Since 1960, when chemical modifications of RNA were
first documented in detail as forms of epistatic modifications,
>170 forms of RNA modifications, which have the role of
maintaining mRNA stability and are involved in precursor shearing,
transport and translation initiation of mRNA (1), have been identified (2). Of these modifications, the
methylation modification of the nitrogen atom at position 6 of the
RNA molecule adenine, i.e., N6-methyladenosine (m6A) modification,
is the most widespread in eukaryotes (3). Although RNA methylation was
discovered >60 years ago, owing to technical limitations,
previous studies on epigenetic modifications in tumors have mostly
focused on DNA methylation and histone modifications (4-7).
It was not until 2012 when Meyer et al (8) first applied m6A-sequencing (seq)
technology to determine the overall m6A levels of human and mouse
genes at the transcriptional level on a large scale, and until
2015, when Linder et al (9) first used m6A
individual-nucleotide-resolution cross-linking and
immunoprecipitation-sequencing technology to achieve single-base
m6A level detection that m6A research was gradually developed.
An increasing number of studies have found that
abnormal m6A methylation is closely associated with the
development, metastatic recurrence and treatment failure of various
tumors (10,11).
Due to the relevance of epigenetic modifications in
the extracellular environment, the complex heterogeneity of tumors
and the potential of immunotherapy, research on m6A and tumors has
predominantly focused on the immune microenvironment and
immunotherapy (12,13). Indeed, only a few studies have
really focused on the relationship between chemoresistance and m6A
methylation, and even less on a specific single cancer entity.
Epigenetic modifications of RNA are associated with
sensitivity to multiple chemotherapeutic agents (17). Therefore, the present study
focused on the relationship between aberrant m6A methylation
modifications and chemotherapeutic response mechanisms in CRC
(18-20) and various potential therapeutic
measures targeting the m6A methylation process to seek novel
strategies to improve the therapeutic effect pertaining to CRC.
Numerous scholars posit that the reversible and
protein-regulatory properties of m6A methylation offer a promising
approach to overcoming the current shortcomings in multiple cancer
therapies (8). However, the real
application of m6A methylation and clinical transformation must
also solve a myriad of issues, including the following problems: i)
M6A methylation as the most extensive RNA modification-how to focus
and target the key molecules; ii) how to screen out the most
accurately targeted drug candidates in each cancer species; and
iii) how to specifically target the regulatory axis involved in m6A
methylation to reverse drug resistance in tumors (21).
The key m6A methylation enzymes can be classified
into methyltransferases, methyl recognition enzymes and
demethylases.
M6A methyltransferases mostly function as complexes
[the m6A methyltransferase complex (MTC). The MTC mainly comprises
methyltransferase-like 3 (METTL3), METTL14, vir like m6A
methyltransferase associated (VIRMA), WT1 associated protein
(WTAP), zinc finger CCCH-type containing 13 (ZC3H13), RNA binding
motif protein 15 and Cbl proto-oncogene like 1 (HAKAI) (22,23). METTL3 was the first m6A
methyltransferase identified and the only catalytic subunit of MTC,
indicating that the presence and activation of METTL3 are the basis
for m6A methylation. Although METTL3 can act independently of other
m6A methyltransferases, its catalytic activity is much weaker than
that of the MTC formed by wrapping it with other transferases
(24). METTL14, as the primary
RNA binding platform, forms a complex with METTL3 through 10
positively charged binding sites (Fig. 1), activating and enhancing METTL3
activity and promoting the recognition of RNA substrates, and thus
enhancing MTC methylation efficiency (25,26). By contrast, WTAP is essential for
promoting the enrichment of METTL3, METTL14 and other
methyltransferases, transporting MTC into the nucleus and
stabilizing MTC activity in the organism (27,28). VIRMA and ZC3H13 are relatively
newly discovered m6A methyltransferases. VIRMA, also known as
KIAA1429, which are the largest molecular weight proteins of MTC
components known to date. VIRMA may be a scaffold for the MTC
structure, connecting the immobilized WTAP, HAKAI and ZC3H13 to
form an envelope structure capable of accommodating the
METTL3-METTL14 complex (29).
ZC3H13 is also a linking protein that bridges WTAP to the
METTL3-METTL14 complex and facilitates the recognition of RNA
substrates.
It was discovered that m6A methyltransferases are
extensively involved in various stages of CRC, including tumor
stemness, microenvironmental remodeling, drug resistance,
metastasis and recurrence. Some of the roles and related pathway
molecules are shown in Table
I.
For a long time, the methyltransferase METTL3 has
been regarded as a pro-oncogene (43,44). However, in recent years, a limited
number of studies have found that under specific conditions, such
as tumor starvation, METTL3 may also inhibit tumor development and
development through the activation of the p38 signaling pathway and
interference with the cell cycle to bring tumor cells into a
dormant state (38,45). More interestingly, it has been
suggested that METTL3 can function not only as a methyltransferase,
but also as a methyl recognition enzyme independent of YTH
N6-methyladenosine RNA binding protein F (YTHDF)1, capturing the
recognition of mRNAs undergoing m6A methylation modification in the
cytoplasm, promoting the recruitment of E74 like ETS transcription
factor 3 (elF3) (46), drive
β-linked protein trans-activation and upregulate c-Myc, VEGF,
cyclin D7, MMP-3, c-Jun and other key genes of intestinal cancer
malignant phenotypes (47).
Of note, m6A methylation is not only related to the
modern medical concept, but also similar to certain Traditional
Chinese Medicine (TCM) concepts (48,49).
In the concept of TCM, the function of various parts
of the body is associated and influenced by the external
environment. This is similar to epigenetics. If the external evil
is more severe than the physical weakness, it is classified as
excessive syndrome (ES) according to TCM concepts; if the two are
of the same magnitude, it is classified as deficiency and excessive
syndrome (DES); and if the physical weakness is more pronounced, it
is defined as deficiency syndrome (DS). The early and early-middle
stages of CRC often appear as ES or DES, while the late stages are
DS (50,51). Under the guidance of these
theories, TCM treatments need to be selected according to the
different manifestations of different syndrome types in patients,
so as to be used correctly (52).
It has been found that patients with CRC with
different syndrome types have differences in gene expression, and
elF3 and the downstream factors it regulates are typical
representatives. High expression of the keratin 19, keratin 18,
keratin 8, ELF3 and serpin family E member 1 genes is a potential
marker to identify the TCM evidence type in CRC and, high
expression of the ELF3 gene in CRC with DES or ES. High expression
of mucin 2 and regenerating family member 4 in DES was mainly
related to cell growth, as well as the MAPK and cyclic adenosine
3′,5′-monophosphate signaling pathways, while high expression of
collagen type I alpha 2 chain and periostin genes in ES was mainly
related to angiogenesis and the PI3K/AKT pathway and the
caveolae-associated protein 2 and glutathione peroxidase 1 genes
were highly expressed in DS and are mainly related to vomiting,
platelet catabolism and endocytosis (50,53).
Whether m6A methyltransferases can function as
methylation recognition enzymes or other epigenetic factors remains
to be elucidated. The combination of classical medical theories,
including TCM and epigenetic modifications, which are emerging
molecular biology concepts, provides new perspectives for antitumor
therapy and warrants further exploration (54,55).
The main function of methyl recognition enzymes is
to recognize bases that undergo modification, thus activating
downstream pathways and participating in biological processes,
including mRNA translation, transcription, fission, and
degradation. The core members include YTHDF1-3 and YTHDC1-3
(56-58). YTHDF2 and YTHDF3 accelerate the
degradation and fission of modified mRNAs by recruiting the C-C
chemokine receptor 4-negative regulator of transcription
deadenosylation complex (59) and
upregulating forkhead box O3 (FOXO3) expression (60), respectively.
Although the exploration of RNA methylases is not
widely performed in clinical practice, it is not only limited to
discovery. There are also some small molecule drugs for methylation
recognition enzymes such as YTHDF1, which is helpful for subsequent
research.
The group of the above-mentioned study also
developed a lipid nanoparticle-encapsulated Rho guanine nucleotide
exchange factor 1 small interfering RNA drug for in vivo
tumor therapy. YTHDF1 also up-regulates the transcription factor
glucocorticoid modulatory element binding protein 2 of the
adhesion-regulating molecule-1/nuclear factor kappa pathway,
thereby activating the pathway to resist apoptosis and drive CRC
progression (62). Ni et
al (63) demonstrated that
YTHDF3 is a novel target for the Yes-associated protein 1 (YAP)
signaling pathway. A long noncoding RNA called growth
arrest-specific transcript 5 (GAS5) binds directly to YAP,
promoting YAP phosphorylation and attenuating YAP-mediated YTHDF3
transcription and allowing YTHDF3 to reversibly and selectively
bind to GAS5, which undergoes m6A-methylation to trigger its decay
and form a negative feedback loop. Although methylation recognition
enzymes have been relatively poorly studied, they are indispensable
for the proper binding of methyltransferases to the modified
site.
Insulin-like growth factor 2 mRNA binding proteins
(IGF2BPs) are also significant methyl recognition enzymes. They can
specifically recognize and then bind directly to the m6A
modification site, subsequently upregulating SOX2 to activate CRC
stem cells (CSCs) (64). IGF2BP2
also can induce chemoresistance in CRC cells by activating the
PI3K/AKT signaling pathway and enhancing aerobic glycolysis
(65). However, these functions
are still inseparable from the upstream regulation of METTL3.
Thus far, the knowledge of demethylases is limited
and the only known demethylases are fat mass and obesity associated
(FTO) and alkylation repair homolog 5 (ALKBH5). Demethylases are
key to the reversibility of m6A methylation (66,67) and are able to complete the
demethylation process by oxidizing m6A to N6-hydroxymethyladenosine
and N6-formyladenosine (68-70). Conventionally, the reversibility
of methylation modifications creates a window for reversing
chemoresistance and increasing demethylase activity to tilt the
reaction rate toward demethylation, thus facilitating the
re-sensitization of drug-resistant cells to chemotherapy.
In brief, the m6A methylation key enzymes are widely
involved in all aspects of CRC development and have considerable
interventional value. Accordingly, it is reasonable to ask what the
role of these enzymes is in the current mainstream chemotherapeutic
benefits pertaining to CRC, and what the potential opportunities
for intervention are.
The mechanisms by which chemoresistance occurs in
CRC are complex and there is no shortage of links involving m6A
methylation enzymes. In particular, the mechanism of chemotherapy
resistance due to an altered tumor microenvironment regulated by
m6A methylation key enzymes has gained attention. The known
molecular mechanisms underlying the m6A methylation of key enzymes
that regulate common chemotherapeutic drug resistance in pan-cancer
are presented in Fig. 2.
As the study of m6A methyltransferase is the most
extensive and because 5-FU and OXA are the most commonly used
chemotherapeutic agents in the treatment of CRC, the resistance
mechanisms associated with m6A methylation were found to be highly
dependent on the regulation of METTL3 (54) (as shown in Fig. 2). Therefore, this study will
further focus on the 5-FU and OXA resistance mechanisms of m6A
methyltransferases, particularly METTL3, in CRC. Due to the high
complexity of the many genes and proteins involved and the
associated mechanisms, the present review categorizes these
mechanistic processes into drug transporter protein-related, stem
cell activity, EMT process and cellular autophagy, as illustrated
in Fig. 3 and below.
Tumor microenvironment refers to the special
survival environment of tumor cells with their surrounding
chemokines, apoptotic factors, immune cells, adhesion proteins,
joint composition of metabolic disorders, defective apoptosis, lack
of oxygen and acidification. Such a special environment is
conducive to the evasion of tumor cells in response to therapy and
epigenetic alterations such as m6A methylation induce the
inevitable adaptation of tumor cells to this special
environment.
Autophagy refers to the formation of autophagic
lysosomes stimulated by various adhesion and apoptotic factors that
transfer intracellular material into lysosomes for degradation.
When the drug-tolerant persister (DTP) state is activated, key
autophagy genes such as unc-51 like autophagy activating kinase 1
and autophagy related 2A are upregulated and when autophagy
inhibitors are administered, cells exit the DTP state and cannot
survive chemotherapy (85,86).
A study from China published in 2020 found that
METTL3 regulates the TGF-β and Snail pathways to affect EMT in
intestinal cancer cells (88).
EMT is also an important mechanism for the development of
chemoresistance in CRC, indicating a decrease in adhesion factors
such as epithelial surface calmodulin, detachment from basement
membrane connections, and cytoskeletal and morphological
convergence to mesenchymal features. Various molecules such as COP9
signalosome subunit 8, runt-related transcription factor 1,
ubiquitin specific peptidase 43, histone deacetylase 2 and tumor
microenvironment-associated fibroblasts can regulate EMT in CRC
cells (89). When EMT occurs, the
malignant phenotype of intestinal cancer becomes more prominent and
resistant to OXA and 5-FU (90).
CSCs are a special class of cells with self-healing
and multi-differentiation potential. The most clinically relevant
feature of CSCs is their ability to metastasize and evade standard
chemotherapy (91). The
differentiation homeostasis of this class of cells is usually
regulated by the Wnt/β-catenin signaling pathway. Sec62 is a key
protein in this pathway, and its increased expression enhances the
sphere-forming ability of CSCs. Liu et al (92) found that Sec62 expression was
positively correlated with METTL3 expression and that the
METTL3-mediated accumulation of m6A methylation, which upregulates
Sec62 levels, competitively disrupted the binding of β-linked
protein and oncogene adenomatosis polyposis coli, leading to 5-FU
resistance. METTL3-mediated m6A methylation accumulation also
drives the methyltransferase recruitment of histone third subunit
IV lysine (H4K3) to the promoter of leucine rich repeat containing
G protein-coupled receptor 5 (LGR5), a colon cancer stem cell
marker, leading to irinotecan and OXA resistance (93). Bai et al (94) found that silencing YTHDF1
downregulated CSC markers, including CD133, CD44, ALDH1,
octamer-binding transcription factor 4 and LGR5, and inhibited
Wnt/β-linked protein pathway activity in ex vivo
experiments. Accordingly, it is proposed that YTHDF1 recognizes and
promotes the translation of m6A-modified frizzled class receptor 9
and Wnt6 mRNAs, leading to aberrant activation of Wnt/β-linked
protein signaling and ultimately affecting tumorigenicity, stem
cell-like activity and response to chemical agents in CRC as a
scientific hypothesis.
In other words, some researchers have found that m6A
methylation key enzymes can regulate the expression of various CSC
markers and, consequently, affect CSC activity and sensitivity to
chemotherapeutic agents. However, the specific mechanisms involved
remain to be further elucidated.
Defects in the DDR pathway are a hallmark of
genomically unstable tumors, and up to 15-20% of CRCs have DDR
pathway defects (95,96). In the past, it was often thought
that DDR pathway defects were mainly associated with peroxisome
proliferators-activated receptors (PPAR) inhibitor efficacy.
Furthermore, defects in the DDR pathway were primarily associated
with the efficacy of PPAR inhibitors. Mechanism of action of common
chemotherapeutic agents, including 5-FU and topoisomerase
inhibitors, is related to the DDR pathway. METTL3 in the
physiological state can promote cell repair after physical damage,
including that caused by UV light. By contrast, METTL3 is
pathologically upregulated in tumors, leading to an excessive rate
of homologous recombination repair (HR), non-homologous end
recombination and failure of chemotherapeutic drugs (97). Li et al (98) found that after METTL3 silencing
treatment of HCT-8/5-FU resistant colon cancer cells, the resistant
cells were able to be re-sensitized to 5-FU, while RAD51-associated
protein 1, a key factor on the HR pathway, was downregulated. Zhang
et al (99) discovered
that METTL3 knockdown in OXA-resistant colon cancer cells also
improved the chemosensitivity of resistant cells, while METTL3
overexpression restored the drug-resistant phenotype. Further
sequencing suggested that the differentially expressed genes were
mainly enriched in classical drug resistance pathways, including
the Hippo and DDR pathways.
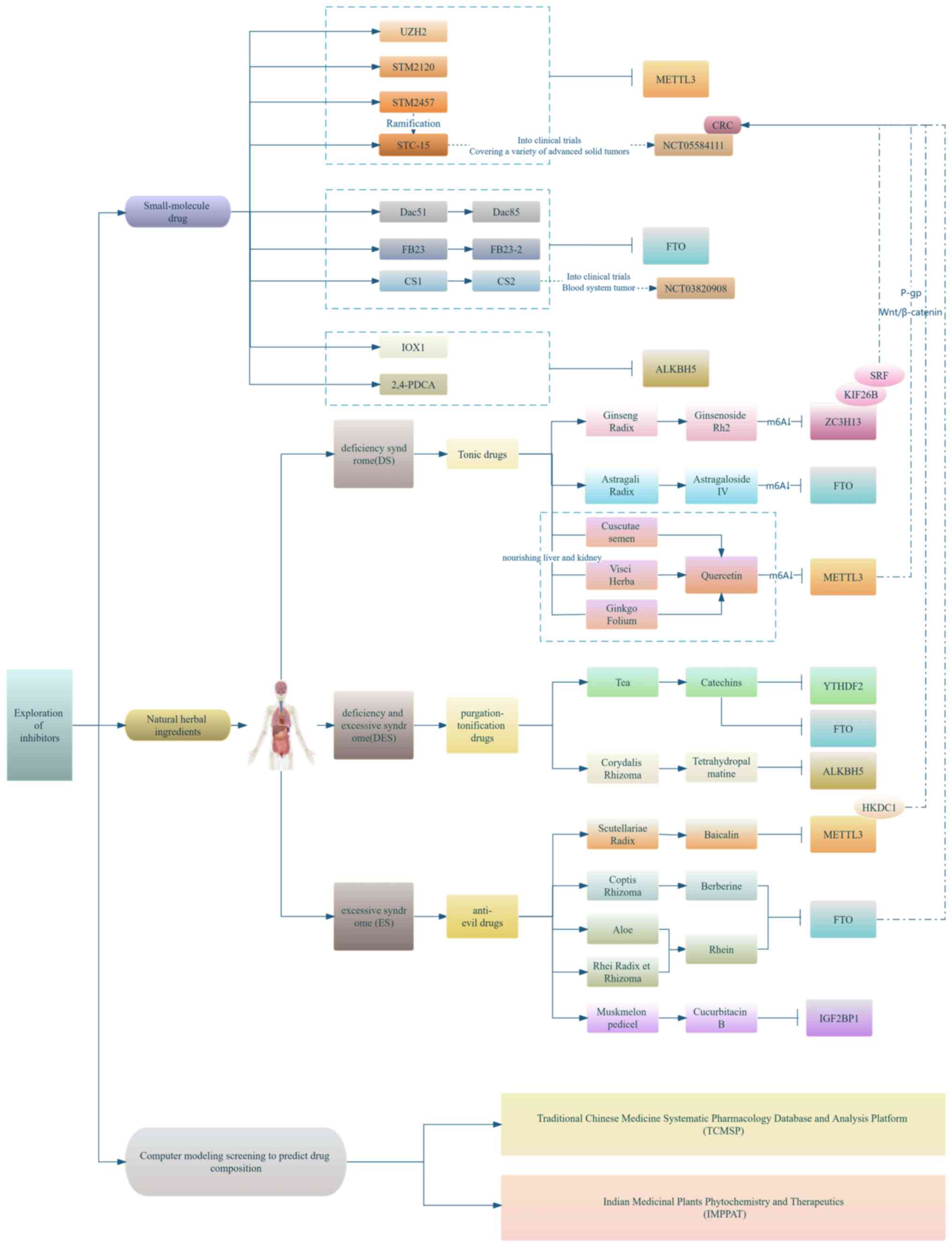
Currently, the exploration of METTL3 inhibitors is
being conducted mainly from the following three perspectives:
Application of natural drug ingredients, small molecule drug
synthesis development and clinical trials, and the combination of
huge data with computer model predictions and screening of drug
targets and pathways (as shown in Fig. 4).
Although m6A methylesterase inhibitors are currently
less used in the treatment of CRC, the METTL3 inhibitor STM2457 has
been reported to exhibit significant therapeutic effects in acute
myeloid leukemia (100) and was
able to reverse chemoresistance in small cell lung cancer (101) However, its derivative STC-15,
the first clinical candidate for an oral agent targeting METTL3, is
in phase I clinical trials for patients with advanced solid tumors
(NCT05584111). Therefore, researchers have begun to explore the
application of key m6A methylation enzyme modulators in solid
tumors from multiple perspectives, including natural drug
components, small molecule targeted drugs and programmed analysis
of potential drug components using computerized big data. CRC has
also received attention as a highly prevalent solid tumor, and an
urgent clinical need to improve the efficacy of its treatments has
emerged.
It has been suggested that most of the natural drug
components that can regulate the action of m6A methylation key
enzymes are polyphenols, alkaloids, flavonoids, anthraquinones and
terpenoids (102). For instance,
curcumin is a phenolic compound extracted from turmeric root that
can reduce the expression of ALKBH5 and enhance the translation of
tumor necrosis factor receptor associated factor 4 (TRAF4),
prompting TRAF4 to bind to YTHDF6, a methyl recognition enzyme with
m6A, and improve the efficiency of m6A methylation modification
(103). Curcumin was also able
to drive the conversion of microtubule-associated protein LC3-I to
LC3-II or upregulate Beclin-1 to induce autophagy in CRC cells,
reduce CSC generation and re-sensitize drug-resistant cells to 5-FU
and OXA (104,105). The combination of curcumin with
another polyphenol, resveratrol, could alter the distribution of
key m6A methylation enzymes such as METTL3 and YTHDF2, reduce the
overall m6A methylation level in the intestine and improve
intestinal mucosal integrity (106).
An increasing number of studies support the
anti-tumor effects of herbal medicines as epigenetic modification
modulators, including turmeric, tannin, yam and Kalanchoe
pinnata (107), which can
target DNA (cytosine-5-)-methyltransferase 1 to inhibit P65 gene
methylation and interfere with CRC cell infiltration and migration
(108). Although the
relationship between Chinese medicine and m6A methylation has not
yet been fully elucidated, some studies have found that herbal
extracts can regulate DNA methylation, including chaihu saponin
(109), quercetin (110) and catechins (111). Flavonoid components such as
chaihu saponin, quercetin and catechin can also increase the
expression of METTL3 and METTL14 and decrease the methyl
recognition proteins such as FTO and ALKBH5 (109,112).
As for small-molecule drug development, besides
STM2457 and its derivative STC-15, modulators targeting methylation
recognition enzymes are also under active development. An inhibitor
of FTO called CS1 inhibited the proliferation of six CRC cell
lines, including HT-29, COLO, HCT-116 and 5-FU-resistant cell lines
(HCT-116/5FU). It also induced G25/M phase cell cycle arrest and
promoted apoptosis of HCT-116 cells by downregulating doublecortin
domain containing 2C (113).
M6A methylation modifications are a bridge between
the tumor microenvironment and phenotypic alterations, including
chemoresistance, the mechanisms of which are complex, and the
knowledge of epigenetics is yet to be refined. Of note, it has been
found that various epigenetic modifications are not completely
isolated from each other and there is a strong correlation between
m6A methylation and DNA methylation, which can specifically lead to
increased DNA methylation at proximal sites when METTL3 is
depleted, resulting in downregulated chromatin binding levels of
fragile X mental retardation, autosomal homolog 1 and tet
methylcytosine dioxygenase 1 (116). Although current studies on m6A
methylation have started from a combination of various
high-throughput screens, molecular deconstruction techniques and
metabolic alteration assays, the specific mechanisms of the
interactions between the various aspects of m6A methylation, how to
accurately achieve a homeostatic balance between methylation and
demethylation, and the specific mechanisms of migration, invasion
resistance and other malignant phenotypes of tumors induced by key
m6A methylation enzymes, as well as the regulation of the tumor
microenvironment remain unresolved. More importantly, some of the
currently developed m6A modification inhibitors and activators have
poor target specificity, therapeutic efficacy, and safety and
pharmacokinetic limitations. The large-scale development and
application of artificial intelligence provide new opportunities to
assist in the preclinical screening of more efficient drug
components. More m6A methylation key enzyme modulators can enter
clinical trials in the near future, providing an effective way to
improve the treatment of CRC.
Of note, the present study had certain limitations.
First, the specific expression of each key m6A methylase was not
summarized and discussed. The expression of these methylation key
enzymes in different cancer species and their close relation to
clinical characteristics and prognosis was not focused on, and may
be discussed in the future. The present review focused on
summarizing some directions of the basic findings of each
methylation key enzyme in CRC chemoresistance and the subsequent
clinical transformation. Furthermore, the relationship between the
complex immune microenvironment of CRC and key m6A methylases was
not summarized and discussed. The present review focused on the
relationship of the immune microenvironment with m6A modification,
and indeed, considerable studies have focused on this aspect. This
is another topic that is not very closely related to the focus of
this paper, but the knowledge system is huge, so it was not
discussed. In the future, a focus will be placed on the role of m6A
in the immune microenvironment.
Not applicable.
SCY and HYJ designed the study. SCY and TXK
consulted the literature on the composition and role of key enzymes
for m6A methylation. SCY, HYY and ZZ collected information on the
relationship between key m6A enzymes and CRC and tracked the
development of m6A-targeted drugs. ZYJ and FMZ categorized the
retrieved materials according to year and differences in the types
of key enzymes involved, and were responsible for the organization
and design of the tables. SCY wrote this manuscript. HX, GT and LM
critically reviewed the manuscript. HX directed and participated in
information gathering, image conception and design, drawing figures
and subsequent revision of the manuscript. All authors have read
and approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This study was supported by the National Natural Science Funds
for Distinguished Young Scholar (grant no. 82204696), Grant of
Nanjing University of Chinese Medicine (grant no. XPT82204696), the
Nanjing Medical Science and Technology Development Project (grant
nos. YKK21199 and YKK21201) and the Nanjing Health Young Talents
Project of Jiangsu Province (grant no. QRX17187).
|
1
|
Gilbert WV, Bell TA and Schaening C:
Messenger RNA modifications: Form, distribution, and function.
Science. 352:1408–1412. 2016.
|
|
2
|
Cohn WE: Pseudouridine, a carbon-carbon
linked ribonucleoside in ribonucleic acids: Isolation, structure,
and chemical characteristics. J Biol Chem. 235:1488–1498. 1960.
|
|
3
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6) A promotes cap-independent translation. Cell. 163:999–1010.
2015.
|
|
4
|
Dawson MA and Kouzarides T: Cancer
epigenetics: from mechanism to therapy. Cell. 150:12–27. 2012.
|
|
5
|
Nishiyama A and Nakanishi M: Navigating
the DNA methylation landscape of cancer. Trends Genet.
37:1012–1027. 2021.
|
|
6
|
Huang W, Li H, Yu Q, Xiao W and Wang DO:
LncRNA-mediated DNA methylation: An emerging mechanism in cancer
and beyond. J Exp Clin Cancer Res. 41:1002022.
|
|
7
|
Esteller M: Cancer epigenomics: DNA
methylomes and histone-modification maps. Nat Rev Genet. 8:286–298.
2007.
|
|
8
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012.
|
|
9
|
Linder B, Grozhik AV, Olarerin-George AO,
Meydan C, Mason CE and Jaffrey SR: Single-nucleotide-resolution
mapping of m6A and m6Am throughout the transcriptome. Nat Methods.
12:767–772. 2015.
|
|
10
|
Sun T, Wu R and Ming L: The role of m6A
RNA methylation in cancer. Biomed Pharmacother. 112:1086132019.
|
|
11
|
Song N, Cui K, Zhang K, Yang J, Liu J,
Miao Z, Zhao F, Meng H, Chen L, Chen C, et al: The role of m6A RNA
methylation in cancer: Implication for nature products anti-cancer
research. Front Pharmacol. 13:9333322022.
|
|
12
|
Li X, Ma S, Deng Y, Yi P and Yu J:
Targeting the RNA m6A modification for cancer
immunotherapy. Mol Cancer. 21:762022.
|
|
13
|
An Y and Duan H: The role of m6A RNA
methylation in cancer metabolism. Mol Cancer. 21:142022.
|
|
14
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
|
|
15
|
Shi JF, Wang L, Ran JC, Wang H, Liu CC,
Zhang HZ, Yang L, Shi SS, Jiang LM, Fan JH, et al: Clinical
characteristics, medical service utilization, and expenditure for
colorectal cancer in China, 2005 to 2014: Overall design and
results from a multicenter retrospective epidemiologic survey.
Cancer. 127:1880–1893. 2021.
|
|
16
|
Alfarouk KO, Stock CM, Taylor S, Walsh M,
Muddathir AK, Verduzco D, Bashir AH, Mohammed OY, Elhassan GO,
Harguindey S, et al: Resistance to cancer chemotherapy: Failure in
drug response from ADME to P-gp. Cancer Cell Int. 15:712015.
|
|
17
|
Liu K, Ouyang QY, Zhan Y, Yin H, Liu BX,
Tan LM, Liu R, Wu W and Yin JY: Pharmacoepitranscriptomic landscape
revealing m6A modification could be a drug-effect biomarker for
cancer treatment. Mol Ther Nucleic Acids. 28:464–476. 2022.
|
|
18
|
Pan J, Liu F, Xiao X, Xu R, Dai L, Zhu M,
Xu H, Xu Y, Zhao A, Zhou W, et al: METTL3 promotes colorectal
carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp
Clin Cancer Res. 41:192022.
|
|
19
|
Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X,
Zhang X, Cao Y, Ma D, Zhu X, et al: m6A-dependent
glycolysis enhances colorectal cancer progression. Mol Cancer.
19:722020.
|
|
20
|
Peng W, Li J, Chen R, Gu Q, Yang P, Qian
W, Ji D, Wang Q, Zhang Z, Tang J and Sun Y: Upregulated METTL3
promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK
signaling pathway. J Exp Clin Cancer Res. 38:3932019.
|
|
21
|
Liu Z, Zou H, Dang Q, Xu H, Liu L, Zhang
Y, Lv J, Li H, Zhou Z and Han X: Biological and pharmacological
roles of m6A modifications in cancer drug resistance.
Mol Cancer. 21:2202022.
|
|
22
|
Yan J, Liu F, Guan Z, Yan X, Jin X, Wang
Q, Wang Z, Yan J, Zhang D, Liu Z, et al: Structural insights into
DNA N6-adenine methylation by the MTA1 complex. Cell
Discov. 9:82023.
|
|
23
|
Yan X, Pei K, Guan Z, Liu F, Yan J, Jin X,
Wang Q, Hou M, Tang C and Yin P: AI-empowered integrative
structural characterization of m6A methyltransferase
complex. Cell Res. 32:1124–1127. 2022.
|
|
24
|
Zhou Z, Lv J, Yu H, Han J, Yang X, Feng D,
Wu Q, Yuan B, Lu Q and Yang H: Mechanism of RNA modification
N6-methyladenosine in human cancer. Mol Cancer. 19:1042020.
|
|
25
|
Wang Z, Pan Z, Adhikari S, Harada BT, Shen
L, Yuan W, Abeywardana T, Al-Hadid Q, Stark JM, He C, et al:
m6 A deposition is regulated by PRMT1-mediated arginine
methylation of METTL14 in its disordered C-terminal region. EMBO J.
40:e1063092021.
|
|
26
|
Liu X, Du Y, Huang Z, Qin H, Chen J and
Zhao Y: Insights into roles of METTL14 in tumors. Cell Prolif.
55:e131682022.
|
|
27
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014.
|
|
28
|
Fan Y, Li X, Sun H, Gao Z, Zhu Z and Yuan
K: Role of WTAP in cancer: From mechanisms to the therapeutic
potential. Biomolecules. 12:12242022.
|
|
29
|
Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang
Z, Cheng T, Gao M, Shu X, Ma H, et al: VIRMA mediates preferential
m6A mRNA methylation in 3′UTR and near stop codon and
associates with alternative polyadenylation. Cell Discov.
4:102018.
|
|
30
|
Huang L, Liang D, Zhang Y, Chen X, Chen J,
Wen C, Liu H, Yang X, Yang X and Lin S: METTL3 promotes colorectal
cancer metastasis by promoting the maturation of pri-microRNA-196b.
J Cancer Res Clin Oncol. 149:5095–5108. 2023.
|
|
31
|
Zhang F, Su T and Xiao M: RUNX3-regulated
circRNA METTL3 inhibits colorectal cancer proliferation and
metastasis via miR-107/PER3 axis. Cell Death Dis. 13:5502022.
|
|
32
|
Shi K, Yang S, Chen C, Shao B, Guo Y, Wu
X, Zhao L, Yang X, Zhang Q, Yuan W and Sun Z: RNA
methylation-mediated LINC01559 suppresses colorectal cancer
progression by regulating the miR-106b-5p/PTEN axis. Int J Biol
Sci. 18:3048–3065. 2022.
|
|
33
|
Xu Q, Lu X, Li J, Feng Y, Tang J, Zhang T,
Mao Y, Lan Y, Luo H, Zeng L, et al: Fusobacterium nucleatum induces
excess methyltransferase-like 3-mediated microRNA-4717-3p
maturation to promote colorectal cancer cell proliferation. Cancer
Sci. 113:3787–3800. 2022.
|
|
34
|
Chen S, Zhang L, Li M, Zhang Y, Sun M,
Wang L, Lin J, Cui Y, Chen Q, Jin C, et al: Fusobacterium nucleatum
reduces METTL3-mediated m6A modification and contributes
to colorectal cancer metastasis. Nat Commun. 13:12482022.
|
|
35
|
Chen H, Gao S, Liu W, Wong CC, Wu J, Wu J,
Liu D, Gou H, Kang W, Zhai J, et al: RNA
N6-methyladenosine methyltransferase METTL3 facilitates
colorectal cancer by activating the m6A-GLUT1-mTORC1
axis and is a therapeutic target. Gastroenterology.
160:1284–1300.e16. 2021.
|
|
36
|
Lu S, Han L, Hu X, Sun T, Xu D, Li Y, Chen
Q, Yao W, He M, Wang Z, et al: N6-methyladenosine reader IMP2
stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect:
Implication in colorectal cancer. J Hematol Oncol. 14:1882021.
|
|
37
|
Sun L, Wan A, Zhou Z, Chen D, Liang H, Liu
C, Yan S, Niu Y, Lin Z, Zhan S, et al: RNA-binding protein RALY
reprogrammes mitochondrial metabolism via mediating miRNA
processing in colorectal cancer. Gut. 70:1698–1712. 2021.
|
|
38
|
Deng R, Cheng Y, Ye S, Zhang J, Huang R,
Li P, Liu H, Deng Q, Wu X, Lan P and Deng Y: m6A
methyltransferase METTL3 suppresses colorectal cancer proliferation
and migration through p38/ERK pathways. Onco Targets Ther.
12:4391–4402. 2019.
|
|
39
|
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B,
Li C, Sun L, Qin J, Xu T, et al: METTL14-mediated
N6-methyladenosine modification of SOX4 mRNA inhibits tumor
metastasis in colorectal cancer. Mol Cancer. 19:1062020.
|
|
40
|
Hou Y, Zhang X, Yao H, Hou L, Zhang Q, Tao
E, Zhu X, Jiang S, Ren Y, Hong X, et al: METTL14 modulates
glycolysis to inhibit colorectal tumorigenesis in p53-wild-type
cells. EMBO Rep. 24:e563252023.
|
|
41
|
Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang
K, Go MY, Ng SC, Chan FK, Sung JJ and Yu J: Carbonic anhydrase IV
inhibits colon cancer development by inhibiting the Wnt signalling
pathway through targeting the WTAP-WT1-TBL1 axis. Gut.
65:1482–1493. 2016.
|
|
42
|
Li Y, He L, Wang Y, Tan Y and Zhang F:
N6-methyladenosine methyltransferase KIAA1429 elevates
colorectal cancer aerobic glycolysis via HK2-dependent manner.
Bioengineered. 13:11923–11932. 2022.
|
|
43
|
Wei X, Huo Y, Pi J, Gao Y, Rao S, He M,
Wei Q, Song P, Chen Y, Lu D, et al: METTL3 preferentially enhances
non-m6A translation of epigenetic factors and promotes
tumourigenesis. Nat Cell Biol. 24:1278–1290. 2022.
|
|
44
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13:1172020.
|
|
45
|
Collignon E, Cho B, Furlan G,
Fothergill-Robinson J, Martin SB, McClymont SA, Ross RL, Limbach PA
and Ramalho-Santos M: m6A RNA methylation orchestrates
transcriptional dormancy during paused pluripotency. Nat Cell Biol.
25:1279–1289. 2023.
|
|
46
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016.
|
|
47
|
Wang JL, Chen ZF, Chen HM, Wang MY, Kong
X, Wang YC, Sun TT, Hong J, Zou W, Xu J and Fang JY: Elf3 drives
β-catenin transactivation and associates with poor prognosis in
colorectal cancer. Cell Death Dis. 5:e12632014.
|
|
48
|
Xiang Y, Guo Z, Zhu P, Chen J and Huang Y:
Traditional Chinese medicine as a cancer treatment: Modern
perspectives of ancient but advanced science. Cancer Med.
8:1958–1975. 2019.
|
|
49
|
Xin T, Zhang Y, Pu X, Gao R, Xu Z and Song
J: Trends in herbgenomics. Sci China Life Sci. 62:288–308.
2019.
|
|
50
|
Wang YN, Zou M, Wang D, Zhang ZK, Qu LP,
Xu J, Shi CD and Gao F: An exploratory study on TCM syndrome
differentiation in preoperative patients with colorectal cancer
assisted by laboratory indicators. Heliyon. 8:e102072022.
|
|
51
|
Wang CY, Ding HZ, Tang X and Li ZG:
Comparative analysis of immune function, hemorheological
alterations and prognosis in colorectal cancer patients with
different traditional Chinese medicine syndromes. Cancer Biomark.
21:701–710. 2018.
|
|
52
|
Zhang X, Wang X, Shi R, Ran X, He X and
Dou D: Effective substances and mechanism of red ginseng on rats
with spleen-deficiency syndrome based on the substance and energy
metabolism as well as the 'brain-gut' axis. J Ethnopharmacol.
311:1164382023.
|
|
53
|
Lu Y, Zhou C, Zhu M, Fu Z, Shi Y, Li M,
Wang W, Zhu S, Jiang B, Luo Y and Su S: Traditional chinese
medicine syndromes classification associates with tumor cell and
microenvironment heterogeneity in colorectal cancer: A single cell
RNA sequencing analysis. Chin Med. 16:1332021.
|
|
54
|
Liu WW, Zhang ZY, Wang F and Wang H:
Emerging roles of m6A RNA modification in cancer therapeutic
resistance. Exp Hematol Oncol. 12:212023.
|
|
55
|
Ma SC, Zhang JQ, Yan TH, Miao MX, Cao YM,
Cao YB, Zhang LC and Li L: Novel strategies to reverse
chemoresistance in colorectal cancer. Cancer Med. 12:11073–11096.
2023.
|
|
56
|
Zou Z, Sepich-Poore C, Zhou X, Wei J and
He C: The mechanism underlying redundant functions of the YTHDF
proteins. Genome Biol. 24:172023.
|
|
57
|
Chen L, Gao Y, Xu S, Yuan J, Wang M, Li T
and Gong J: N6-methyladenosine reader YTHDF family in biological
processes: Structures, roles, and mechanisms. Front Immunol.
14:11626072023.
|
|
58
|
Sarraf G and Chhabra R: Emerging role of
mRNA methylation in regulating the hallmarks of cancer. Biochimie.
206:61–72. 2023.
|
|
59
|
Chang G, Shi L, Ye Y, Shi H, Zeng L,
Tiwary S, Huse JT, Huo L, Ma L, Ma Y, et al: YTHDF3 Induces the
translation of m6A-enriched gene transcripts to promote
breast cancer brain metastasis. Cancer Cell. 38:857–871. 2020.
|
|
60
|
Hao W, Dian M, Zhou Y, Zhong Q, Pang W, Li
Z, Zhao Y, Ma J, Lin X, Luo R, et al: Autophagy induction promoted
by m6A reader YTHDF3 through translation upregulation of
FOXO3 mRNA. Nat Commun. 13:58452022.
|
|
61
|
Wang S, Gao S, Zeng Y, Zhu L, Mo Y, Wong
CC, Bao Y, Su P, Zhai J, Wang L, et al: N6-methyladenosine reader
YTHDF1 promotes ARHGEF2 translation and RhoA signaling in
colorectal cancer. Gastroenterology. 162:1183–1196. 2022.
|
|
62
|
Ning Z, Wu Z, Zhang F, Yang M, Lu Z, Yu B,
Long F, Guo Y, Yang K, Hu G, et al: GMEB2 promotes the growth of
colorectal cancer by activating ADRM1 transcription and NF-κB
signalling and is positively regulated by the m6A reader
YTHDF1. Cancers (Basel). 14:60462022.
|
|
63
|
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou
A, Liu J, Che L and Li J: Long noncoding RNA GAS5 inhibits
progression of colorectal cancer by interacting with and triggering
YAP phosphorylation and degradation and is negatively regulated by
the m6A reader YTHDF3. Mol Cancer. 18:1432019.
|
|
64
|
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN,
Chen ZH, Zeng ZL, Wang F, Zheng J, et al: METTL3 facilitates tumor
progression via an m6A-IGF2BP2-dependent mechanism in
colorectal carcinoma. Mol Cancer. 18:1122019.
|
|
65
|
Liu X, He H, Zhang F, Hu X, Bi F, Li K, Yu
H, Zhao Y, Teng X, Li J, et al: m6A methylated EphA2 and VEGFA
through IGF2BP2/3 regulation promotes vasculogenic mimicry in
colorectal cancer via PI3K/AKT and ERK1/2 signaling. Cell Death
Dis. 13:4832022.
|
|
66
|
Zhao Y and Peng H: The role of
N6-methyladenosine (m6A) methylation
modifications in hematological malignancies. Cancers (Basel).
14:3322022.
|
|
67
|
Qu X and Shi Y: Bile reflux and bile acids
in the progression of gastric intestinal metaplasia. Chin Med J
(Engl). 135:1664–1672. 2022.
|
|
68
|
Li H, Wu H, Wang Q, Ning S, Xu S and Pang
D: Dual effects of N6-methyladenosine on cancer
progression and immunotherapy. Mol Ther Nucleic Acids. 24:25–39.
2021.
|
|
69
|
Liu C, Yang S, Zhang Y, Wang C, Du D, Wang
X, Liu T and Liang G: Emerging roles of N6-methyladenosine
demethylases and its interaction with environmental toxicants in
digestive system cancers. Cancer Manag Res. 13:7101–7114. 2021.
|
|
70
|
Uddin MB, Wang Z and Yang C:
Dysregulations of functional RNA modifications in cancer, cancer
stemness and cancer therapeutics. Theranostics. 10:3164–3189.
2020.
|
|
71
|
Relier S, Ripoll J, Guillorit H, Amalric
A, Achour C, Boissière F, Vialaret J, Attina A, Debart F, Choquet
A, et al: FTO-mediated cytoplasmic m6Am
demethylation adjusts stem-like properties in colorectal cancer
cell. Nat Commun. 12:17162021.
|
|
72
|
Ruan DY, Li T, Wang YN, Meng Q, Li Y, Yu
K, Wang M, Lin JF, Luo LZ, Wang DS, et al: FTO downregulation
mediated by hypoxia facilitates colorectal cancer metastasis.
Oncogene. 40:5168–5181. 2021.
|
|
73
|
Ballester V, Taylor WR, Slettedahl SW,
Mahoney DW, Yab TC, Sinicrope FA, Boland CR, Lidgard GP,
Cruz-Correa MR, Smyrk TC, et al: Novel methylated DNA markers
accurately discriminate Lynch syndrome associated colorectal
neoplasia. Epigenomics. 12:2173–2187. 2020.
|
|
74
|
Li N, Kang Y, Wang L, Huff S, Tang R, Hui
H, Agrawal K, Gonzalez GM, Wang Y, Patel SP and Rana TM: ALKBH5
regulates anti-PD-1 therapy response by modulating lactate and
suppressive immune cell accumulation in tumor microenvironment.
Proc Natl Acad Sci USA. 117:20159–20170. 2020.
|
|
75
|
Wang YQ, Li HZ, Gong WW, Chen YY, Zhu C,
Wang L, Zhong JM and Du LB: Cancer incidence and mortality in
Zhejiang Province, Southeast China, 2016: A population-based study.
Chin Med J (Engl). 134:1959–1966. 2021.
|
|
76
|
Chen HM, Lin CC, Chen WS, Jiang JK, Yang
SH, Chang SC, Ho CL, Yang CC, Huang SC, Chao Y, et al: Insulin-like
growth factor 2 mRNA-binding protein 1 (IGF2BP1) is a prognostic
biomarker and associated with chemotherapy responsiveness in
colorectal cancer. Int J Mol Sci. 22:69402021.
|
|
77
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022.
|
|
78
|
Yang W, Wang Y, Tao C, Li Y, Cao S and
Yang X: CRNDE silencing promotes apoptosis and enhances cisplatin
sensitivity of colorectal carcinoma cells by inhibiting the
Akt/mTORC1-mediated Warburg effect. Oncol Lett. 23:702022.
|
|
79
|
Wei TT, Lin YT, Tang SP, Luo CK, Tsai CT,
Shun CT and Chen CC: Metabolic targeting of HIF-1α potentiates the
therapeutic efficacy of oxaliplatin in colorectal cancer. Oncogene.
39:414–427. 2020.
|
|
80
|
Peng L, Jiang J, Chen HN, Zhou L, Huang Z,
Qin S, Jin P, Luo M, Li B, Shi J, et al: Redox-sensitive
cyclophilin A elicits chemoresistance through realigning cellular
oxidative status in colorectal cancer. Cell Rep. 37:1100692021.
|
|
81
|
Zhang K, Zhang T, Yang Y, Tu W, Huang H,
Wang Y, Chen Y, Pan K and Chen Z:
N6-methyladenosine-mediated LDHA induction potentiates
chemoresistance of colorectal cancer cells through metabolic
reprogramming. Theranostics. 12:4802–4817. 2022.
|
|
82
|
Yang Z, Quan Y, Chen Y, Huang Y, Huang R,
Yu W, Wu D, Ye M, Min Z and Yu B: Knockdown of RNA
N6-methyladenosine methyltransferase METTL3 represses Warburg
effect in colorectal cancer via regulating HIF-1α. Signal Transduct
Target Ther. 6:892021.
|
|
83
|
Han S, Zhu L, Zhu Y, Meng Y, Li J, Song P,
Yousafzai NA, Feng L, Chen M, Wang Y, et al: Targeting
ATF4-dependent pro-survival autophagy to synergize glutaminolysis
inhibition. Theranostics. 11:8464–8479. 2021.
|
|
84
|
Chen P, Liu XQ, Lin X, Gao LY, Zhang S and
Huang X: Targeting YTHDF1 effectively re-sensitizes
cisplatin-resistant colon cancer cells by modulating GLS-mediated
glutamine metabolism. Mol Ther Oncolytics. 20:228–239. 2021.
|
|
85
|
Rehman SK, Haynes J, Collignon E, Brown
KR, Wang Y, Nixon AML, Bruce JP, Wintersinger JA, Singh Mer A, Lo
EBL, et al: Colorectal Cancer cells enter a diapause-like DTP state
to survive chemotherapy. Cell. 184:226–242.e21. 2021.
|
|
86
|
Wang Y, Yang L, Zhang J, Zhou M, Shen L,
Deng W, Liang L, Hu R, Yang W, Yao Y, et al: Radiosensitization by
irinotecan is attributed to G2/M phase arrest, followed by enhanced
apoptosis, probably through the ATM/Chk/Cdc25C/Cdc2 pathway in
p53-mutant colorectal cancer cells. Int J Oncol. 53:1667–1680.
2018.
|
|
87
|
Lin Z, Wan AH, Sun L, Liang H, Niu Y, Deng
Y, Yan S, Wang QP, Bu X, Zhang X, et al: N6-methyladenosine
demethylase FTO enhances chemo-resistance in colorectal cancer
through SIVA1-mediated apoptosis. Mol Ther. 31:517–534. 2023.
|
|
88
|
Li J, Chen F, Peng Y, Lv Z, Lin X, Chen Z
and Wang H: N6-methyladenosine regulates the expression and
secretion of TGFbeta1 to affect the epithelial-mesenchymal
transition of cancer cells. Cells. 9:2962020.
|
|
89
|
Yang M, Sun M and Zhang H: The interaction
between epigenetic changes, EMT, and exosomes in predicting
metastasis of colorectal cancers (CRC). Front Oncol.
12:8798482022.
|
|
90
|
Sabouni E, Nejad MM, Mojtabavi S, Khoshduz
S, Mojtabavi M, Nadafzadeh N, Nikpanjeh N, Mirzaei S, Hashemi M,
Aref AR, et al: Unraveling the function of epithelial-mesenchymal
transition (EMT) in colorectal cancer: Metastasis, therapy
response, and revisiting molecular pathways. Biomed Pharmacother.
160:1143952023.
|
|
91
|
Oskarsson T, Batlle E and Massagué J:
Metastatic stem cells: Sources, niches, and vital pathways. Cell
Stem Cell. 14:306–321. 2014.
|
|
92
|
Liu X, Su K, Sun X, Jiang Y, Wang L, Hu C,
Zhang C, Lu M, Du X and Xing B: Sec62 promotes stemness and
chemoresistance of human colorectal cancer through activating
Wnt/β-catenin pathway. J Exp Clin Cancer Res. 40:1322021.
|
|
93
|
Zhang Y, Kang M, Zhang B, Meng F, Song J,
Kaneko H, Shimamoto F and Tang B: m6A
modification-mediated CBX8 induction regulates stemness and
chemosensitivity of colon cancer via upregulation of LGR5. Mol
Cancer. 18:1852019.
|
|
94
|
Bai Y, Yang C, Wu R, Huang L, Song S, Li
W, Yan P, Lin C, Li D and Zhang Y: YTHDF1 regulates tumorigenicity
and cancer stem cell-like activity in human colorectal carcinoma.
Front Oncol. 9:3322019.
|
|
95
|
Mauri G, Arena S, Siena S, Bardelli A and
Sartore-Bianchi A: The DNA damage response pathway as a land of
therapeutic opportunities for colorectal cancer. Ann Oncol.
31:1135–1147. 2020.
|
|
96
|
Catalano F, Borea R, Puglisi S, Boutros A,
Gandini A, Cremante M, Martelli V, Sciallero S and Puccini A:
Targeting the DNA damage response pathway as a novel therapeutic
strategy in colorectal cancer. Cancers (Basel). 14:13882022.
|
|
97
|
Zhang C, Chen L, Peng D, Jiang A, He Y,
Zeng Y, Xie C, Zhou H, Luo X, Liu H, et al: METTL3 and
N6-methyladenosine promote homologous recombination-mediated repair
of DSBs by modulating DNA-RNA hybrid accumulation. Mol Cell.
79:425–442.e7. 2020.
|
|
98
|
Li M, Xia M, Zhang Z, Tan Y, Li E, Guo Z,
Fang M, Zhu Y and Hu Z: METTL3 antagonizes 5-FU chemotherapy and
confers drug resistance in colorectal carcinoma. Int J Oncol.
61:1062022.
|
|
99
|
Zhang SH QQLP: METTL3-mediated m6A
modification in oxaliplatin resistance in colorectal cancer. J
Chongqing Med Univ. 47:pp. 941–947. 2022, (In Chinese). https://kns.cnki.net/kcms/detail/detail.aspx?FileName=ZQYK202208010&DbName=DKFX2022.
View Article : Google Scholar
|
|
100
|
Yankova E, Blackaby W, Albertella M, Rak
J, De Braekeleer E, Tsagkogeorga G, Pilka ES, Aspris D, Leggate D,
Hendrick AG, et al: Small-molecule inhibition of METTL3 as a
strategy against myeloid leukaemia. Nature. 593:597–601. 2021.
|
|
101
|
Sun Y, Shen W, Hu S, Lyu Q, Wang Q, Wei T,
Zhu W and Zhang J: METTL3 promotes chemoresistance in small cell
lung cancer by inducing mitophagy. J Exp Clin Cancer Res.
42:652023.
|
|
102
|
Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M,
Lyu WY, Qi Q, Tiwari AK, Chen JX, Zhang DM and Chen ZS: m6A
modification: recent advances, anticancer targeted drug discovery
and beyond. Mol Cancer. 21:522022.
|
|
103
|
Chen Y, Wu R, Chen W, Liu Y, Liao X, Zeng
B, Guo G, Lou F, Xiang Y, Wang Y and Wang X: Curcumin prevents
obesity by targeting TRAF4-induced ubiquitylation in m6
A-dependent manner. EMBO Rep. 22:e521462021.
|
|
104
|
Weng W and Goel A: Curcumin and colorectal
cancer: An update and current perspective on this natural medicine.
Semin Cancer Biol. 80:73–86. 2022.
|
|
105
|
Su P, Yang Y, Wang G, Chen X and Ju Y:
Curcumin attenuates resistance to irinotecan via induction of
apoptosis of cancer stem cells in chemoresistant colon cancer
cells. Int J Oncol. 53:1343–1353. 2018.
|
|
106
|
Gan Z, Wei W, Wu J, Zhao Y, Zhang L, Wang
T and Zhong X: Resveratrol and curcumin improve intestinal mucosal
integrity and decrease m6A RNA methylation in the
intestine of weaning piglets. ACS Omega. 4:17438–17446. 2019.
|
|
107
|
Hernández-Caballero ME, Sierra-Ramírez JA,
Villalobos-Valencia R and Seseña-Méndez E: Potential of
Kalanchoe pinnata as a cancer treatment adjuvant and an
epigenetic regulator. Molecules. 27:64252022.
|
|
108
|
Zhu D, Li A, Lv Y and Fan Q: Traditional
Chinese medicine: A class of potentially reliable epigenetic drugs.
Front Pharmacol. 13:9070312022.
|
|
109
|
Sun K, Du Y, Hou Y, Zhao M, Li J, Du Y,
Zhang L, Chen C, Yang H, Yan F and Su R: Saikosaponin D exhibits
anti-leukemic activity by targeting FTO/m6A signaling.
Theranostics. 11:5831–5846. 2021.
|
|
110
|
Xu W, Xie S, Chen X, Pan S, Qian H and Zhu
X: Effects of quercetin on the efficacy of various chemotherapeutic
drugs in cervical cancer cells. Drug Des Devel Ther. 15:577–588.
2021.
|
|
111
|
Wu R, Yao Y, Jiang Q, Cai M, Liu Q, Wang Y
and Wang X: Epigallocatechin gallate targets FTO and inhibits
adipogenesis in an mRNA m6A-YTHDF2-dependent manner. Int
J Obes (Lond). 42:1378–1388. 2018.
|
|
112
|
Jiao Y, Williams A and Wei N: Quercetin
ameliorated insulin resistance via regulating METTL3-mediated
N6-methyladenosine modification of PRKD2 mRNA in skeletal muscle
and C2C12 myocyte cell line. Nutr Metab Cardiovasc Dis.
32:2655–2668. 2022.
|
|
113
|
Phan T, Nguyen VH, Su R, Li Y, Qing Y, Qin
H, Cho H, Jiang L, Wu X, Chen J, et al: Targeting fat mass and
obesity-associated protein mitigates human colorectal cancer growth
in vitro and in a murine model. Front Oncol. 13:10876442023.
|
|
114
|
Du Y, Yuan Y, Xu L, Zhao F, Wang W, Xu Y
and Tian X: Discovery of METTL3 small molecule inhibitors by
virtual screening of natural products. Front Pharmacol.
13:8781352022.
|
|
115
|
Manna S, Samal P, Basak R, Mitra A, Roy
AK, Kundu R, Ahir A, Roychowdhury A and Hazra D: Amentoflavone and
methyl hesperidin, novel lead molecules targeting epitranscriptomic
modulator in acute myeloid leukemia: In silico drug screening and
molecular dynamics simulation approach. J Mol Model. 29:92022.
|
|
116
|
Deng S, Zhang J, Su J, Zuo Z, Zeng L, Liu
K, Zheng Y, Huang X, Bai R, Zhuang L, et al: RNA m6A
regulates transcription via DNA demethylation and chromatin
accessibility. Nat Genet. 54:1427–1437. 2022.
|