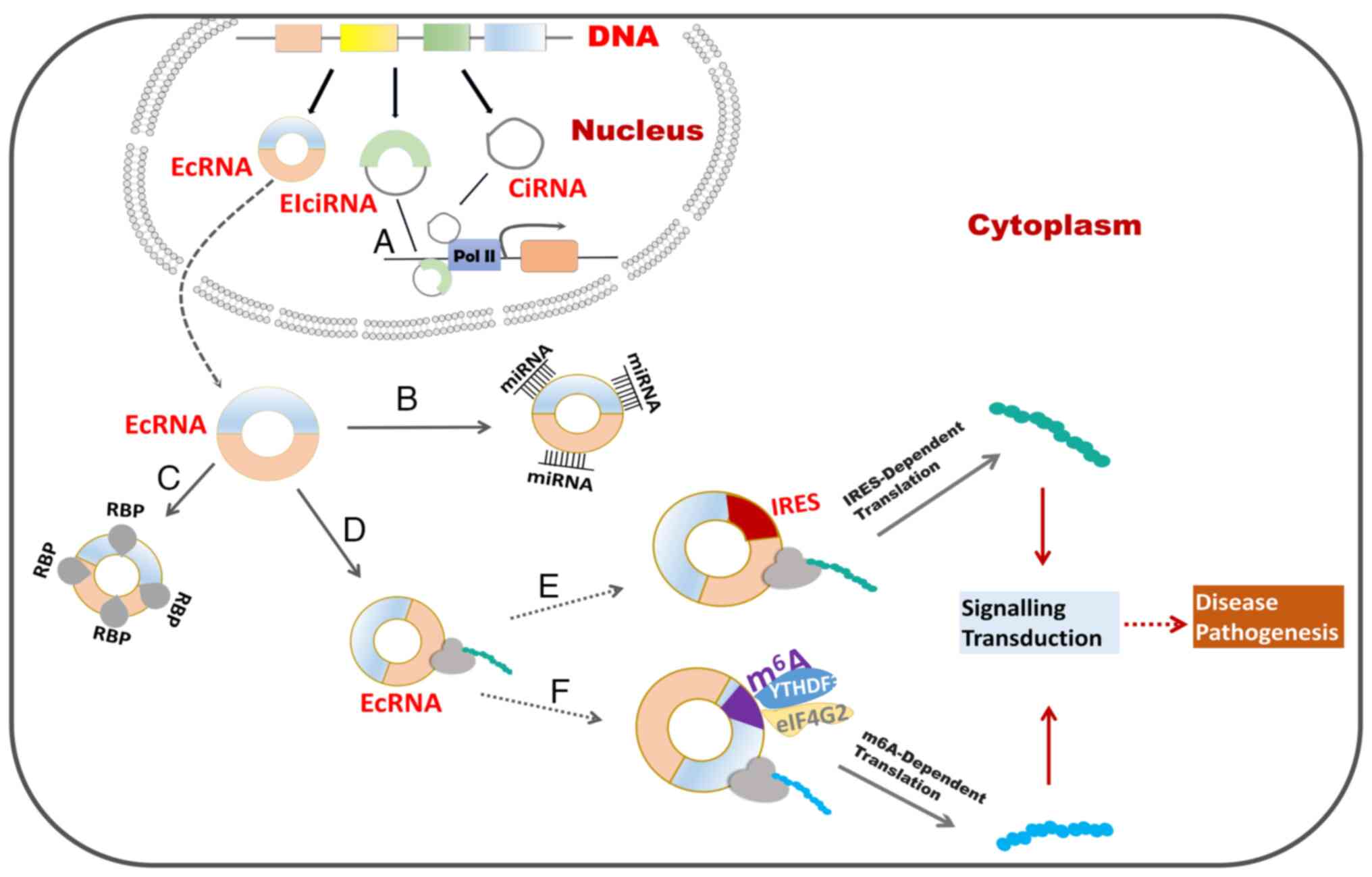
Circular RNAs (circRNAs) are distinctive noncoding
RNAs characterized by a covalently closed circular structure that
lack the typical translation initiation elements (5′ cap and 3′
polyA tail) of linear RNAs (1).
Originally identified in plant viruses (2), circRNAs were initially presumed to
lack functional roles (3-5). Subsequently, circRNAs were found in
multiple eukaryotes ranging from yeast to humans (6,7).
With advances in technology and experimental methods, previous
studies have confirmed the multifunctional roles of circRNAs in
various physiological and pathological processes (8-10).
CircRNAs are widely involved in different types of coronary artery
diseases, such as myocardial infarction and angiogenesis (11,12). They also have pivotal roles in
various cancer types, such as non-small cell lung cancer (1,13),
gastric cancer (14,15) and glioma (16,17).
CircRNAs can function by sponging microRNAs (miRNAs)
or RNA binding proteins (RBPs) and modulating parental gene
expression (18). Remarkably,
circRNAs can encode proteins or peptides that directly influence
relevant signalling pathways (19-21). CircRNAs were first found to have
protein-coding ability in viruses and prokaryotes, such as
hepatitis D virus (22)
and Escherichia coli (23). Subsequently, additional circRNAs
that encode proteins or peptides were identified (24-26). For instance, circ-ZNF609 promotes
myoblast proliferation in patients with Duchenne muscular dystrophy
and can be translated into a protein in a splicing-dependent manner
(27). CircMBL can be translated
into a protein that may exert synaptic functions dependent on
forkhead box (FOX)O activity (28). CircRNA-encoded proteins or
peptides are closely associated with cancer pathogenesis (29-32). The present review offers a
succinct summary of the properties and functional mechanisms of
circRNAs, with a particular emphasis on the mechanisms underlying
circRNA translation. Furthermore, the functions of circRNA-encoded
proteins or peptides in the pathogenesis of various cancer types
are explored.
There are three types of circRNAs produced by
backsplicing: i) Exonic circRNAs (ecRNAs) (33), ii) exon-intron circRNAs (EIciRNAs)
(34) and iii) circular intronic
RNAs (ciRNAs) (33). CircRNAs are
produced via four different mechanisms (35). The lariat-driven circularization
model, involving the activities of splice donors and acceptors,
exclusively produces ecRNAs (9,36).
The intron pairing-driven circularization model relies on RNA base
motif pairing (e.g., Alu repeats) in introns, resulting in the
generation of ecRNAs or EIciRNAs (9). In the RBP-mediated circularization
model, RBPs bridge with pre-mRNAs, leading to the formation of
ecRNAs or EIciRNAs. CiRNAs are produced by the binding of C-rich
elements near the branch and GU-rich elements close to the 5′splice
site of introns (1).
Translation initiation is the key step in protein
generation. In this step, ribosomes and transfer RNAs interact with
mRNAs to form an initiation complex. This intricate process
involves the assembly of initiation factors into various complexes,
such as the eukaryotic translation initiation factor (eIF)4E
complex (comprising eIF4E, eIF4G and eIF4A) and the eIF3 complex
(including eIF4G2, eIF4A and eIF4B). These complexes facilitate and
ensure an accurate cap-dependent translation (44). In contrast to linear RNAs,
circRNAs undergo cap-independent translation. Initiation factors
bind to the IRES or m6A to facilitate the formation of
the initiation complex (45)
(Fig. 1E and F).
Various bioinformatics tools have been developed to
facilitate the identification and exploration of circRNAs with
translational potential (Table
I). CircBase and CircNet offer comprehensive information,
including genome sequences, annotation, expression profile and
possible downstream factors of circRNAs (54,55). CircCode, CircPro and CircRNADb can
predict and identify circRNAs with coding potential (56-58). CircRNADb additionally provides
genome sequences, IRES information and open reading frame (ORF)
details (56). ORF Finder
searches for possible ORFs and predicts corresponding amino acid
(aa) sequences (59). BLASTp
assesses the conservation of these aa sequences, providing
reference information for functional research. IRESite, IRESbase,
DeepM6ASeq and M6APred-EL can predict IRES/m6A
modifications and provide evidence of their existence (60-63).
In addition to bioinformatic tools, experimental
evidence is crucial to detect the translation of circRNAs.
High-throughput techniques such as ribosome profiling, ribosome
immunoprecipitation and ribosome affinity purification can identify
circRNAs with encoding function (64). Mass spectrometry offers direct
evidence through identifying specific peptides (65). Biochemical molecular techniques,
including western blotting, enzyme immunoassays and dual luciferase
reporting assays, enable the detection of putative peptides or
proteins (66,67), screening of the targeted proteins
and evaluation of IRES/m6A activity (29,47).
Glioma, a malignant brain tumour, has a high
mortality rate and is categorized into four grades (I, II, III, IV)
according to the World Health Organization classification (71), with a higher grade indicating
greater malignancy. Glioma frequently recurs after surgery and
survival rates are markedly low. Among gliomas, glioblastoma (GBM),
classified as a grade IV glioma, represents the most malignant
subtype (72,73). GBM develops rapidly; it usually
progresses from the initial stage to the late stage within a short
period of time (only 3-6 months). CircRNAs have been demonstrated
to affect glioma progression by encoding functional proteins or
peptides (Fig. 2).
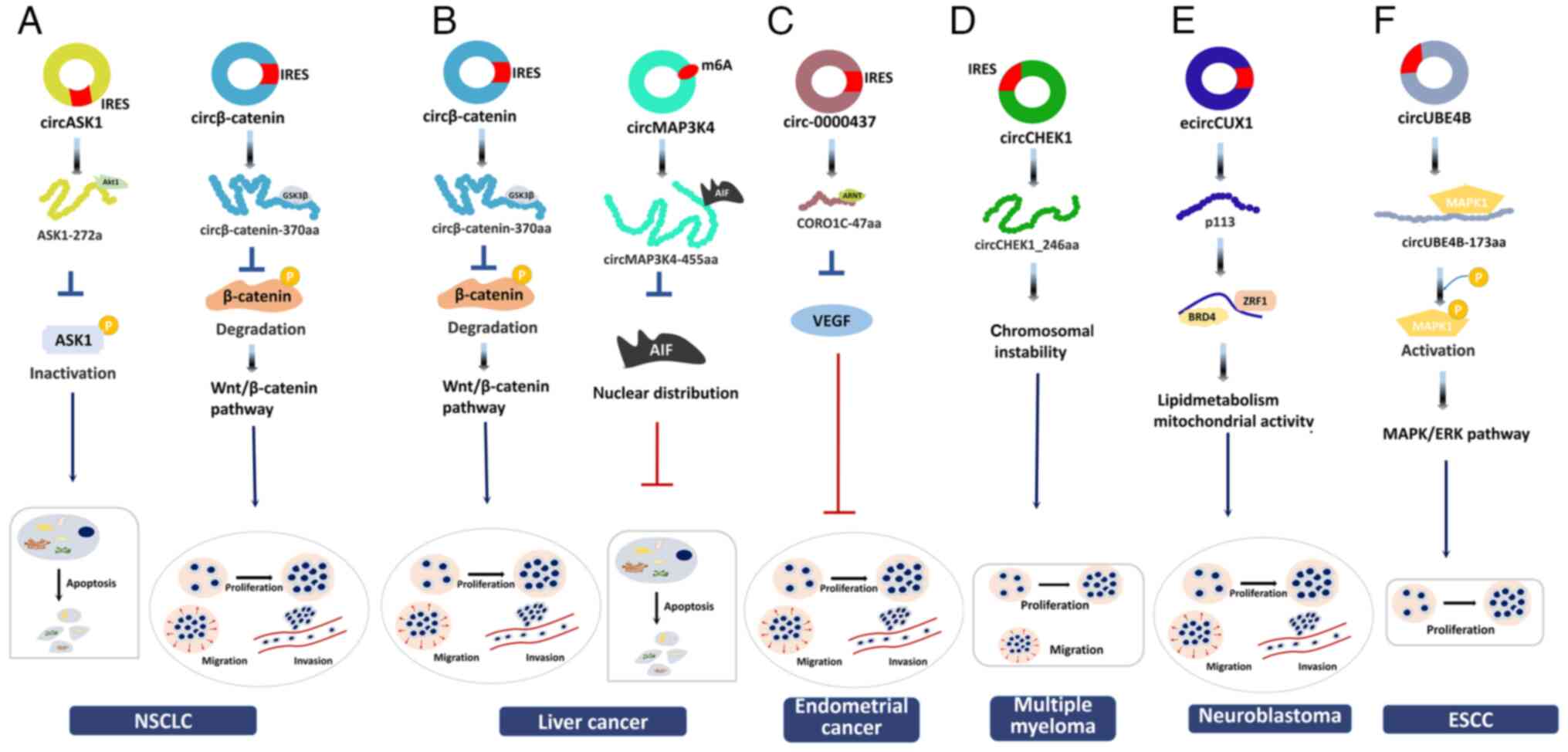
CircRNA F-box and WD repeat domain containing 7
(circFBXW7) generated from exons 3 and 4 of the FBXW7 gene,
was identified by deep RNA sequencing (RNA-seq) of clinical samples
from patients with GBM (16).
IRES-dependent translation of circFBXW7 generates a 185-aa protein
named FBXW7-185aa. FBXW7-185aa overexpression induces cell cycle
arrest and represses glioma cell proliferation, whereas knockdown
of FBXW7-185aa exacerbates tumour symptoms. FBXW7-185aa
competitively interacts with ubiquitin specific peptidase 28
(USP28) to degrade c-Myc via the ubiquitination pathway. The
expression levels of circFBXW7 and FBXW7-185aa are both decreased
in GBM tissues. High levels of circFBXW7 and FBXW7-185aa are
associated with a more favourable prognosis. Altogether, these
observations indicate the inhibitory role of the protein encoded by
circFBXW7 in glioma progression by destabilization of c-Myc
(16).
Long intergenic non-protein-coding RNA p53-induced
transcript (LINC-PINT) is a long non-coding RNA with
tumor-suppressive properties (74). Zhang et al (75) found a circRNA, circPINTexon2, that
was generated from exon 2 of LINC-PINT. CircPINTexon2 has an active
open reading frame (ORF) and contains an IRES. CircPINTexon2 can be
translated to an 87-aa peptide named PINT87aa. Both PINT87aa and
circPINTexon2 levels are lower in glioma tissues than in normal
tissues. PINT87aa overexpression induces cell cycle arrest and
inhibits tumour cell proliferation. Further experiments have
revealed that circPINTexon2 acted as a tumor suppressor by encoding
PINT87aa. Polymerase-associated factor complex (PAF1) participates
in the recruitment of RNA Pol II and modulates the transcriptional
elongation of multiple genes (76). PINT87aa directly interacts with
PAF1 and suppresses the transcriptional elongation of numerous
oncogenes. PINT87aa expression is negatively associated with the
clinical prognosis of patients with glioma. Collectively, these
results suggest that PINT87aa may function as a tumour suppressor
and can serve as a therapeutic target (75).
The receptor tyrosine
kinase/phosphatidylinositol-4,5-bisphosphate 3-kinase
(PI3K)/protein kinase B (AKT) pathway has a pivotal role in GBM
progression (80,81). Circ-AKT3 is significantly
downregulated in GBM tissues (82). Circ-AKT3 is an ecRNA originating
from exons 3-7 of the AKT3 gene. Circ-AKT3 has an ORF and an
IRES that can initiate the translation of a 174-aa protein named
AKT3-174aa. AKT3-174aa upregulation suppresses GBM cell
proliferation, radioresistance and tumorigenicity, thus indicating
its inhibitory role. The expression level of AKT3-174aa is
positively associated with the overall survival rate of patients
with GBM. AKT3-174aa interacts with activated (phosphorylated)
3-phosphoinositide-dependent kinase 1 and then inhibits the
activation of AKT by reducing its phosphorylation at Thr308.
Conclusively, these findings emphasize the antitumour role of
AKT3-174aa in GBM pathogenesis by positively modulating the
PI3K/AKT signalling pathway (82).
The epidermal growth factor receptor (EGFR)
signalling pathway is important for GBM progression (83,84). Gao et al (85) identified a circRNA capable of
encoding a protein that facilitates EGFR signalling pathway
activity. By RNA-seq analysis, the authors identified circular
E-cadherin (circ-E-Cad) RNA as the most upregulated circRNA.
Circ-E-Cad RNA can be translated into a 254-aa protein named
circRNA-encoded E-cadherin (C-E-Cad). C-E-Cad is highly expressed
in cells and tissues from patients with GBM compared with control
subjects. In addition, the C-E-Cad levels are negatively associated
with the prognosis of patients with GBM. C-E-Cad worsens the
symptoms of malignant GBM. Both C-E-Cad and circ-E-Cad levels are
positively correlated with GBM cell stemness. C-E-Cad is soluble
and can activate the EGFR signalling pathway by binding to the CR2
domain of EGFR, thereby promoting GBM tumorigenicity. EGFR-targeted
treatments for GBM are ineffective, but the removal of C-E-Cad
significantly enhances the antitumour activity of conventional
anti-EGFR therapeutic strategies in GBM. Taken together, these
findings indicate that C-E-Cad has an oncogenic role in GBM
pathogenesis by activating the EGFR signalling pathway, thus paving
a new way for GBM treatment (85).
Breast carcinomas (BC) are heterogeneous tumours.
TNBC is the subtype of BC with the poorest prognosis (89). Studies have shown that circRNAs
may be translated into important proteins or peptides that
influence TNBC progression (31,66,90) (Fig.
3A).
Human epidermal growth factor receptor 2 (HER2) is
an important target for TNBC treatment. Li et al (90) identified a circular RNA named
circ-HER2, which is produced from exons 3-7 of the HER2 gene
by backsplicing. Circ-HER2 has an ORF that can be translated into a
103-aa protein driven by an IRES. This protein is denoted as
HER2-103. Circ-HER2 and HER2-103 are expressed in ~30% of TNBC
tissues and their levels are negatively correlated with prognostic
efficacy. HER2-103 knockout suppresses TNBC cell proliferation,
invasion and tumorigenicity. HER2-103 can directly interact with
EGFR and HER3 to promote the formation of EGFR/EGFR homodimers or
EGFR/HER3 heterodimers. This binding subsequently activates
downstream signalling cascades to promote tumorigenesis. The
majority of the HER2-103 sequences are identical to the HER2 CR1
domain, which can be targeted by pertuzumab, an antibody against
HER2. Addition of pertuzumab reverses the tumour-promoting effect
of HER2-103 overexpression, suggesting that HER2-103 is a possible
clinical target for TNBC treatment (90).
GC is a common malignancy with high morbidity and
mortality. The pathological progression of GC is notably slow,
often making early detection challenging (91). Therefore, patients with GC are
frequently diagnosed at advanced stages, implying a generally poor
prognosis. Consequently, the exploration of effective therapeutic
targets and regulators is of great importance. Recent studies have
shown that circRNAs can encode functional proteins to modulate GC
pathogenesis (15,20,42) (Fig.
3B).
Adenomatous polyposis coli (APC)/AXIN is a
regulatory complex that inhibits the Wnt/β-catenin signalling
pathway (92). AXIN dysfunction
results in the abnormal accumulation of β-catenin (92). CircAXIN1 is derived from exon 2 of
the AXIN gene (20). Peng
et al (20) found
upregulated circAXIN1 expression in GC tissues. CircAXIN1
overexpression promotes GC cell proliferation, migration and
invasion, whereas circAXIN1 downregulation suppresses gastric
tumorigenesis. CircAXIN1 is able to encode a 295-aa protein
(AXIN1-295aa) in a process dependent on an IRES. AXIN1-295aa
directly interacts with the APC protein to block the interaction
between AXIN1 and APC, thus abolishing the suppressive effect of
the AXIN/APC complex on the Wnt/β-catenin pathway. This leads to
the activation of the Wnt/β-catenin pathway, which, in turn,
promotes gastric tumorigenesis. Therefore, circAXIN1 may promote
gastric tumorigenesis via AXIN1-295aa (20).
The mitogen-activated protein kinase (MAPK)
signalling pathway is a classical signal transduction pathway that
participates in cell proliferation, differentiation, metastasis and
other processes (96). CircMAPK1,
generated from exons 2-4 of the MAPK1 gene, is downregulated
in GC tissues and cell lines (15). A low circMAPK1 level suggests poor
survival in patients with GC. CircMAPK1 may suppress GC cell
proliferation and invasion in vivo and in vitro. With
an ORF and an IRES, circMAPK1 can be translated into a 109-aa
protein, MAPK1-109aa. MAPK1-109aa competitively interacts with MAPK
kinase 1, an upstream kinase in the MAPK signaling pathway, and
then inhibits MAPK1 phosphorylation, leading to the inactivation of
MAPK1 and the downstream signalling pathway. Altogether, these
observations indicate that circMAPK1 has an antitumour role in GC
progression via its encoded protein MAPK1-109aa (15).
Fibronectin type III domain-containing protein 3B
(FNDC3B) is associated with cell migration (99). CircFNDC3B, generated from exons 5
and 6 of the FNDC3B gene, has been implicated in various
cancer types (14,100). CircFNDC3B can encode a protein
(14), although the specific
functions of this protein are still unknown. Pan et al
(26) reported the role of
circFNDC3B as a translation template in CRC development. CircFNDC3B
is downregulated in CRC tissues and cell lines. Lower levels of
circFNDC3B are associated with poor overall survival.
Overexpression of circFNDC3B suppresses CRC cell proliferation,
migration and invasion. CircFNDC3B may be translated into a 218-aa
protein, called circFNDC3B-218aa, which exerts inhibitory effects
on CRC cell proliferation, invasion and migration. Snail is a
transcription factor known to induce epithelial-mesenchymal
transition (EMT) by reducing fructose-1,6-bisphosphatase 1 (FBP1)
expression (24,101). Mechanistic exploration has shown
that circFNDC3B-218aa can suppress the expression of the Snail
protein, resulting in FBP1 upregulation and EMT inhibition in CRC
cells. Overall, these observations suggest that circFNDC3B-218aa
may have a tumour-suppressive role in CRC pathogenesis through the
Snail-FBP-EMT axis (26).
Non-small cell lung cancer (NSCLC) is the most
prevalent and malignant type of lung cancer, with high mortality
and morbidity due to its high metastasis and recurrence rates
(107). CircRNAs are important
regulators of NSCLC pathogenesis (1). Recent studies have revealed that the
proteins or peptides encoded by circRNAs also participate in NSCLC
progression (13,108) (Fig. 4A).
Liver cancer is a devastating cancer type with a
poor prognosis. The lack of techniques for early diagnosis, the
limited therapeutic options and chemotherapeutic resistance make
the treatment of liver cancer difficult. CircRNAs and their encoded
proteins or peptides have emerged as important regulators in liver
cancer progression (110,111)
(Fig. 4B).
Hepatocellular carcinoma (HCC) is a type of primary
liver cancer. CircMAP3K4, generated from exon 3 of the MAPK kinase
kinase 4 (MAP3K4) gene, is upregulated in HCC (110). Higher levels of circMAP3K4
indicate poor overall survival. CircMAP3K4 can be translated into a
455-aa peptide, circMAP3K4-455aa via m6A modification.
CircMAP3K4-455aa promotes HCC development in vivo and in
vitro. Apoptosis-inducing factor (AIF), a regulator of cell
survival and death, can interact with circMAP3K4-455aa. Such
interaction reduces the nuclear distribution of AIF and suppresses
cisplatin-induced HCC cell apoptosis. Therefore, circMAP3K4-455aa
enhances HCC progression by influencing AIF activity (110).
Circβ-catenin and its encoded protein
circβ-catenin-370aa can promote the progression of liver cancer
(111). Circβ-catenin is
significantly upregulated in liver cancer tissues.
Circβ-catenin-370aa interacts with GSK3β to abolish the
GSK3β-mediated degradation of β-catenin, thereby promoting
Wnt/β-catenin signalling. Activation of the Wnt/β-catenin
signalling pathway enhances liver cancer progression (111).
EC is a common malignant tumour in women that
accounts for 1-2% of cancer-associated mortalities (107,112). Li et al (19) conducted deep RNA-seq on human EC
tissue samples and identified a circRNA (circ-0000437) that was
involved in EC progression (Fig.
4C). Circ-0000437 is significantly downregulated in EC samples
and does not function as a miRNA sponge. Circ-0000437 encodes a
47-aa peptide (named CORO1C-47aa) in the presence of an IRES.
CORO1C-47aa overexpression suppresses angiogenesis in EC through
inhibiting the proliferation, migration and differentiation of
endothelial cells. CORO1C-47aa interacts with the aryl hydrocarbon
receptor nuclear translocator (ARNT) protein, which is related to
hypoxia (113). CORO1C-47aa
competes with transforming acidic coiled-coil-containing protein 3,
a co-activator of the vascular endothelial growth factor (VEGF)
promoter, for binding to ARNT to suppress VEGF activation, thereby
inhibiting angiogenesis in EC. Thus, CORO1C-47aa has an antitumour
role in EC pathogenesis by decreasing VEGF expression (19).
MM originates in the bone marrow. MM is different
from solid tumours, as it is a plasma cell malignancy. MM is
difficult to cure and the mortality rate is markedly high (114). Checkpoint kinase 1 (CHEK1)
facilitates MM development (115,116). CHEK1 overexpression promotes MM
cell proliferation and macrophage-osteoclast differentiation, and
enhances the drug resistance of MM cells, thus having an oncogenic
role (32). CircCHEK1 is derived
from the CHEK1 gene and can be translated into a functional
protein called circCHEK1_246aa in an IRES-dependent manner
(Fig. 4D). Chromosomal
instability is related to cell proliferation. CircCHEK1_246aa
overexpression in MM cells results in chromosomal instability,
thereby promoting MM cell proliferation. Furthermore,
macrophage-osteoclast differentiation is enhanced. Taken together,
these findings indicate that CHEK1 and circCHEK1_246aa are
promising therapeutic targets in MM (32).
NB is a sympathetic nervous system tumour that
originates at the neural crest and is one of the most malignant
life-threatening tumours in children. CUT-like homeobox 1 (CUX1) is
a homeodomain transcription factor that regulates the pathogenesis
of numerous tumours (117). CUX1
can produce a circRNA, termed ecircCUX1 (118) (Fig. 4E). In an IRES-mediated manner,
ecircCUX1 is translated into a 113-aa protein (p113) that is
upregulated in NB samples. p113 facilitates lipid metabolism,
increases mitochondrial activity and promotes NB cell
proliferation, migration and metastasis. EcircCUX1 exacerbates NB
cell tumour symptoms via p113. p113 functions by directly binding
to Zuotin-related factor 1 (ZRF1) and bromodomain protein 4 (BRD4)
to form a transcriptional regulatory complex. The activity of the
p113/ZRF1/BRD4 complex increases the levels of lipid
metabolism-related proteins to induce lipid metabolic reprogramming
and mitochondrial complex I activity, thereby increasing the
tumorigenesis and aggressiveness of NB cells. Disassociation of the
p113-ZRF1 complex attenuates NB pathogenesis. High levels of p113,
ZRF1 and BRD4 indicate poor overall survival in patients with NB.
Overall, these results indicate that the ecircCUX1-encoded protein
has an oncogenic role in NB through the formation of the
p113/ZRF1/BRD4 complex (118).
The characteristics of circRNAs, such as their
unique conformation and stability, have attracted numerous
researchers to develop technologies based on circRNAs (124,125). The high stability and specific
expression of circRNAs make them promising candidate diagnostic and
prognostic biomarkers. CircRNAs can be employed in the development
of circRNA-based aptamers or sensors to modulate various
intracellular pathways (126).
CircRNAs containing IRESs/m6A modifications are
translatable, implying their potential as expression vectors to
initiate the production of diverse proteins. There have been some
patents that confirm the roles of circRNAs as effective
vectors/carriers for vaccines. In comparison to linear RNA (mRNA)
vaccines, circRNA vaccines exhibit higher stability and reduced
susceptibility to degradation. Of note, during the COVID-19
pandemic, scientists developed circRNA vaccines aimed at enabling
enhanced and more enduring antigen production (127).
CircRNA-encoded proteins/peptides hold significant
potential for extensive clinical applications in cancer, such as
early diagnosis, drug development, prognosis and treatment. Based
on the antigen-antibody interaction principle, a rapid test strip
method has been developed for early diagnosis, including early
pregnancy, COVID-19 detection and H1N1 detection strips. This
method offers non-invasive, fast and convenient (only several drops
of urine or nasal mucus are needed) diagnostic options,
facilitating a quick preliminary diagnosis at home. The prospect of
constructing distinct circRNA-encoded protein/peptide strips for
diagnosing various cancer types seems feasible. In the realm of
cancer therapy, precise treatment and targeting are crucial. The
limited number of effective target drugs is mainly due to low
target specificity, which highlights the need for more precise
approaches. Compared with nucleotide targets, protein targets are
more specific. Developing precision antitumour drugs targeting
proteins or peptides translated from circRNAs may yield highly
selective and minimally toxic therapeutic effects. Despite their
potential, challenges must be addressed before clinical
implementation.
First, the rapid degradation of multiple
circRNA-encoded proteins or peptides results in low abundance
(48). This adds to the
difficulty of their clinical application. Second, the understanding
of circRNA translation mechanisms is still insufficient. Many
underlying mechanisms are unclear and need further exploration.
Third, there is a lack of a uniform naming method for proteins or
peptides translated from circRNAs, making research findings across
different laboratories confusing. Therefore, more efforts should be
made to overcome these limitations. Future studies should provide
new insight and ideas for clinical applications.
In summary, circRNA-encoded proteins or peptides
have crucial roles in numerous signalling pathways and are thus
important factors in cancer pathogenesis. The aforementioned
findings open up new avenues for cancer diagnosis and therapeutic
strategies.
Not applicable.
LZ drafted the manuscript. HG and XL edited the
manuscript. FY revised the manuscript. LZ and PL conceived the idea
of the review and performed the final proofreading. All authors
have read and approved the final manuscript. Data authentication is
not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was funded by the Natural Science Foundation of
Shandong Province, China (grant no. ZR2020QH016) and Integrated
Project of Major Research Plan of the National Natural Science
Foundation of China (grant no. 92249303).
|
1
|
Liu Y, Ao X, Yu W, Zhang Y and Wang J:
Biogenesis, functions, and clinical implications of circular RNAs
in non-small cell lung cancer. Mol Ther Nucleic Acids. 27:50–72.
2021.
|
|
2
|
Kolakofsky D: Isolation and
characterization of Sendai virus DI-RNAs. Cell. 8:547–555.
1976.
|
|
3
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993.
|
|
4
|
Cocquerelle C, Daubersies P, Majerus MA,
Kerckaert JP and Bailleul B: Splicing with inverted order of exons
occurs proximal to large introns. EMBO J. 11:1095–1098. 1992.
|
|
5
|
Nigro JM, Cho KR, Fearon ER, Kern SE,
Ruppert JM, Oliner JD, Kinzler KW and Vogelstein B: Scrambled
exons. Cell. 64:607–613. 1991.
|
|
6
|
Schroeder R, Breitenbach M and Schweyen
RJ: Mitochondrial circular RNAs are absent in sporulating cells of
Saccharomyces cerevisiae. Nucleic Acids Res. 11:1735–1746.
1983.
|
|
7
|
Cocquerelle C, Mascrez B, Hetuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993.
|
|
8
|
Zhou X, Ao X, Jia Z, Li Y, Kuang S, Du C,
Zhang J, Wang J and Liu Y: Non-coding RNA in cancer drug
resistance: Underlying mechanisms and clinical applications. Front
Oncol. 12:9518642022.
|
|
9
|
Zhang L, Zhang Y, Yu F, Li X, Gao H and Li
P: The circRNA-miRNA/RBP regulatory network in myocardial
infarction. Front Pharmacol. 13:9411232022.
|
|
10
|
Wang M, Yu F, Li P and Wang K: Emerging
Function and clinical significance of Exosomal circRNAs in cancer.
Mol Ther Nucleic Acids. 21:367–383. 2020.
|
|
11
|
Zhang L, Zhang Y, Wang Y, Zhao Y, Ding H
and Li P: Circular RNAs: Functions and clinical significance in
cardiovascular disease. Front Cell Dev Biol. 8:5840512020.
|
|
12
|
Ye XM, Hang YW, Lu Y, Li D, Shen F, Guan
P, Dong J, Shi L and Hu W: CircRNA circ-NNT mediates myocardial
ischemia/reperfusion injury through activating pyroptosis by
sponging miR-33a-5p and regulating USP46 expression. Cell Death
Discov. 7:3702021.
|
|
13
|
Zhao W, Zhang Y and Zhu Y: Circular RNA
circbeta-catenin aggravates the malignant phenotype of
non-small-cell lung cancer via encoding a peptide. J Clin Lab Anal.
35:e239002021.
|
|
14
|
Hong YL, Qin HF, Li Y, Zhang Y, Zhuang X,
Liu L, Lu K, Li L, Deng X, Liu F, et al: FNDC3B circular RNA
promotes the migration and invasion of gastric cancer cells via the
regulation of E-cadherin and CD44 expression. J Cell Physiol.
234:19895–19910. 2019.
|
|
15
|
Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S,
Xuan Z, Xie L, Qiu S, He Z, et al: A novel protein encoded by
circMAPK1 inhibits progression of gastric cancer by suppressing
activation of MAPK signaling. Mol Cancer. 20:662021.
|
|
16
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel Role of FBXW7
Circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018.
|
|
17
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018.
|
|
18
|
Wang M, Yu F, Zhang Y, Zhang L, Chang W
and Wang K: The Emerging roles of circular RNAs in the
chemoresistance of gastrointestinal cancer. Front Cell Dev Biol.
10:8216092022.
|
|
19
|
Li F, Cai Y, Deng S, Yang L, Liu N, Chang
X, Jing L, Zhou Y and Li H: A peptide CORO1C-47aa encoded by the
circular noncoding RNA circ-0000437 functions as a negative
regulator in endometrium tumor angiogenesis. J Biol Chem.
297:1011822021.
|
|
20
|
Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo
X, Hossain MT, Zhu X, Du K, Hu F, et al: A novel protein
AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin
signaling pathway to promote gastric cancer progression. Mol
Cancer. 20:1582021.
|
|
21
|
Wu X, Xiao S, Zhang M, Yang L, Zhong J, Li
B, Li F, Xia X, Li X, Zhou H, et al: A novel protein encoded by
circular SMO RNA is essential for Hedgehog signaling activation and
glioblastoma tumorigenicity. Genome Biology. 22:332021.
|
|
22
|
Kos A, Dijkema R, Arnberg AC, van der
Meide PH and Schellekens H: The hepatitis delta (delta) virus
possesses a circular RNA. Nature. 323:558–560. 1986.
|
|
23
|
Perriman R and Ares M: Circular mRNA can
direct translation of extremely long repeating-sequence proteins in
vivo. RNA. 4:1047–1054. 1998.
|
|
24
|
Liu GM, Li Q, Zhang PF, Shen SL, Xie WX,
Chen B, Wu J, Hu WJ, Huang XY and Peng BG: Restoration of FBP1
suppressed Snail-induced epithelial to mesenchymal transition in
hepatocellular carcinoma. Cell Death Dis. 9:11322018.
|
|
25
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019.
|
|
26
|
Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang
J, Xia L, Yin Q, Zou B, Zheng J, et al: A novel protein encoded by
circFNDC3B inhibits tumor progression and EMT through regulating
Snail in colon cancer. Mol Cancer. 19:712020.
|
|
27
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 Is a Circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017.
|
|
28
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017.
|
|
29
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017.
|
|
30
|
Quintanal-Villalonga A, Molina-Pinelo S,
Cirauqui C, Ojeda-Márquez L, Marrugal Á, Suarez R, Conde E,
Ponce-Aix S, Enguita AB, Carnero A, et al: FGFR1 cooperates with
EGFR in lung cancer oncogenesis, and their combined inhibition
shows improved efficacy. J Thorac Oncol. 14:641–655. 2019.
|
|
31
|
Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou
X, Xie X and Tang H: circFBXW7 inhibits malignant progression by
sponging miR-197-3p and encoding a 185-aa protein in
triple-negative breast cancer. Mol Ther Nucleic Acids. 18:88–98.
2019.
|
|
32
|
Gu C, Wang W, Tang X, Xu T, Zhang Y, Guo
M, Wei R, Wang Y, Jurczyszyn A and Janz S: CHEK1 and
circCHEK1_246aa evoke chromosomal instability and induce bone
lesion formation in multiple myeloma. Mol Cancer. 20:842021.
|
|
33
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014.
|
|
34
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015.
|
|
35
|
Wang M, Yu F and Li PF: Noncoding RNAs as
an emerging resistance mechanism to immunotherapies in cancer:
Basic evidence and therapeutic implications. Front Immunol.
14:12687452023.
|
|
36
|
Zhang L, Wang Y, Zhang Y, Zhao YF and Li
PF: Pathogenic mechanisms and the potential clinical value of
circFoxo3 in cancers. Mol Ther-Nucl Acids. 23:908–917. 2021.
|
|
37
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
|
|
38
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013.
|
|
39
|
Liang ZX, Liu HS, Xiong L, Yang X, Wang
FW, Zeng ZW, He XW, Wu XR and Lan P: A novel NF-κB regulator
encoded by circPLCE1 inhibits colorectal carcinoma progression by
promoting RPS3 ubiquitin-dependent degradation. Mol Cancer.
20:1032021.
|
|
40
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013.
|
|
41
|
Sun G, Shen JF, Wei XF and Qi GX: Circular
RNA Foxo3 relieves myocardial Ischemia/Reperfusion injury by
suppressing autophagy via inhibiting HMGB1 by repressing KAT7 in
myocardial infarction. J Inflamm Res. 14:6397–6407. 2021.
|
|
42
|
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li
X, Yang W, Zhang C, Yang Q, Yee A, et al: A circular RNA binds to
and activates AKT phosphorylation and nuclear localization reducing
apoptosis and enhancing cardiac repair. Theranostics. 7:3842–3855.
2017.
|
|
43
|
Zhang Y, Jiang J, Zhang J, Shen H, Wang M,
Guo Z, Zang X, Shi H, Gao J, Cai H, et al: CircDIDO1 inhibits
gastric cancer progression by encoding a novel DIDO1-529aa protein
and regulating PRDX2 protein stability. Mol Cancer. 20:1012021.
|
|
44
|
Das S, Vera M, Gandin V, Singer RH and
Tutucci E: Intracellular mRNA transport and localized translation.
Nat Rev Mol Cell Biol. 22:483–504. 2021.
|
|
45
|
Prats AC, David F, Diallo LH, Roussel E,
Tatin F, Garmy-Susini B and Lacazette E: Circular RNA, the key for
translation. Int J Mol Sci. 21:85912020.
|
|
46
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020.
|
|
47
|
Choi JH, Wang W, Park D, Kim SH, Kim KT
and Min KT: IRES-mediated translation of cofilin regulates axonal
growth cone extension and turning. EMBO J. 37:2018.
|
|
48
|
Fan XJ, Yang Y, Chen CY and Wang ZF:
Pervasive translation of circular RNAs driven by short IRES-like
elements. Nat Commun. 13:37512022.
|
|
49
|
Yang Y and Wang Z: IRES-mediated
cap-independent translation, a path leading to hidden proteome. J
Mol Cell Biol. 11:911–919. 2019.
|
|
50
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′UTR
m(6)A promotes Cap-independent translation. Cell. 163:999–1010.
2015.
|
|
51
|
Zou Q, Xing P, Wei L and Liu B: Gene2vec:
Gene subsequence embedding for prediction of mammalian
N6-methyladenosine sites from mRNA. Rna. 25:205–218. 2019.
|
|
52
|
Gu Y, Wu X, Zhang J, Fang Y, Pan Y, Shu Y
and Ma P: The evolving landscape of N6-methyladenosine
modification in the tumor microenvironment. Mol Ther. 29:1703–1715.
2021.
|
|
53
|
Su T, Huang M, Liao J, Lin S, Yu P, Yang
J, Cai Y, Zhu S, Xu L, Peng Z, et al: Insufficient radiofrequency
ablation promotes hepatocellular carcinoma metastasis through
N6-Methyladenosine mRNA Methylation-Dependent mechanism.
Hepatology. 74:1339–1356. 2021.
|
|
54
|
Glazar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670.
2014.
|
|
55
|
Liu YC, Li JR, Sun CH, Andrews E, Chao RF,
Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al: CircNet: A
database of circular RNAs derived from transcriptome sequencing
data. Nucleic Acids Res. 44:D209–D215. 2016.
|
|
56
|
Chen XP, Han P, Zhou T, Guo XJ, Song XF
and Li Y: circRNADb: A comprehensive database for human circular
RNAs with protein-coding annotations. Sci Rep. 6:349852016.
|
|
57
|
Meng XW, Chen Q, Zhang PJ and Chen M:
CircPro: An integrated tool for the identification of circRNAs with
protein-coding potential. Bioinformatics. 33:3314–3316. 2017.
|
|
58
|
Sun PS and Li GL: CircCode: A powerful
tool for identifying circRNA coding ability. Front Genetics.
10:9812019.
|
|
59
|
Rombel IT, Sykes KF, Rayner S and Johnston
SA: ORF-FINDER: A vector for high-throughput gene identification.
Gene. 282:33–41. 2002.
|
|
60
|
Mokrejs M, Masek T, Vopalensky V, Hlubucek
P, Delbos P and Pospisek M: IRESite-a tool for the examination of
viral and cellular internal ribosome entry sites. Nucleic Acids
Res. 38(Database Issue): D131–D136. 2010.
|
|
61
|
Wei LY, Chen HR and Su R: M6APred-EL: A
sequence-based predictor for identifying N6-methyladenosine sites
using ensemble learning. Mol Ther Nucleic Acids. 12:635–644.
2018.
|
|
62
|
Zhang YQ and Hamada M: DeepM6ASeq:
Prediction and characterization of m6A-containing sequences using
deep learning. BMC Bioinformatics. 19(Suppl 19): S5242018.
|
|
63
|
Zhao J, Li Y, Wang C, Zhang H, Zhang H,
Jiang B, Guo X and Song X: IRESbase: A comprehensive database of
experimentally validated internal ribosome entry sites. Genomics
Proteomics Bioinformatics. 18:129–139. 2020.
|
|
64
|
Wu P, Mo YZ, Peng M, Tang T, Zhong Y, Deng
X, Xiong F, Guo C, Wu X, Li Y, et al: Emerging role of
tumor-related functional peptides encoded by lncRNA and circRNA.
Mol Cancer. 19:222020.
|
|
65
|
Liu Y, Li Z, Zhang M, Zhou H, Wu X, Zhong
J, Xiao F, Huang N, Yang X, Zeng R, et al: Rolling-translated EGFR
variants sustain EGFR signaling and promote glioblastoma
tumorigenicity. Neuro Oncol. 23:743–756. 2021.
|
|
66
|
Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang
H, Song X, Han D, Wang X, Liu Y, et al: circ-EIF6 encodes
EIF6-224aa to promote TNBC progression via stabilizing MYH9 and
activating the Wnt/beta-catenin pathway. Mol Ther. 30:415–430.
2022.
|
|
67
|
Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu
CH, Shen MJ and Huang Q: Circular RNA ciRS-7 promotes the
proliferation and metastasis of pancreatic cancer by regulating
miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat
Dis Int. 18:580–586. 2019.
|
|
68
|
Zhao HD, Zhou QJ and Li XT: Protein bait
hypothesis: CircRNA-Encoded proteins competitively inhibit cognate
functional isoforms. Trends Genet. 37:616–624. 2021.
|
|
69
|
Wang L, Zhou J, Zhang C, Chen R, Sun Q,
Yang P, Peng C, Tan Y, Jin C, Wang T, et al: A novel tumour
suppressor protein encoded by circMAPK14 inhibits progression and
metastasis of colorectal cancer by competitively binding to MKK6.
Clin Transl Med. 11:e6132021.
|
|
70
|
Song J, Zheng J, Liu X, Dong W, Yang C,
Wang D, Ruan X, Zhao Y, Liu L, Wang P, et al: A novel protein
encoded by ZCRB1-induced circHEATR5B suppresses aerobic glycolysis
of GBM through phosphorylation of JMJD5. J Exp Clin Cancer Res.
41:1712022.
|
|
71
|
McKinnon C, Nandhabalan M, Murray SA and
Plaha P: Glioblastoma: Clinical presentation, diagnosis, and
management. BMJ. 374:n15602021.
|
|
72
|
Broekman ML, Maas SLN, Abels ER, Mempel
TR, Krichevsky AM and Breakefield XO: Multidimensional
communication in the microenvirons of glioblastoma. Nat Rev Neurol.
14:482–495. 2018.
|
|
73
|
Caragher SP, Hall RR, Ahsan R and Ahmed
AU: Monoamines in glioblastoma: Complex biology with therapeutic
potential. Neuro Oncol. 20:1014–1025. 2018.
|
|
74
|
Marin-Bejar O, Marchese FP, Athie A,
Sánchez Y, González J, Segura V, Huang L, Moreno I, Navarro A,
Monzó M, et al: Pint lincRNA connects the p53 pathway with
epigenetic silencing by the Polycomb repressive complex 2. Genome
Biol. 14:R1042013.
|
|
75
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018.
|
|
76
|
Chen FX, Woodfin AR, Gardini A, Rickels
RA, Marshall SA, Smith ER, Shiekhattar R and Shilatifard A: PAF1, a
molecular regulator of promoter-proximal pausing by RNA Polymerase
II. Cell. 162:1003–1015. 2015.
|
|
77
|
Unk I, Hajdu I, Fatyol K, Szakál B,
Blastyák A, Bermudez V, Hurwitz J, Prakash L, Prakash S and
Haracska L: Human SHPRH is a ubiquitin ligase for
Mms2-Ubc13-dependent polyubiquitylation of proliferating cell
nuclear antigen. Proc Natl Acad Sci USA. 103:18107–18112. 2006.
|
|
78
|
Chen X, Sun N, Li R, Sang X, Li X, Zhao J,
Han J, Yang J and Ikezoe T: Targeting HLA-F suppresses the
proliferation of glioma cells via a reduction in hexokinase
2-dependent glycolysis. Int J Biol Sci. 17:1263–1276. 2021.
|
|
79
|
Wang HJ, Hsieh YJ, Cheng WC, Lin CP, Lin
YS, Yang SF, Chen CC, Izumiya Y, Yu JS, Kung HJ and Wang WC: JMJD5
regulates PKM2 nuclear translocation and reprograms
HIF-1alpha-mediated glucose metabolism. Proc Natl Acad Sci USA.
111:279–284. 2014.
|
|
80
|
Chin YR, Yuan X, Balk SP and Toker A:
PTEN-Deficient tumors depend on AKT2 for maintenance and survival.
Cancer Discov. 4:942–955. 2014.
|
|
81
|
Zhao HF, Wang J, Shao W, Wu CP, Chen ZP,
To ST and Li WP: Recent advances in the use of PI3K inhibitors for
glioblastoma multiforme: Current preclinical and clinical
development. Mol Cancer. 16:1002017.
|
|
82
|
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H,
Huang N, Yang X, Xiao F, Liu D, et al: A novel tumor suppressor
protein encoded by circular AKT3 RNA inhibits glioblastoma
tumorigenicity by competing with active phosphoinositide-dependent
Kinase-1. Mol Cancer. 18:1312019.
|
|
83
|
Furnari FB, Cloughesy TF, Cavenee WK and
Mischel PS: Heterogeneity of epidermal growth factor receptor
signalling networks in glioblastoma. Nat Rev Cancer. 15:302–310.
2015.
|
|
84
|
Reifenberger G, Wirsching HG,
Knobbe-Thomsen CB and Weller M: Advances in the molecular genetics
of Gliomas-Implications for classification and therapy. Nat Rev
Clin Oncol. 14:434–452. 2017.
|
|
85
|
Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X,
Zhong J, Zhao Z, Zhao K, Liu D, et al: Circular RNA-encoded
oncogenic E-cadherin variant promotes glioblastoma tumorigenicity
through activation of EGFR-STAT3 signalling. Nat Cell Biol.
23:278–291. 2021.
|
|
86
|
Filbin MG, Dabral SK, Pazyra-Murphy MF,
Ramkissoon S, Kung AL, Pak E, Chung J, Theisen MA, Sun Y,
Franchetti Y, et al: Coordinate activation of Shh and PI3K
signaling in PTEN-deficient glioblastoma: New therapeutic
opportunities. Nat Med. 19:1518–1523. 2013.
|
|
87
|
Kool M, Jones DT, Jäger N, Northcott PA,
Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, et
al: Genome sequencing of SHH medulloblastoma predicts
genotype-related response to smoothened inhibition. Cancer Cell.
25:393–405. 2014.
|
|
88
|
Xie J: Hedgehog signaling pathway:
Development of antagonists for cancer therapy. Curr Oncol Rep.
10:107–113. 2008.
|
|
89
|
Bianchini G, Balko JM, Mayer IA, Sanders
ME and Gianni L: Triple-negative breast cancer: Challenges and
opportunities of a heterogeneous disease. Nat Rev Clin Oncol.
13:674–690. 2016.
|
|
90
|
Li J, Ma M, Yang X, Zhang M, Luo J, Zhou
H, Huang N, Xiao F, Lai B, Lv W and Zhang N: Circular HER2 RNA
positive triple negative breast cancer is sensitive to Pertuzumab.
Mol Cancer. 19:1422020.
|
|
91
|
Du Y, Zhang JY, Gong LP, Feng ZY, Wang D,
Pan YH, Sun LP, Wen JY, Chen GF, Liang J, et al: Hypoxia-induced
ebv-circLMP2A promotes angiogenesis in EBV-associated gastric
carcinoma through the KHSRP/VHL/HIF1 α/VEGFA pathway. Cancer Lett.
526:259–272. 2022.
|
|
92
|
Ji L, Jiang B, Jiang X, Charlat O, Chen A,
Mickanin C, Bauer A, Xu W, Yan X and Cong F: The SIAH E3 ubiquitin
ligases promote Wnt/β-catenin signaling through mediating
Wnt-induced Axin degradation. Genes Dev. 31:904–915. 2017.
|
|
93
|
Futterer A, de Celis J, Navajas R,
Almonacid L, Gutiérrez J, Talavera-Gutiérrez A, Pacios-Bras C,
Bernascone I, Martin-Belmonte F and Martinéz-A C: DIDO as a
switchboard that regulates Self-Renewal and differentiation in
embryonic stem cells. Stem Cell Rep. 8:1062–1075. 2017.
|
|
94
|
Kang DH, Lee DJ, Lee S, Lee SY, Jun Y, Kim
Y, Kim Y, Lee JS, Lee DK, Lee S, et al: Interaction of tankyrase
and peroxiredoxin II is indispensable for the survival of
colorectal cancer cells. Nat Commun. 8:402017.
|
|
95
|
Park YH, Kim SU, Kwon TH, Kim JM, Song IS,
Shin HJ, Lee BK, Bang DH, Lee SJ, Lee DS, et al: Peroxiredoxin II
promotes hepatic tumorigenesis through cooperation with
Ras/Forkhead box M1 signaling pathway. Oncogene. 35:3503–3513.
2016.
|
|
96
|
Fang JY and Richardson BC: The MAPK
signalling pathways and colorectal cancer. Lancet Oncol. 6:322–327.
2005.
|
|
97
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.
|
|
98
|
Yin F, Dong J, Kang LI and Liu X:
Hippo-YAP signaling in digestive system tumors. Am J Cancer Res.
11:2495–2507. 2021.
|
|
99
|
Cai C, Rajaram M, Zhou X, Liu Q, Marchica
J, Li J, Powers RS, et al: Activation of multiple cancer pathways
and tumor maintenance function of the 3q amplified oncogene FNDC3B.
Cell Cycle. 11:1773–1781. 2012.
|
|
100
|
Liu HW, Bi JM, Dong W, Yang M, Shi J,
Jiang N, Lin T and Huang J: Invasion-related circular RNA
circFNDC3B inhibits bladder cancer progression through the
miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 17:1612018.
|
|
101
|
Dong C, Yuan T, Wu Y, Wang Y, Fan TW,
Miriyala S, Lin Y, Yao J, Shi J, Kang T, et al: Loss of FBP1 by
Snail-Mediated repression provides metabolic advantages in
Basal-like breast cancer. Cancer Cell. 23:316–331. 2013.
|
|
102
|
Gupta J, del Barco Barrantes I, Igea A,
Sakellariou S, Pateras IS, Gorgoulis VG and Nebreda AR: Dual
function of p38α MAPK in colon cancer: Suppression of
colitis-associated tumor initiation but requirement for cancer cell
survival. Cancer Cell. 25:484–500. 2014.
|
|
103
|
Luo JL, Maeda S, Hsu LC, Yagita H and
Karin M: Inhibition of NF-kappa B in cancer cells converts
inflammation-induced tumor growth mediated by TNF alpha to
TRAIL-mediated tumor regression. Cancer Cell. 6:297–305. 2004.
|
|
104
|
Clemo NK, Collard TJ, Southern SL, Edwards
KD, Moorghen M, Packham G, Hague A, Paraskeva C and Williams AC:
BAG-1 is up-regulated in colorectal tumour progression and promotes
colorectal tumour cell survival through increased NF-kappa B
activity. Carcinogenesis. 29:849–857. 2008.
|
|
105
|
Hodgson A, Wier EM, Fu K, Sun X, Yu H,
Zheng W, Sham HP, Johnson K, Bailey S, Vallance BA and Wan F:
Metalloprotease NleC suppresses host NF-κB/inflammatory responses
by cleaving p65 and interfering with the p65/RPS3 interaction. PLoS
Pathog. 11:e10047052015.
|
|
106
|
Kim TS, Jang CY, Kim HD, Lee JY, Ahn BY
and Kim J: Interaction of Hsp90 with ribosomal proteins protects
from ubiquitination and proteasome-dependent degradation. Mol Biol
Cell. 17:824–833. 2006.
|
|
107
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
|
|
108
|
Wang T, Liu Z, She Y, Deng J, Zhong Y,
Zhao M, Li S, Xie D, Sun X, Hu X and Chen C: A novel protein
encoded by circASK1 ameliorates gefitinib resistance in lung
adenocarcinoma by competitively activating ASK1-dependent
apoptosis. Cancer Lett. 520:321–331. 2021.
|
|
109
|
Ichijo H, Nishida E, Irie K, ten Dijke P,
Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K and Gotoh
Y: Induction of apoptosis by ASK1, a mammalian MAPKKK that
activates SAPK/JNK and p38 signaling pathways. Science. 275:90–94.
1997.
|
|
110
|
Duan JL, Chen W, Xie JJ, Zhang ML, Nie RC,
Liang H, Mei J, Han K, Xiang ZC, Wang FW, et al: A novel peptide
encoded by N6-methyladenosine modified circMAP3K4 prevents
apoptosis in hepatocellular carcinoma. Mol Cancer. 21:932022.
|
|
111
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM and Zhang JF: Translation
of the circular RNA circ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
|
|
112
|
Lee TY, Martinez-Outschoorn UE, Schilder
RJ, Kim CH, Richard SD, Rosenblum NG and Johnson JM: Metformin as a
therapeutic target in endometrial cancers. Front Oncol.
8:3412018.
|
|
113
|
Kewley RJ, Whitelaw ML and Chapman-Smith
A: The mammalian basic helix-loop-helix/PAS family of
transcriptional regulators. Int J Biochem Cell Biol. 36:189–204.
2004.
|
|
114
|
Pawlyn C and Morgan GJ: Evolutionary
biology of high-risk multiple myeloma. Nat Rev Cancer. 17:543–556.
2017.
|
|
115
|
de Boussac H, Bruyer A, Jourdan M, Maes A,
Robert N, Gourzones C, Vincent L, Seckinger A, Cartron G, Hose D,
et al: Kinome expression profiling to target new therapeutic
avenues in multiple myeloma. Haematologica. 105:784–795. 2020.
|
|
116
|
Pei XY, Dai Y, Youssefian LE, Chen S,
Bodie WW, Takabatake Y, Felthousen J, Almenara JA, Kramer LB, Dent
P and Grant S: Cytokinetically quiescent (G0/G1) human multiple
myeloma cells are susceptible to simultaneous inhibition of Chk1
and MEK1/2. Blood. 118:5189–5200. 2011.
|
|
117
|
Michl P, Ramjaun AR, Pardo OE, Warne PH,
Wagner M, Poulsom R, D'Arrigo C, Ryder K, Menke A, Gress T and
Downward J: CUTL1 is a target of TGF(beta) signaling that enhances
cancer cell motility and invasiveness. Cancer Cell. 7:521–532.
2005.
|
|
118
|
Yang F, Hu A, Guo Y, Wang J, Li D, Wang X,
Jin S, Yuan B, Cai S, Zhou Y, et al: p113 isoform encoded by CUX1
circular RNA drives tumor progression via facilitating ZRF1/BRD4
transactivation. Mol Cancer. 20:1232021.
|
|
119
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019.
|
|
120
|
Hu X, Wu D, He X, Zhao H, He Z, Lin J,
Wang K, Wang W, Pan Z, Lin H and Wang M: circGSK3β promotes
metastasis in esophageal squamous cell carcinoma by augmenting
β-catenin signaling. Mol Cancer. 18:1602019.
|
|
121
|
Li CJ, Li DG, Liu EJ and Jiang GZ:
Circ8199 encodes a protein that inhibits the activity of OGT by
JAK2-STAT3 pathway in esophageal squamous cell carcinoma. Am J
Cancer Res. 13:1107–1117. 2023.
|
|
122
|
Lyu YC, Tan BH, Li L, Liang R, Lei K, Wang
K, Wu D, Lin H and Wang M: A novel protein encoded by circUBE4B
promotes progression of esophageal squamous cell carcinoma by
augmenting MAPK/ERK signaling. Cell Death Dis. 14:3462023.
|
|
123
|
De Luca A, Maiello MR, D'Alessio A,
Pergameno M and Normanno N: The RAS/RAF/MEK/ERK and the PI3K/AKT
signalling pathways: Role in cancer pathogenesis and implications
for therapeutic approaches. Expert Opin Ther Tar. 16(Suppl 2):
S17–S27. 2012.
|
|
124
|
Liu Y, Li XG, Zhou XH, Wang JX and Ao X:
FADD as a key molecular player in cancer progression. Mol Med.
28:1322022.
|
|
125
|
Liu Y, Wang Y, Li X, Jia Y, Wang J and Ao
X: FOXO3a in cancer drug resistance. Cancer Lett.
540:2157242022.
|
|
126
|
Litke JL and Jaffrey SR: Highly efficient
expression of circular RNA aptamers in cells using autocatalytic
transcripts. Nat Biotechnol. 37:667–675. 2019.
|
|
127
|
Qu L, Yi ZY, Shen Y, Lin L, Chen F, Xu Y,
Wu Z, Tang H, Zhang X, Tian F, et al: Circular RNA vaccines against
SARS-CoV-2 and emerging variants. Cell. 185:1728–1744. 2022.
|