitochondrial-anchored protein ligase) signaling
(21). Moreover, CHMP3 is
markedly associated with several biological processes in immunity
(for example, cellular response to interleukin-1 or
interferon-gamma) and tumor development (for example, primary
immunodeficiency) (22). CHMP3
was found to co-occur with mutations in CD8A, and the chromosomal
location of the two genes is very similar, which may be
cross-synergistic in the process of tumorigenesis (23). There are significant differences
in CHMP3 gene expression between multiple myeloma and healthy
individuals (24). CHMP3 as a
tumor susceptibility gene has been little studied in liver
cancer.
In studies related to pyroptosis and HCC, it has
been recently reported that CHMP3 may be involved in the pyroptotic
process in liver cancer, but no relevant experiments have been
conducted to explore this (25).
Overall, a growing number of studies have identified a key function
of pyroptosis in the development of HCC and in the antitumor
process. Therefore, the main aim of the present study was to assess
CHMP3 expression in HCC tissues and to demonstrate the effect of
CHMP3 on liver cancer cells proliferation, migration and invasion
and the possible contribution of pyroptosis.
Materials and methods
Database
RNA sequencing data were derived from the The Cancer
Genome Atlas (TCGA) repository for 374 patients with HCC and 50
normal liver samples with corresponding clinicopathological
features on 4 August 2021.
Construction of a prognostic model on the
basis of differentially expressed genes (DEGs) involved in
pyroptosis
A selection of 53 genes associated with pyroptosis
were analysed from the available reviews (26,27). The 'limma' data package is
available for screening DEGs. To construct a protein-protein
interaction (PPI) network, the Search Tool for the Retrieval of
Interacting Genes website (https://string-db.org/) was used. The R (v4.1.3;
Robert Gentleman and Ross Ihaka) package 'glmnet' (2.0.16) can be
used to narrow down the range of genes to be screened. After
central normalization of TCGA dataset ('scale' function in R is
applied), risk scores were calculated. The risk score formula is as
follows: Risk score=∑8iXixYi (X: coefficient, Y: gene expression
level). Kaplan-Meier analysis was used to analyze the overall
survival (OS) times of patients with hepatocellular carcinoma
obtained from the TCGA repository based on the log-rank test with a
cut-off value of P<0.05. The R package 'prcomp' was used to
conduct principal component analysis. Univariate and multivariate
Cox regression models were performed to analyze the clinical
characteristics of cases in the TCGA cohort.
Survival and expression analysis by Gene
Expression Profiling Interactive Analysis (GEPIA) and Human Protein
Atlas (HPA)
Survival curves of differentially expressed CHMP3
were analyzed by GEPIA (28) to
find out whether this gene's expression affects the survival of HCC
patients. In addition, the staging plot was analyzed to compare
CHMP3 expression in different pathological stages. The expression
of CHMP3 was further compared by means of HPA between normal liver
and HCC tissues with immunohistochemical (IHC) images.
Clinical materials and sample
preparation
Between September 2020 and March 2022, all clinical
samples were collected after surgical resection and stored
immediately at −80°C for subsequent experiments at the Department
of General Surgery of Shengjing Hospital of China Medical
University. All specimens were acquired from patients with HCC
confirmed by pathologists. Written informed consent was obtained by
all patients whose tissue samples were collected prior to
enrolment. Human studies (approval no. 2022PS785K) were approved by
the Ethics Committee of Shengjing Hospital of China Medical
University (Shenyang, China). Clinicopathological characteristics
of patients (including age and sex distribution) are listed in
Table I.
 | Table IRelationship between CHMP3 expression
and the clinical characteristics of 65 patients with hepatocellular
carcinoma. |
Table I
Relationship between CHMP3 expression
and the clinical characteristics of 65 patients with hepatocellular
carcinoma.
| Clinicopathological
variables | Total number | Expression level of
charged multivesicular body protein 3
| P-value |
|---|
| Low/moderate | High |
|---|
| All cases | 65 | 21 | 44 | |
| Age, years | | | | 0.952 |
| <55 | 22 | 7 | 15 | |
| ≥55 | 43 | 14 | 29 | |
| Sex | | | | 0.401 |
| Male | 45 | 16 | 29 | |
| Female | 20 | 5 | 15 | |
| Tumor size
(cm) | | | | |
| <5 | 26 | 13 | 13 | 0.013 |
| ≥5 | 39 | 8 | 31 | |
| Preoperative level
of alpha-fetoprotein | | | | 0.294 |
| <200
μg/ml | 25 | 10 | 15 | |
| ≥200
μg/ml | 40 | 11 | 29 | |
| Hepatitis B
virus | | | | 0.915 |
| Negative | 22 | 9 | 13 | |
| Positive | 43 | 17 | 26 | |
| Invasiveness | | | | 0.766 |
| No | 17 | 5 | 12 | |
| Yes | 48 | 16 | 32 | |
IHC staining
The collected tissue was fixed in an appropriate
volume of 4% paraformaldehyde at 4°C for 24 h, then procedurally
dehydrated and embedded in paraffin. Tissue sections of 3-μm
thickness were produced using a microtome, followed by
de-paraffinization using xylene and microwave heating. Sections
were placed in the configured antigen repair solution (Citrate
antigen retrieval solution; Beyotime Institute of Biotechnology)
and boiled for 5 min. Primary antibody against CHMP3 (1:100; cat.
no. 15472-1-AP; Proteintech Group, Inc.) was added to the sections
after washing with PBS and incubated at 4°C overnight. The
appropriate proportion of HRP-conjugated secondary antibody (ready
to use; cat. no. PR30011; Proteintech Group, Inc.) was added, and
incubated at 37°C for 1 h. Antibodies were diluted with TBST, and
Tween-20 was used in a dosage of 0.5 ml/l. Finally, chromogenic
detection was carried out under the light microscope using DAB. The
final score is the percentage of cells multiplied by the staining
intensity (29).
Cell culture
The human liver cancer cell lines HepG2 (Procell
Life Science & Technology Co., Ltd.) and Huh-7 (Nanjing KeyGen
Biotech Co., Ltd.) cell lines were purchased in August 2021. Cells
were expanded and stored at -80°C and only early passages
(<passage 5) within 6 months of these cell lines were used in
the present study. HepG2 cells were cultured in MEM (Hyclone;
Cytiva). Huh-7 cells were cultured in DMEM (Hyclone; Cytiva).
Ac-YVAD-CMK (cat. no. GC42721) was purchased form GLPBIO.
Cell line authentication statement
HepG2 and Huh-7 were tested for genotyping of the
STR locus and Amelogenin locus using the 20-STR amplification
protocol. The results showed that no cross-contamination of human
cells was detected in the cell lines and a 100% match to their
cytotyping could be found in the cell bank, named HepG2 and Huh-7,
respectively.
CHMP3 small interfering (si)RNA and CHMP3
plasmid transfection
Negative control siRNA (siRNA-NC) (UUG TCC GAA CGU
CTC AAG UTT), small interfering (siRNA)-CHMP3 (UGU GAA GAU UCC AGA
GAU UTT), the plasmid vector and plasmid-CHMP3 were purchased from
Shanghai GenePharma Co., Ltd. Transfection was carried out at room
temperature with Lipofectamine 3000 (GlpBio) following the
manufacturer's instructions. Cells were transfected with 2.5
μg RNA/DNA added to 3.75 μl of reagent, and
serum-free medium was replaced with complete medium 6 h later.
Transfection efficiency was verified by western blotting at 24 h
post-transfection. ImageJ (v1.52) (National Institutes of Health)
was used to analyze the banding results.
Colony formation
Groups of treated cells were seeded in six-well
plates at a density of 1,000 cells/well and cultured in
serum-containing medium. The medium was changed every three days
and culture was stopped when colony formation was observed with the
naked eye. Ac-YVAN-CMK (40 μM, AYC) (GLPBIO) was added to
the cells after transfection. Lastly, colonies were fixed with 4%
paraformaldehyde for 2 h at room temperature as well as stained
with crystal violet solution for 30 min (Beyotime Institute of
Biotechnology). More than 50 cells were counted as one colony and
then the total number of colonies per well was counted.
Wound healing assay
Groups of treated cells were scratched with a
200-μl pipette tip. The plates were washed with PBS wells to
remove residual cells, then serum-free medium was added. Images of
the wound shape were captured with a light microscope at a specific
point in time (magnification, ×200). Wound healing rate=(0 h wound
area-48 h wound area)/0 h wound area ×100%.
Transwell invasion assay
The Matrigel (Corning, Inc.) was removed from −20°C,
placed at 4°C to thaw and melt into liquid form, and then the
pre-cooled tip was used to spread the Matrigel evenly on the bottom
of upper chamber, followed by placing it in a 37°C incubator for
2-4 h to wait for the Matrigel to completely solidify. The lower
chamber of the Transwell plate was added with 700 μl of
serum-containing medium, and the upper chamber was filled with
groups of cells resuspended in 200-μl of serum-free medium.
The upper chamber was placed in the Transwell plate and incubated
for 24 h at 37°C. Cells from the upper chamber outer membrane were
then fixed in 4% paraformaldehyde for 1 h and stained with crystal
violet for 30 min at room temperature. Matrigel was wiped from the
inner membrane of the upper chamber with a cotton sab and the upper
chamber was gently rinsed with PBS to wash away excess color. The
invasive cells were observed and images were captured with a light
microscope.
Cell Counting Kit-8 (CCK-8) assay
The cell proliferation capacity was assayed
according to the manufacturer's instructions of the CCK-8 assay kit
(Epizyme; http://www.epizyme.cn/). HepG2 and Huh-7
(3×103 cells/well) were inoculated into 96-well plates.
Cells were transfected with si-CHMP3 and incubated at 37°C for 24 h
before adding AYC (40 μM). A total of 10 μl CCK-8 was
then added at each point (24, 48, 72 and 96 h) and incubated at
37°C for 4 h. The absorbance of the cells at 450 nm was measured
using a microplate reader.
Western blotting
Tissue and cell samples were lysed in ice-cold RIPA
lysis buffer and PMSF (both from Beyotime, Institute of
Biotechnology). The protein concentration was calculated according
to the BCA assay kit instructions (Epizyme). Gel electrophoresis
separation was performed by 10%- or 15%-PAGE Gel Rapid Preparation
Kit (Epizyme) and then transferred to polyvinylidene difluoride
membranes (Epizyme). Each lane contained 20 μg of protein.
The bands were blocked with 5% skim milk blocking solution prepared
in TBST for 2 h at room temperature. After blocking was completed,
incubation was carried out overnight at 4°C using the primary
antibodies. The membranes were subsequently incubated for 2 h at
room temperature using the HRP-conjugated Affinipure Goat
Anti-Mouse/Rabbit secondary antibody (cat. nos. SA00001-1 and
SA00001-2, Proteintech Group, Inc.). Protein bands were visualized
using an ECL kit (Epizyme). The information of primary antibodies
is provided in Table SI.
Transmission electron microscopy
(TEM)
Huh-7 cells that had been treated differently were
collected. The cell pellets were then pre-fixed in 2.5%
glutaraldehyde overnight at 4°C. They were fixed in 1.0% osmic acid
for 2 h. The samples were dehydrated and soaked with a 1:1 mixture
of epoxy resin and acetone overnight at room temperature. The
samples were incubated in 1% uranyl acetate staining solution for 1
h. After 4 days, the samples were sectioned with an ultrathin
sectioning machine. Finally, staining was carried out with uranyl
acetate and lead citrate. Examination was carried out using
TEM.
Xenograft mouse model
In vivo tumor formation was investigated by
constructing a xenograft nude mouse model. A total of 18 male
BALB/c nude mice (4 weeks old, weighing ~20 g) were used in the
animal experiments and were purchased from Beijing HFK Bioscience
co. Ltd. All mice were housed under specific pathogen-free
conditions (temperature 22°C; humidity 50%; light/dark cycle 12/12
h), with free access to food and water, and padding changed twice
every three days. Huh-7 cells at a density of 1×106 were
resuspended in 100 μl PBS to form a cell suspension. Each
mouse received a gentle, slow subcutaneous inoculation of 100
μl of cell suspension in the right axilla. When the tumors
grew to an optimal volume, the tumor-bearing mice were randomly
divided into three groups. siRNA-CHMP3 in vivo (10
μg/μl) was incubated with Lipofectamine 3000 for 30
min at room temperature to form siRNA-lipofectamine complexes,
which were injected intratumorally every 3 days for 4 weeks. AYC
(0.1 mg/kg/day) was injected intraperitoneally every 24 h. Tumor
volume (mm3) was calculated using the following formula:
Volume=(width)2 × length/2. Tumor size was measured
every three days. Animal experiments (approval no. 2022PS779K) were
approved by the Ethics Committee of Shengjing Hospital, China
Medical University (Shenyang, China).
Statistical analysis
The experimental values are shown as the mean ± SEM
of at least three independent experiments. One-way analysis of
variance (ANOVA) followed by Tukey's multiple comparison test was
used to analyze the data with GraphPad Prism (version 9.0.0;
Dotmatics). The chi-square test was used to statistically analyze
the relationship between different clinical characteristics and
CHMP3 expression. Differences between groups were considered
statistically significant when P<0.05. All experiments were
repeated three times.
Results
Identification of DEGs between normal and
tumor tissues
The workflow was summarized in a flowchart (Fig. S1). 39 DEGs (all P<0.05) were
recognized through comparing 53 pyroptosis-associated genes'
expression in TCGA downloadable data from patients with HCC. From
these, 33 genes were upregulated in tumor tissues, while the six
other genes exhibited low expression (Fig. S2A). PPI network and an
association network containing 20 pyroptosis-associated genes are
demonstrated in Fig. S2B and C
(red: positive association). When the clustering variable k=2, the
intra-group relevance is higher and the inter-group relevance is
lower (Fig. S2D). The heat map
showed little difference in clinical characteristics between the
two clusters except for the degree of tumor differentiation
(P<0.001) and OS (P<0.05) (Fig. S2E and F).
Establishment of prognostic gene model in
TCGA cohort
Univariate Cox regression was used to find 8 genes
that met the criteria of P<0.5 and hazard ratios (HRs) >1 for
the next analysis (Fig. S3A). A
seven-gene signature was constructed on the basis of the optimal λ
values derived from least absolute shrinkage and selection operator
Cox regression analysis (Fig. S3B
and C). Risk scores were calculated and 424 patients with HCC
were divided into low and high-risk groups based on median scores
(Fig. S3D). As demonstrated in
Fig. S3E-G, it could be
concluded that patients in the divided high-risk group clearly had
higher mortality and shorter survival compared with the low-risk
group. The area under the receiver operating characteristic curve
varied from 0.7-0.85, indicating that the prediction model was
effective (Fig. S3H).
Independent prognostic value of risk
model
Univariate (HR=7.222, 95% Confidence Interval (CI):
4.322-12.066; Fig. S4A) and
multifactorial (HR=6.315, 95% CI: 3.638-10.964; Fig. S4B) Cox regression analyses
indicated that risk score could be used as an independent predictor
of poor survival. Finally, a heat map of clinical features
(Fig. S4C) revealed a distinct
patient distribution by sex and tumor differentiation in different
subgroups (P<0.01). The relationship between CHMP3 and HCC among
these seven genes (BAK1, BAX, CHMP3, GSDME, CASP8, GSDMC and
SCAF11) remains unstudied. Therefore, the role of CHMP3 in the
progression of liver cancer was investigated in the present
study.
The relationship between CHMP3 expression
and caspase-1 and HCC
As demonstrated in Fig. 1A, CHMP3 was highly expressed in
LIHC. The curves of OS and disease-free survival indicated that
patients with high expression of CHMP3 presented lower survival
rate (Fig. 1B and C). It was also
identified that CHMP3 expression varies across pathological stages
(Pr (>F) i.e., P<0.05) (Fig.
1D). IHC results obtained from HPA revealed that the expression
of CHMP3 was markedly higher in HCC compared with normal tissue
(Fig. 1E and F). The database
results illustrated two indications: i) CHMP3 participates in the
advancement of HCC and, ii) it might be related to pyroptosis.
The CHMP3 expression in HCC tissues
To further validate the results of the public
database, the expression of CHMP3 was determined by IHC based on
previous predictions. It was found that CHMP3 was apparently
overexpressed in the HCC samples compared with the corresponding
para-cancer tissues (Fig. 2A-D).
The high/low expression is based on tissue type. Another five pairs
of carcinoma and para-cancerous tissues were collected to verify
the CHMP3 expression and pyroptosis-related proteins by western
blot assay, for detecting the level of pyroptosis in HCC. A
previous study demonstrated reduced caspase-1 activation and IL-1β
expression for some cancers (12). It was then observed that
caspase-1, IL-1, IL-18 and GSDMD were significantly downregulated,
suggesting the same trend (Fig. 2E
and F). Analysis of the clinical characteristics of 65 patients
with HCC revealed a significant association between CHMP3
expression and tumor size using the chi-square test
(P<0.05).
The CHMP3 expression promotes the
proliferation of liver cancer cell lines
According to the aforementioned bioinformatics
analysis and the results of IHC and western blotting, CHMP3 was
highly expressed in liver cancer. Therefore, CHMP3 was knocked down
and overexpressed to observe the effect on the proliferative
capacity of liver cancer. The results of western blot analysis
showed a significant decrease in CHMP3 expression after
transfection with si-CHMP3 in HepG2 and Huh-7 cells (Fig. 3A and B). Colony formation assays
revealed that knocking down CHMP3 reduced the number of colonies in
liver cancer cell lines, which suggests that suppression of CHMP3
impaired the proliferation ability in liver cancer (Fig. 3C-E). On the contrary, cells
transfected with plasmid CHMP3 had significantly enhanced colony
formation capacity compared with cells transfected with vector
(Fig. 3F-J). The transfection
efficiency of CHMP3 overexpression was detected by western
blotting.
 | Figure 3Effect of CHMP3 on the proliferative
capacity of liver cancer cells, and its relationship with
pyroptosis. (A and B) Western blot analysis of CHMP3 levels
following transfection with si-CHMP3. (C-E) Colony formation assays
demonstrated the proliferation of HepG2 and Huh-7 cells. (F and G)
Overexpressing CHMP3 was evaluated by western blotting. (H-J) The
proliferative abilities of the cells overexpressing CHMP3 were
higher compared with those of the NC or vector group. (K)
Transmission electron microscopy images of cell morphological
alterations. (L and M) Western blot analysis of pro caspase-1,
cleaved caspase-1, pro IL-1β, IL-1β, pro IL-18, IL-18 and cleaved
N-terminal GSDMD levels. *P<0.05 and
***P<0.001. CHMP3, charged multivesicular body
protein 3; GSDMD, gasdermin; si-, small interfering; NC, negative
control. |
CHMP3 inhibition induces cell membrane
blistering and cytoplasm leakage
To explore whether CHMP3 leads to pyroptosis in
Huh-7 cells, cellular alterations were observed after knockdown
using transmission electron microscopy. It was observed that the
transfected cells exhibited typical membrane rupture and
cytoplasmic leakage (red arrows) as demonstrated in Fig. 3K. The expression of several
proteins was apparently increased after CHMP3 knockdown, including
cleaved caspase-1, IL-1β, IL-18 and N-terminal GSDMD (Fig. 3L and M). However, no changes were
observed regarding their corresponding precursor forms of
expression. Thus, it can be assumed that CHMP3 might promote liver
cancer progression through pyroptosis mediated via the
caspase-1/IL-1β pathway.
CHMP3 inhibition activates caspase-1
dependent pyroptosis in vitro
To explore whether CHMP3 inhibition-induced
pyroptosis is modulated by caspase-1, Ac-YVAD-CMK (AYC, an
inhibitor of caspase-1) was used in the present study. The changes
in proliferative capacity of liver cancer was examined after the
administration of AYC by colony formation and CCK-8 assays.
Knocking down CHMP3 with si-CHMP3 significantly decreased the
proliferation of HepG2 and Huh-7 cells, while the application of
AYC reversed the ability of si-CHMP3 to reduce cell proliferation
(Fig. 4A-F). By carefully
examining the data, it was identified that AYC diminished CHMP3
knockdown-induced caspase-1 activation and reduced IL-1β, IL-18 and
GSDMD cleavage in liver cancer cells (Fig. 4G and H). Briefly, these data
supported the notion that CHMP3 inhibits pyroptosis by caspase-1,
and this effect can be reversed by caspase-1 inhibitor.
CHMP3 inhibition activates caspase-1
dependent pyroptosis in vivo
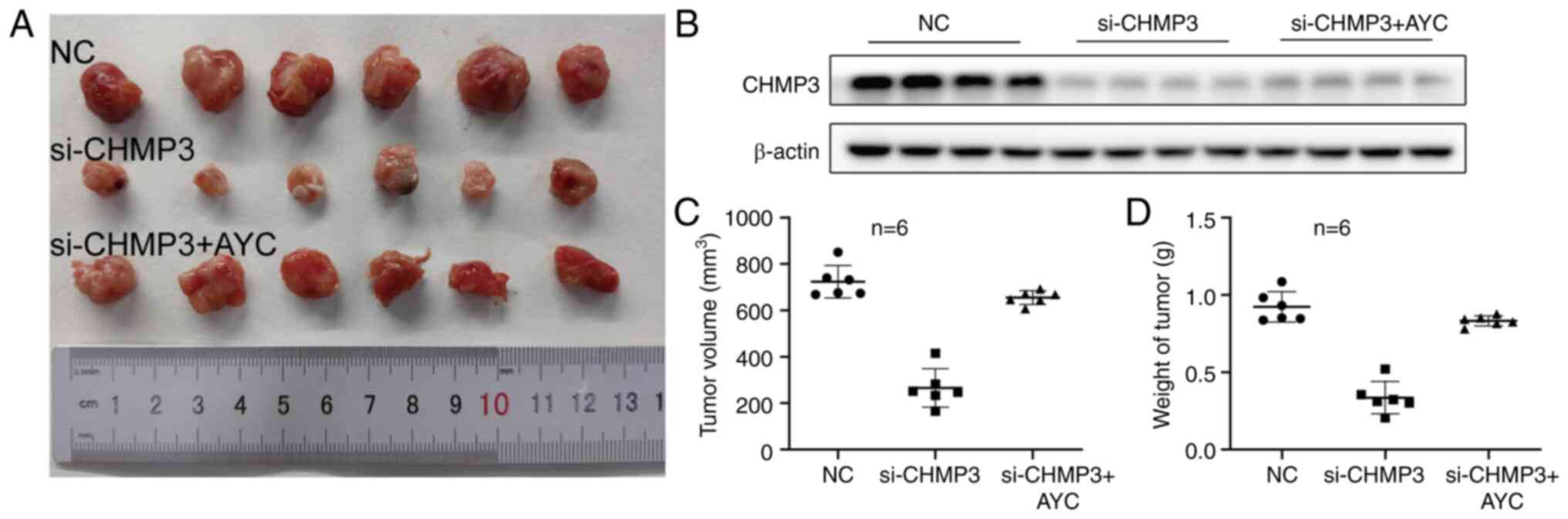
The effect of CHMP3 in tumor formation was then
examined in vivo. Knockdown of CHMP3 significantly inhibited
the growth of subcutaneous tumor, and the volume and weight of the
tumor were lower than those of the control group. The addition of
AYC reversed this inhibition, indicating that low-expression CHMP3
also inhibited the growth of HCC in vivo (Fig. 5A-D).
The relationship between CHMP3 expression
and migration and invasion in liver cancer cells
There is a well-known fact that HCC is a highly
aggressive and metastatic cancer. In the wound healing assay, the
capacity of cell migration was impaired after CHMP3 silencing
(Fig. 6A-C) and enhanced after
overexpression (Fig. 7A-C).
Transwell invasion assay revealed that si-CHMP3 reduced the number
of invasive cells (Fig. 6D-F),
while CHMP3 overexpression in the plasmid-CHMP3 group increased the
number of HepG2 and Huh-7 cells crossing Matrigel (Fig. 7D-F). To provide further evidence
of changes in invasion and migration capacity, expression of
EMT-related proteins was next investigated in both cells by western
blotting. Knocking down CHMP3 caused an significant decrease of
N-cadherin, matrix metalloproteinase 9 (MMP9) and vimentin
expression, and an increase of E-cadherin expression in liver
cancer cells (Fig. 6G and H).
Discussion
Emerging evidence suggests that the impacts of
pyroptosis appear to play a different role in liver diseases
(11,30). In HCC, a significant reduction in
NLRP3 expression suggested a reduced level of pyroptosis (31). NEK7 was found to enhance
pathological proliferation of HCC cells in vivo and in
vitro, while its downregulation inhibited cancer-stromal
interactions by causing cancer cell pyroptosis (32).
In the beginning of the present study, the mRNA
levels of 53 genes associated with pyroptosis were examined and
found to be differentially expressed in both HCC and normal
tissues. CHMP3 was then selected as a target for study from the
seven gene signatures that had been established. The remaining six
genes have been studied with respect to pyroptosis (33-36). However, relatively little research
has been conducted on CHMP3 in cancer and cell death. Therefore,
exploring the relationship between CHMP3 and the clinical features
of HCC can provide an improved understanding of whether CHMP3 can
influence the progression of HCC. As expected, patients with higher
CHMP3 level showed higher tumor grade and poorer survival
conditions. The role of CHMP3 in promoting liver cancer tumor
growth at the cellular level was also demonstrated, and its
inhibition induced cancer cells to undergo pyroptosis thereby
inhibiting liver cancer progression.
Over the past decade, pyroptosis and
pyroptosis-activated inflammatory factors in human cancers have
been increasingly investigated (37-39). The caspase-1/IL-1β pathway affects
cell proliferation and other behaviors, and their activation and
cleavage enhance cellular pyroptosis and attenuates other
pro-oncogenic events (40,41).
There is a critical role for this pathway in malignancy invasion,
angiogenesis and tumor-immune system interactions (42-44). However, the role of IL-1β in HCC
remains controversial, meaning that its expression may either
promote or inhibit tumor development. He et al (45) and Dang et al (46) reported that blocking IL-1β
signaling potentially suppressed HCC invasion and metastasis and
improved the tumor microenvironment. Nevertheless, a study by Hage
et al (47) found that
dual blockade of IL-1β and IL-18 completely eliminated the anti-HCC
effects of sorafenib. Metformin suppressed the development of HCC
by cleaving IL-1β and IL-18 through activation of the pyroptosis
signaling molecule caspase-1 (48). In addition, it has been
demonstrated that CHMP3 knockdown significantly enhanced cellular
pyroptosis and IL-1 release (49). In the present study, it was found
that the pathway was aberrantly inhibited and that its inhibition
may be an oncogenic signaling pathway in liver cancer. When
knocking down CHMP3, the caspase-1 precursor did not change, yet
its cleaved form increased. Meanwhile electron microscopy revealed
changes in the liver cancer cells experiencing pyroptosis. It was
hypothesized that knockdown of CHMP3 may be related to caspase-1
activation. Cleavage of GSDMD disrupted the integrity of the cell
membrane, which caused the occurrence of cellular pyroptosis. The
mechanisms involved in the present study are illustrated in
Fig. 8. Therefore, CHMP3 may
control the inflammasome activation and inflammatory factor
release.
The pyroptosis is able to inhibit tumor migration
and invasion to some extent (40,50). It was found that liver cancer
cells with different expression levels of CHMP3 also differed in
their ability to migrate and invade. Highly aggressive and
metastatic are the most important causes of death for HCC (51). The expression of epithelial
calreticulin E-cadherin was reduced, while the expression of
N-cadherin, MMP9 and Vimentin was increased when tumor cells
metastasized (52-54). N-cadherin is able to increase MMP9
expression which then degrades the extracellular matrix, the
vascular basement membrane and the N-cadherin-catenin complex,
thereby enabling the promotion of aggressive metastasis of tumor
cells (55,56). It was identified that CHMP3 is
involved in the migration and invasion of liver cancer by
regulating the expression of E-cadherin, N-cadherin, MMP9 and
vimentin.
Certain limitations should be considered in the
interpretation of the present study. The expression levels of
pyroptosis-related genes in resected tumor tissues by western
blotting were only detected. It is necessary to continue to explore
and improve the relevant experiments in future studies. It is worth
considering that the increase in the number of liver cancer cells
observed after CHMP3 overexpression may be largely attributable to
a decrease in cell death. To improve understanding of the
mechanisms involved, the expression of genes known to directly
regulate cell proliferation, including CDK, Cyclin D, Rb and E2F,
could be investigated in subsequent studies. Evaluating the
expression levels of these genes would provide valuable insights
into the possible involvement of CHMP3 in the regulation of cell
proliferation in liver cancer. Since CHMP3 regulates
caspase-1-mediated pyroptosis, which may directly affect tumor
immune checkpoint mechanisms, the reduction in tumor size in mice
resulting from knockdown of CHMP3 can be attributed, at least in
part, to this, rather than to the mere inhibition of cell
proliferation.
In conclusion, the function and mechanism of action
of CHMP3 in liver cancer was demonstrated. Therefore, CHMP3 may
serve as a new prognostic biomarker and therapeutic candidate.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ wrote the manuscript. YZ, SY, WD and JW performed
the experiments and generated the figures. SW, SB, XZ, ZZ and YS
collected the public and clinic data. JK participated in analyzing
and interpreting the data and revised and reviewed the manuscript.
All authors made critical revisions, read and approved the final
manuscript. All authors confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Written informed consent was obtained by all
patients whose tissue samples were collected prior to enrolment.
Human studies (approval no. 2022PS785K) and animal experiments
(approval no. 2022PS779K) were approved by the Ethics Committee of
Shengjing Hospital, China Medical University (Shenyang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
CHMP3
|
charged multivesicular body protein
3
|
|
ESCRT
|
endosomal sorting complexes required
for transport
|
|
HCC
|
hepatocellular carcinoma
|
|
DEGs
|
differentially expressed genes
|
|
TCGA
|
The Cancer Genome Atlas
|
|
IL
|
interleukin
|
|
NLRP3
|
NOD-like receptor thermal protein
domain associated protein 3
|
|
GSDMD
|
Gasdermin D
|
|
GEPIA
|
Gene Expression Profiling Interactive
Analysis
|
|
HPA
|
Human Protein Atlas
|
|
siRNA
|
small interfering RNA
|
|
PPI
|
protein-protein interaction
|
|
OS
|
overall survival
|
|
AYC
|
Ac-YVAN-CMK
|
|
MMP9
|
matrix metalloproteinase-9
|
|
TEM
|
transmission electron microscopy
|
|
TNBC
|
triple-negative breast cancer
|
Acknowledgments
Not applicable.
Funding
The present study was supported by the Liaoning Science and
Technology Plan Project (grant no. 2021JH2/10300118) and the 345
Talent Project Program of China Medical University Shengjing
Hospital (grant no. 2022-50A).
References
|
1
|
Chapiro J, Wood LD, Lin M, Duran R,
Cornish T, Lesage D, Charu V, Schernthaner R, Wang Z, Tacher V, et
al: Radiologic-pathologic analysis of contrast-enhanced and
diffusion-weighted MR imaging in patients with HCC after TACE:
Diagnostic accuracy of 3D quantitative image analysis. Radiology.
273:746–758. 2014.
|
|
2
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
|
|
3
|
Tao S, Liang S, Zeng T and Yin D:
Epigenetic modification-related mechanisms of hepatocellular
carcinoma resistance to immune checkpoint inhibition. Front
Immunol. 13:10436672023.
|
|
4
|
Glantzounis GK, Paliouras A, Stylianidi
MC, Milionis H, Tzimas P, Roukos D, Pentheroudakis G and Felekouras
E: The role of liver resection in the management of intermediate
and advanced stage hepatocellular carcinoma. A systematic review.
Eur J Surg Oncol. 44:195–208. 2018.
|
|
5
|
Cammarota A, Zanuso V, Manfredi GF, Murphy
R, Pinato DJ and Rimassa L: Immunotherapy in hepatocellular
carcinoma: How will it reshape treatment sequencing? Ther Adv Med
Oncol. 15:175883592211480292023.
|
|
6
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
|
|
7
|
Fang Y, Tian S, Pan Y, Li W, Wang Q, Tang
Y, Yu T, Wu X, Shi Y, Ma P and Shu Y: Pyroptosis: A new frontier in
cancer. Biomed Pharmacother. 121:1095952020.
|
|
8
|
Ouyang X, Zhou J, Lin L, Zhang Z, Luo S
and Hu D: Pyroptosis, inflammasome, and gasdermins in tumor
immunity. Innate Immun. 29:3–13. 2023.
|
|
9
|
Shi J, Zhao Y, Wang K, Shi X, Wang Y,
Huang H, Zhuang Y, Cai T, Wang F and Shao F: Cleavage of GSDMD by
inflammatory caspases determines pyroptotic cell death. Nature.
526:660–665. 2015.
|
|
10
|
Vande Walle L and Lamkanfi M: Pyroptosis.
Curr Biol. 26:R568–R572. 2016.
|
|
11
|
Yu P, Zhang X, Liu N, Tang L, Peng C and
Chen X: Pyroptosis: Mechanisms and diseases. Signal Transduct
Target Ther. 6:1282021.
|
|
12
|
Kang R, Zeng L, Zhu S, Xie Y, Liu J, Wen
Q, Cao L, Xie M, Ran Q, Kroemer G, et al: Lipid peroxidation drives
gasdermin D-mediated pyroptosis in lethal polymicrobial sepsis.
Cell Host Microbe. 24:97–108.e4. 2018.
|
|
13
|
Pilla DM, Hagar JA, Haldar AK, Mason AK,
Degrandi D, Pfeffer K, Ernst RK, Yamamoto M, Miao EA and Coers J:
Guanylate binding proteins promote caspase-11-dependent pyroptosis
in response to cytoplasmic LPS. Proc Natl Acad Sci USA.
111:6046–6051. 2014.
|
|
14
|
Wei Q, Guo P, Mu K, Zhang Y, Zhao W, Huai
W, Qiu Y, Li T, Ma X, Liu Y, et al: Estrogen suppresses
hepatocellular carcinoma cells through ERβ-mediated upregulation of
the NLRP3 inflammasome. Lab Invest. 95:804–816. 2015.
|
|
15
|
Lata S, Schoehn G, Solomons J, Pires R,
Göttlinger HG and Weissenhorn W: Structure and function of
ESCRT-III. Biochem Soc Trans. 37:156–160. 2009.
|
|
16
|
Babst M, Katzmann DJ, Estepa-Sabal EJ,
Meerloo T and Emr SD: Escrt-III: An endosome-associated
heterooligomeric protein complex required for mvb sorting. Dev
Cell. 3:271–282. 2002.
|
|
17
|
Babst M, Wendland B, Estepa EJ and Emr SD:
The Vps4p AAA ATPase regulates membrane association of a Vps
protein complex required for normal endosome function. Embo J.
17:2982–2993. 1998.
|
|
18
|
Bishop N and Woodman P: ATPase-defective
mammalian VPS4 localizes to aberrant endosomes and impairs
cholesterol trafficking. Mol Biol Cell. 11:227–239. 2000.
|
|
19
|
Saksena S, Wahlman J, Teis D, Johnson AE
and Emr SD: Functional reconstitution of ESCRT-III assembly and
disassembly. Cell. 136:97–109. 2009.
|
|
20
|
Teis D, Saksena S and Emr SD: Ordered
assembly of the ESCRT-III complex on endosomes is required to
sequester cargo during MVB formation. Dev Cell. 15:578–589.
2008.
|
|
21
|
Wang Z and Wang X: miR-122-5p promotes
aggression and epithelial-mesenchymal transition in triple-negative
breast cancer by suppressing charged multivesicular body protein 3
through mitogen-activated protein kinase signaling. J Cell Physiol.
235:2825–2835. 2020.
|
|
22
|
Zhou Y, Zheng J, Bai M, Gao Y and Lin N:
Effect of pyroptosis-related genes on the prognosis of breast
cancer. Front Oncol. 12:9481692022.
|
|
23
|
Niu D, Chen Y, Mi H, Mo Z and Pang G: The
epiphany derived from T-cell-inflamed profiles: Pan-cancer
characterization of CD8A as a biomarker spanning clinical
relevance, cancer prognosis, immunosuppressive environment, and
treatment responses. Front Genet. 13:9744162022.
|
|
24
|
Li C, Liang H, Bian S, Hou X and Ma Y:
Construction of a prognosis model of the pyroptosis-related gene in
multiple myeloma and screening of core genes. ACS Omega.
7:34608–34620. 2022.
|
|
25
|
Li Y, Li Y, Zhang X, Duan X, Feng H, Yu Z
and Gao Y: A novel association of pyroptosis-related gene signature
with the prognosis of hepatocellular carcinoma. Front Oncol.
12:9868272022.
|
|
26
|
Man SM and Kanneganti TD: Regulation of
inflammasome activation. Immunol Rev. 265:6–21. 2015.
|
|
27
|
Kovacs SB and Miao EA: Gasdermins:
Effectors of pyroptosis. Trends Cell Biol. 27:673–684. 2017.
|
|
28
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.
|
|
29
|
Liu L, Li Y, Cao D, Qiu S, Li Y, Jiang C,
Bian R, Yang Y, Li L, Li X, et al: SIRT3 inhibits gallbladder
cancer by induction of AKT-dependent ferroptosis and blockade of
epithelial-mesenchymal transition. Cancer Lett. 510:93–104.
2021.
|
|
30
|
Jia C, Chen H, Zhang J, Zhou K, Zhuge Y,
Niu C, Qiu J, Rong X, Shi Z, Xiao J, et al: Role of pyroptosis in
cardiovascular diseases. Int Immunopharmacol. 67:311–318. 2019.
|
|
31
|
Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X,
Zhao W, Huai W, Guo P and Han L: Deregulation of the NLRP3
inflammasome in hepatic parenchymal cells during liver cancer
progression. Lab Invest. 94:52–62. 2014.
|
|
32
|
Yan Z, Da Q, Li Z, Lin Q, Yi J, Su Y, Yu
G, Ren Q, Liu X, Lin Z, et al: Inhibition of NEK7 suppressed
hepatocellular carcinoma progression by mediating cancer cell
pyroptosis. Front Oncol. 12:8126552022.
|
|
33
|
Chen W, Quan Y, Fan S, Wang H, Liang J,
Huang L, Chen L, Liu Q, He P and Ye Y: Exosome-transmitted circular
RNA hsa_circ_0051443 suppresses hepatocellular carcinoma
progression. Cancer Lett. 475:119–128. 2020.
|
|
34
|
Zhang Q, Chen L, Gao M, Wang S, Meng L and
Guo L: Molecular docking and in vitro experiments verified that
kaempferol induced apoptosis and inhibited human HepG2 cell
proliferation by targeting BAX, CDK1, and JUN. Mol Cell Biochem.
478:767–780. 2023.
|
|
35
|
Sun X, Zhong X, Ma W, Feng W, Huang Q, Ma
M, Lv M, Hu R, Han Z, Li J and Zhou X: Germacrone induces
caspase-3/GSDME activation and enhances ROS production, causing
HepG2 pyroptosis. Exp Ther Med. 24:4562022.
|
|
36
|
Hou J, Zhao R, Xia W, Chang CW, You Y, Hsu
JM, Nie L, Chen Y, Wang YC, Liu C, et al: PD-L1-mediated gasdermin
C expression switches apoptosis to pyroptosis in cancer cells and
facilitates tumour necrosis. Nat Cell Biol. 22:1264–1275. 2020.
|
|
37
|
Zhang T, Li Y, Zhu R, Song P, Wei Y, Liang
T and Xu G: Transcription factor p53 suppresses tumor growth by
prompting pyroptosis in non-small-cell lung cancer. Oxid Med Cell
Longev. 2019:87468952019.
|
|
38
|
Tan Y, Chen Q, Li X, Zeng Z, Xiong W, Li
G, Li X, Yang J, Xiang B and Yi M: Pyroptosis: A new paradigm of
cell death for fighting against cancer. J Exp Clin Cancer Res.
40:1532021.
|
|
39
|
Hsu SK, Li CY, Lin IL, Syue WJ, Chen YF,
Cheng KC, Teng YN, Lin YH, Yen CH and Chiu CC: Inflammation-related
pyroptosis, a novel programmed cell death pathway, and its
crosstalk with immune therapy in cancer treatment. Theranostics.
11:8813–8835. 2021.
|
|
40
|
Cui J, Zhou Z, Yang H, Jiao F, Li N, Gao
Y, Wang L, Chen J and Quan M: MST1 suppresses pancreatic cancer
progression via ROS-induced pyroptosis. Mol Cancer Res.
17:1316–1325. 2019.
|
|
41
|
Teng JF, Mei QB, Zhou XG, Tang Y, Xiong R,
Qiu WQ, Pan R, Law BY, Wong VK, Yu CL, et al: Polyphyllin VI
induces caspase-1-mediated pyroptosis via the induction of
ROS/NF-κB/NLRP3/GSDMD signal axis in non-small cell lung cancer.
Cancers (Basel). 12:1932020.
|
|
42
|
Rébé C and Ghiringhelli F: Interleukin-1β
and cancer. Cancers (Basel). 12:17912020.
|
|
43
|
Yan W, Chang Y, Liang X, Cardinal JS,
Huang H, Thorne SH, Monga SP, Geller DA, Lotze MT and Tsung A:
High-mobility group box 1 activates caspase-1 and promotes
hepatocellular carcinoma invasiveness and metastases. Hepatology.
55:1863–1875. 2012.
|
|
44
|
Lopez-Pastrana J, Ferrer LM, Li YF, Xiong
X, Xi H, Cueto R, Nelson J, Sha X, Li X, Cannella AL, et al:
Inhibition of caspase-1 activation in endothelial cells improves
angiogenesis: A NOVEL THERAPEUTIC POTENTIAL FOR ISCHEMIA. J Biol
Chem. 290:17485–17494. 2015.
|
|
45
|
He Q, Liu M, Huang W, Chen X, Zhang B,
Zhang T, Wang Y, Liu D, Xie M, Ji X, et al: IL-1β-induced elevation
of solute carrier family 7 member 11 promotes hepatocellular
carcinoma metastasis through up-regulating programmed death ligand
1 and colony-stimulating factor 1. Hepatology. 74:3174–3193.
2021.
|
|
46
|
Dang Y, Chen J, Feng W, Qiao C, Han W, Nie
Y, Wu K, Fan D and Xia L: Interleukin 1β-mediated HOXC10
overexpression promotes hepatocellular carcinoma metastasis by
upregulating PDPK1 and VASP. Theranostics. 10:3833–3848. 2020.
|
|
47
|
Hage C, Hoves S, Strauss L, Bissinger S,
Prinz Y, Pöschinger T, Kiessling F and Ries CH: Sorafenib induces
pyroptosis in macrophages and triggers natural killer cell-mediated
cytotoxicity against hepatocellular carcinoma. Hepatology.
70:1280–1297. 2019.
|
|
48
|
Shen Z, Zhou H, Li A, Wu T, Ji X, Guo L,
Zhu X, Zhang D and He X: Metformin inhibits hepatocellular
carcinoma development by inducing apoptosis and pyroptosis through
regulating FOXO3. Aging (Albany NY). 13:22120–22133. 2021.
|
|
49
|
Rühl S, Shkarina K, Demarco B, Heilig R,
Santos JC and Broz P: ESCRT-dependent membrane repair negatively
regulates pyroptosis downstream of GSDMD activation. Science.
362:956–960. 2018.
|
|
50
|
Tang Q, Li W, Zheng X, Ren L, Liu J, Li S,
Wang J and Du G: MELK is an oncogenic kinase essential for
metastasis, mitotic progression, and programmed death in lung
carcinoma. Signal Transduct Target Ther. 5:2792020.
|
|
51
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014.
|
|
52
|
Kuphal S and Bosserhoff AK: Influence of
the cytoplasmic domain of E-cadherin on endogenous N-cadherin
expression in malignant melanoma. Oncogene. 25:248–259. 2006.
|
|
53
|
Mondal S, Adhikari N, Banerjee S, Amin SA
and Jha T: Matrix metalloproteinase-9 (MMP-9) and its inhibitors in
cancer: A minireview. Eur J Med Chem. 194:1122602020.
|
|
54
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011.
|
|
55
|
Walker A, Frei R and Lawson KR: The
cytoplasmic domain of N-cadherin modulates MMP-9 induction in oral
squamous carcinoma cells. Int J Oncol. 45:1699–1706. 2014.
|
|
56
|
Cao ZQ, Wang Z and Leng P: Aberrant
N-cadherin expression in cancer. Biomed Pharmacother.
118:1093202019.
|