|
1
|
Anderson AS, Key TJ, Norat T, Scoccianti
C, Cecchini M, Berrino F, Boutron-Ruault MC, Espina C, Leitzmann M,
Powers H, et al: European code against cancer 4th edition: Obesity,
body fatness and cancer. Cancer Epidemiol. 39(Suppl 1): S34–S45.
2015.
|
|
2
|
Lachance JA, Darus CJ and Rice LW:
Surgical management and postoperative treatment of endometrial
carcinoma. Rev Obstet Gynecol. 1:97–105. 2008.
|
|
3
|
Makker V, Colombo N, Casado Herráez A,
Santin AD, Colomba E, Miller DS, Fujiwara K, Pignata S, Baron-Hay
S, Ray-Coquard I, et al: Lenvatinib plus pembrolizumab for advanced
endometrial cancer. N Engl J Med. 386:437–448. 2022.
|
|
4
|
Hanahan D: Hallmarks of cancer: New
dimensions. Cancer Discov. 12:31–46. 2022.
|
|
5
|
Zhang L, Lu Q and Chang C: Epigenetics in
health and disease. Adv Exp Med Biol. 1253:3–55. 2020.
|
|
6
|
Margueron R, Trojer P and Reinberg D: The
key to development: Interpreting the histone code? Curr Opin Genet
Dev. 15:163–176. 2005.
|
|
7
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000.
|
|
8
|
Varier RA and Timmers HT: Histone lysine
methylation and demethylation pathways in cancer. Biochim Biophys
Acta. 1815:75–89. 2011.
|
|
9
|
Hamamoto R, Saloura V and Nakamura Y:
Critical roles of non-histone protein lysine methylation in human
tumorigenesis. Nat Rev Cancer. 15:110–124. 2015.
|
|
10
|
Sone K, Piao L, Nakakido M, Ueda K,
Jenuwein T, Nakamura Y and Hamamoto R: Critical role of lysine 134
methylation on histone H2AX for γ-H2AX production and DNA repair.
Nat Commun. 5:56912014.
|
|
11
|
Oki S, Sone K, Oda K, Hamamoto R, Ikemura
M, Maeda D, Takeuchi M, Tanikawa M, Mori-Uchino M, Nagasaka K, et
al: Oncogenic histone methyltransferase EZH2: A novel prognostic
marker with therapeutic potential in endometrial cancer.
Oncotarget. 8:40402–40411. 2017.
|
|
12
|
Kojima M, Sone K, Oda K, Hamamoto R,
Kaneko S, Oki S, Kukita A, Machino H, Honjoh H, Kawata Y, et al:
The histone methyltransferase WHSC1 is regulated by EZH2 and is
important for ovarian clear cell carcinoma cell proliferation. BMC
Cancer. 19:4552019.
|
|
13
|
Kukita A, Sone K, Oda K, Hamamoto R,
Kaneko S, Komatsu M, Wada M, Honjoh H, Kawata Y, Kojima M, et al:
Histone methyltransferase SMYD2 selective inhibitor LLY-507 in
combination with poly ADP ribose polymerase inhibitor has
therapeutic potential against high-grade serous ovarian carcinomas.
Biochem Biophys Res Commun. 513:340–346. 2019.
|
|
14
|
Kojima M, Sone K, Oda K, Hamamoto R,
Kaneko S, Oki S, Kukita A, Kawata A, Honjoh H, Kawata Y, et al: The
histone methyltransferase SMYD2 is a novel therapeutic target for
the induction of apoptosis in ovarian clear cell carcinoma cells.
Oncol Lett. 20:1532020.
|
|
15
|
Wada M, Kukita A, Sone K, Hamamoto R,
Kaneko S, Komatsu M, Takahashi Y, Inoue F, Kojima M, Honjoh H, et
al: Epigenetic modifier SETD8 as a therapeutic target for
high-grade serous ovarian cancer. Biomolecules. 10:16862020.
|
|
16
|
Blanc RS and Richard S: Arginine
methylation: The coming of age. Mol Cell. 65:8–24. 2017.
|
|
17
|
Bedford MT and Clarke SG: Protein arginine
methylation in mammals: Who, what, and why. Mol Cell. 33:1–13.
2009.
|
|
18
|
Yang Y and Bedford MT: Protein arginine
methyltransferases and cancer. Nat Rev Cancer. 13:37–50. 2013.
|
|
19
|
Inoue F, Sone K, Toyohara Y, Tanimoto S,
Takahashi Y, Kusakabe M, Kukita A, Honjoh H, Nishijima A, Taguchi
A, et al: Histone arginine methyltransferase CARM1 selective
inhibitor TP-064 induces apoptosis in endometrial cancer. Biochem
Biophys Res Commun. 601:123–128. 2022.
|
|
20
|
Dowhan DH, Harrison MJ, Eriksson NA,
Bailey P, Pearen MA, Fuller PJ, Funder JW, Simpson ER, Leedman PJ,
Tilley WD, et al: Protein arginine methyltransferase 6-dependent
gene expression and splicing: Association with breast cancer
outcomes. Endocr Relat Cancer. 19:509–526. 2012.
|
|
21
|
Almeida-Rios D, Graça I, Vieira FQ,
Ramalho-Carvalho J, Pereira-Silva E, Martins AT, Oliveira J,
Gonçalves CS, Costa BM, Henrique R and Jerónimo C: Histone
methyltransferase PRMT6 plays an oncogenic role of in prostate
cancer. Oncotarget. 7:53018–53028. 2016.
|
|
22
|
Avasarala S, Wu PY, Khan SQ, Yanlin S, Van
Scoyk M, Bao J, Di Lorenzo A, David O, Bedford MT, Gupta V, et al:
PRMT6 promotes lung tumor progression via the alternate activation
of tumor-associated macrophages. Mol Cancer Res. 18:166–178.
2020.
|
|
23
|
Bouchard C, Sahu P, Meixner M, Nötzold RR,
Rust MB, Kremmer E, Feederle R, Hart-Smith G, Finkernagel F,
Bartkuhn M, et al: Genomic location of PRMT6-dependent H3R2
methylation is linked to the transcriptional outcome of associated
genes. Cell Rep. 24:3339–3352. 2018.
|
|
24
|
Jiang N, Li QL, Pan W, Li J, Zhang MF, Cao
T, Su SG and Shen H: PRMT6 promotes endometrial cancer via AKT/mTOR
signaling and indicates poor prognosis. Int J Biochem Cell Biol.
120:1056812020.
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
26
|
Ohtani H, Liu M, Zhou W, Liang G and Jones
PA: Switching roles for DNA and histone methylation depend on
evolutionary ages of human endogenous retroviruses. Genome Res.
28:1147–1157. 2018.
|
|
27
|
Maruyama R, Choudhury S, Kowalczyk A,
Bessarabova M, Beresford-Smith B, Conway T, Kaspi A, Wu Z,
Nikolskaya T, Merino VF, et al: Epigenetic regulation of cell
type-specific expression patterns in the human mammary epithelium.
PLoS Genet. 7:e10013692011.
|
|
28
|
Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills
AE, Gupta R and Baker J: Chromatin and transcriptional signatures
for nodal signaling during endoderm formation in hESCs. Dev Biol.
357:492–504. 2011.
|
|
29
|
Chang CC, Huang RL, Wang HC, Liao YP, Yu
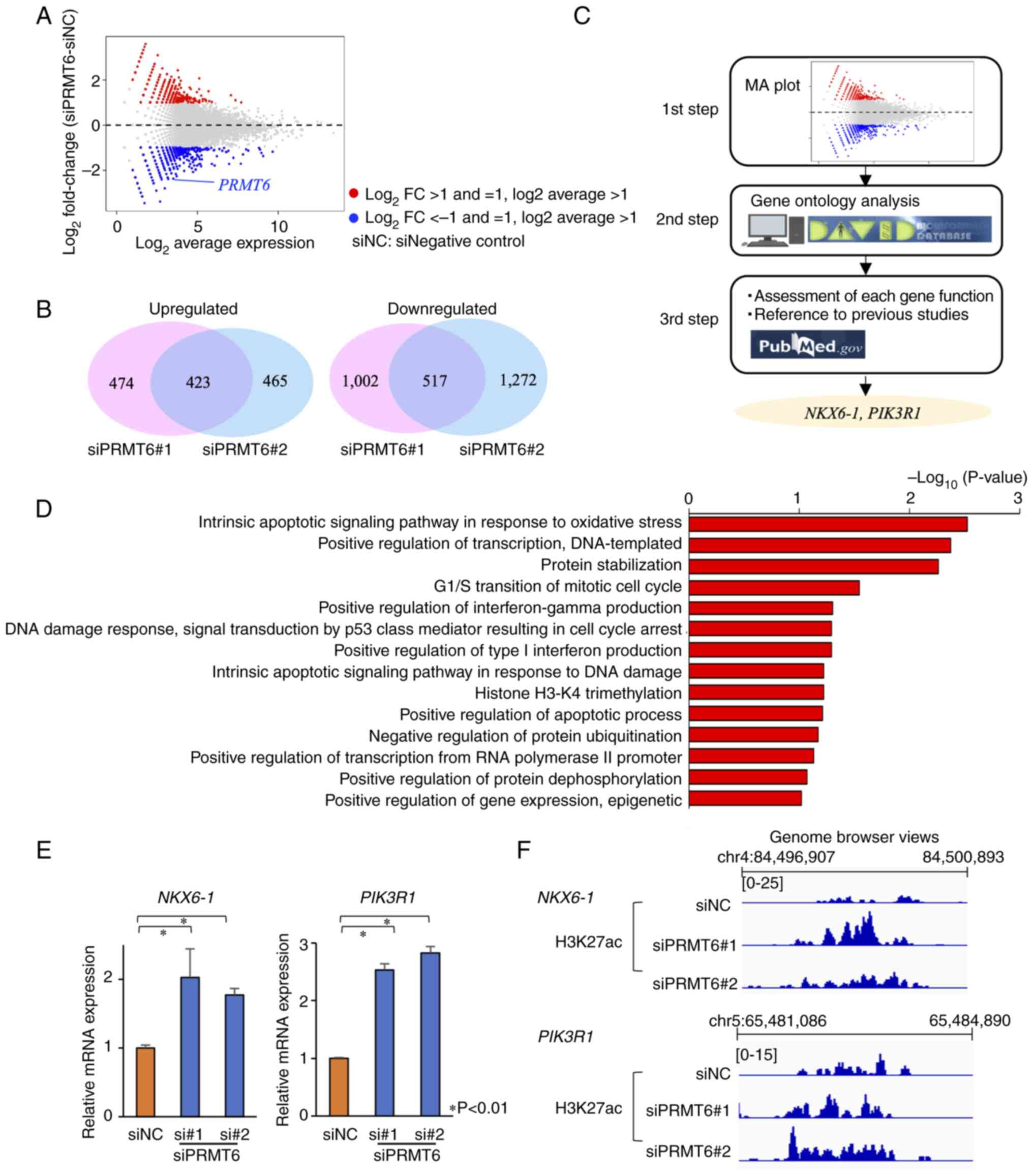
MH and Lai HC: High methylation rate of LMX1A, NKX6-1, PAX1, PTPRR,
SOX1, and ZNF582 genes in cervical adenocarcinoma. Int J Gynecol
Cancer. 24:201–209. 2014.
|
|
30
|
Li HJ, Yu PN, Huang KY, Su HY, Hsiao TH,
Chang CP, Yu MH and Lin YW: NKX6.1 functions as a metastatic
suppressor through epigenetic regulation of the
epithelial-mesenchymal transition. Oncogene. 35:2266–2278.
2016.
|
|
31
|
Chung HH, Lee CT, Hu JM, Chou YC, Lin YW
and Shih YL: NKX6.1 represses tumorigenesis, metastasis and
chemoresistance in colorectal cancer. J Mol Sci. 21:51062020.
|
|
32
|
Cheung LW and Mills GB: Targeting
therapeutic liabilities engendered by PIK3R1 mutations for cancer
treatment. Pharmacogenomics. 17:297–307. 2016.
|
|
33
|
Ishiguro K, Kitajima H, Niinuma T,
Maruyama R, Nishiyama N, Ohtani H, Sudo G, Toyota M, Sasaki H,
Yamamoto E, et al: Dual EZH2 and G9a inhibition suppresses multiple
myeloma cell proliferation by regulating the interferon signal and
IRF4-MYC axis. Cell Death Discov. 7:72021.
|
|
34
|
Roulois D, Loo Yau H, Singhania R, Wang Y,
Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, et al:
DNA-demethylating agents target colorectal cancer cells by inducing
viral mimicry by endogenous transcripts. Cell. 162:961–973.
2015.
|
|
35
|
Lim Y, Yu S, Yun JA, Do IG, Cho L, Kim YH
and Kim HC: The prognostic significance of protein arginine
methyltransferase 6 expression in colon cancer. Oncotarget.
9:9010–9020. 2018.
|
|
36
|
Okuno K, Akiyama Y, Shimada S, Nakagawa M,
Tanioka T, Inokuchi M, Yamaoka S, Kojima K and Tanaka S: Asymmetric
dimethylation at histone H3 arginine 2 by PRMT6 in gastric cancer
progression. Carcinogenesis. 40:15–26. 2019.
|
|
37
|
Stein C, Riedl S, Rüthnick D, Nötzold RR
and Bauer UM: The arginine methyltransferase PRMT6 regulates cell
proliferation and senescence through transcriptional repression of
tumor suppressor genes. Nucleic Acids Res. 40:9522–9533. 2012.
|
|
38
|
Kleinschmidt MA, de Graaf P, van Teeffelen
HA and Timmers HT: Cell cycle regulation by the PRMT6 arginine
methyltransferase through repression of cyclin-dependent kinase
inhibitors. PLoS One. 7:e414462012.
|
|
39
|
Rao A, Luo C and Hogan PG: Transcription
factors of the NFAT family: Regulation and function. Annu Rev
Immunol. 15:707–747. 1997.
|
|
40
|
Reppert S, Zinser E, Holzinger C, Sandrock
L, Koch S and Finotto S: NFATc1 deficiency in T cells protects mice
from experimental autoimmune encephalomyelitis. Eur J Immunol.
45:1426–1440. 2015.
|
|
41
|
Zhang Y, Alexander PB and Wang XF: TGF-β
family signaling in the control of cell proliferation and survival.
Cold Spring Harb Perspect Biol. 9:a0221452017.
|
|
42
|
Ribatti D, Tamma R and Annese T:
Epithelial-mesenchymal transition in cancer: A historical overview.
Transl Oncol. 13:1007732020.
|
|
43
|
Kriseman M, Monsivais D, Agno J, Masand
RP, Creighton CJ and Matzuk MM: Uterine double-conditional
inactivation of Smad2 and Smad3 in mice causes endometrial
dysregulation, infertility and uterine cancer. Proc Natl Acad Sci
USA. 116:3873–3882. 2019.
|
|
44
|
Hennessy BT, Smith DL, Ram PT, Lu Y and
Mills GB: Exploiting the PI3K/AKT pathway for cancer drug
discovery. Nat Rev Drug Discov. 4:988–1004. 2005.
|
|
45
|
European Bioinformatics Institute; Ewan B,
Nick G, Arkadiusz K, Emmanuel M, Alistair RG, Guy S, Arne S, Abel
UV, Simon W, et al: Initial sequencing and comparative analysis of
the mouse genome. Nature. 420:520–562. 2002.
|
|
46
|
Goodier JL: Restricting retrotransposons:
A review. Mob DNA. 7:162016.
|
|
47
|
Chiappinelli KB, Strissel PL, Desrichard
A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et
al: Inhibiting DNA methylation causes an interferon response in
cancer via dsRNA including endogenous retroviruses. Cell.
169:3612017.
|