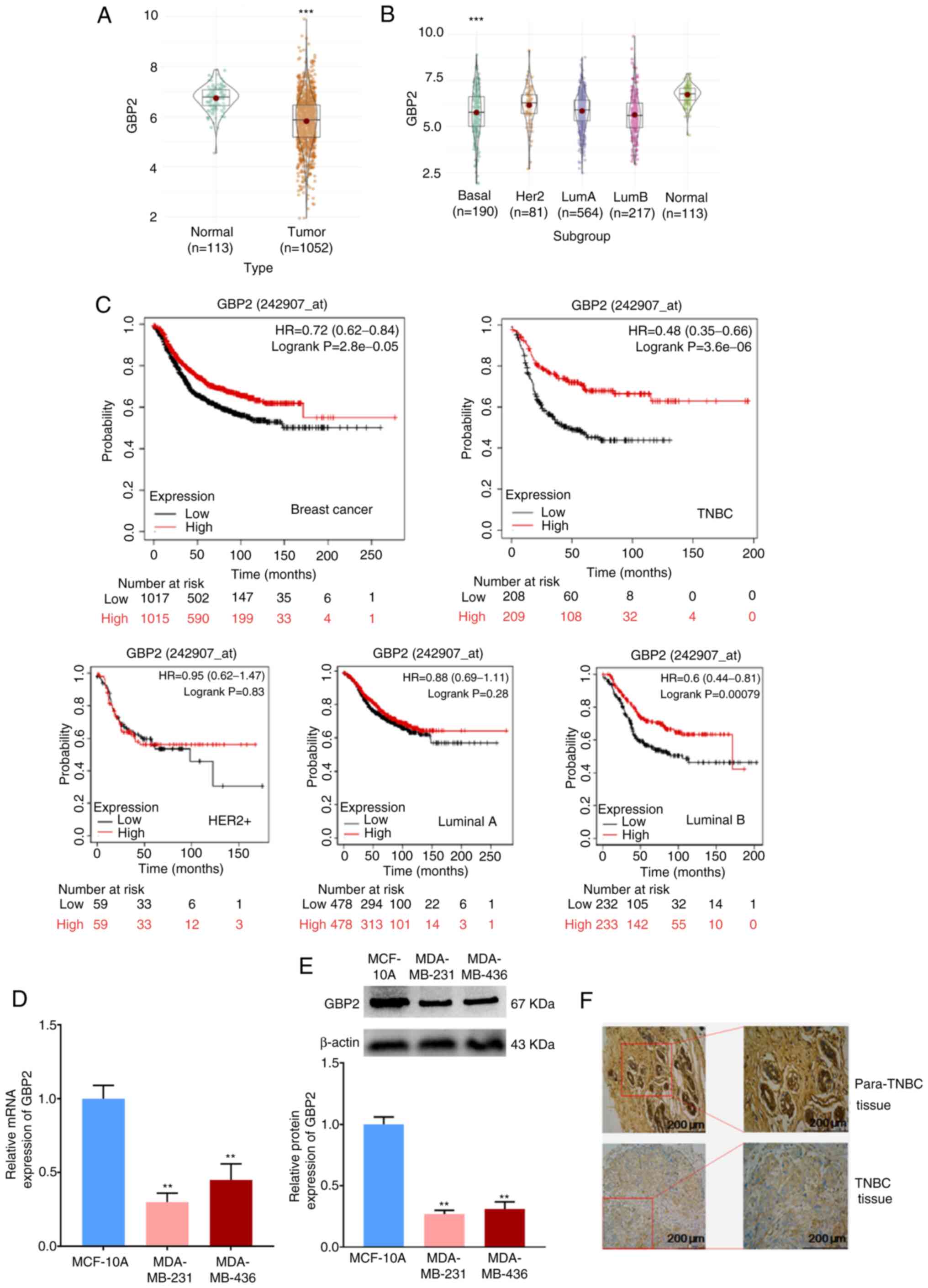
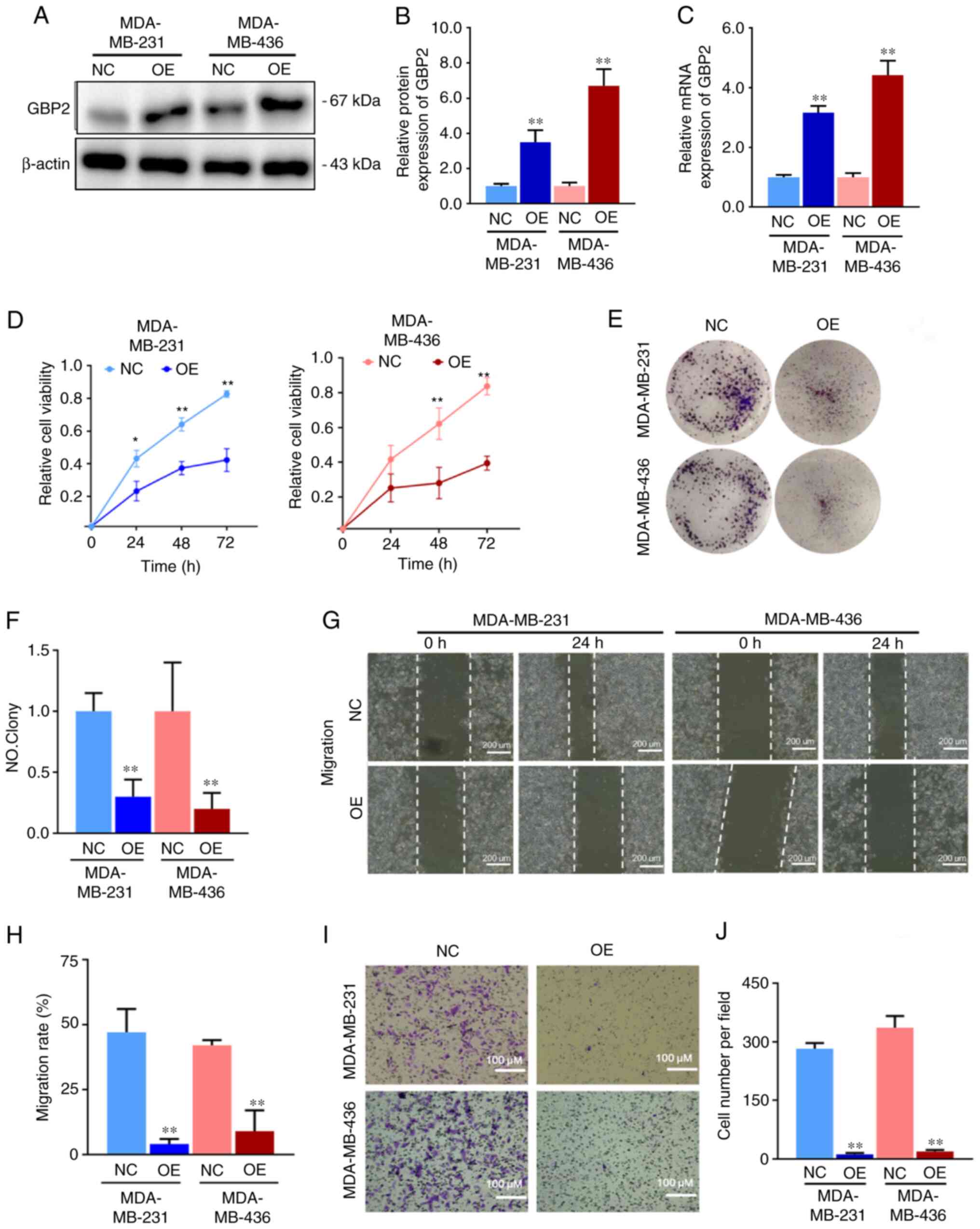
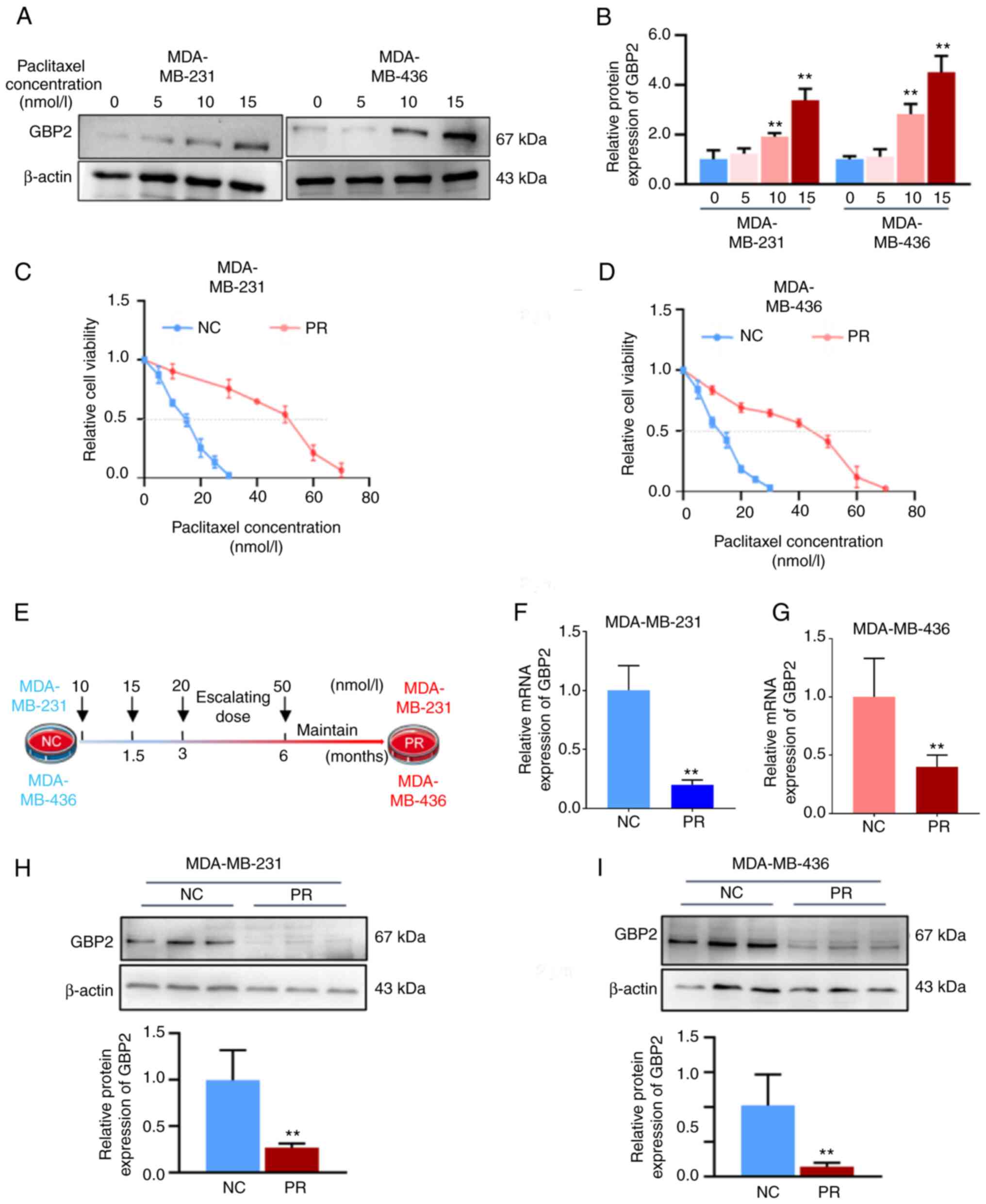
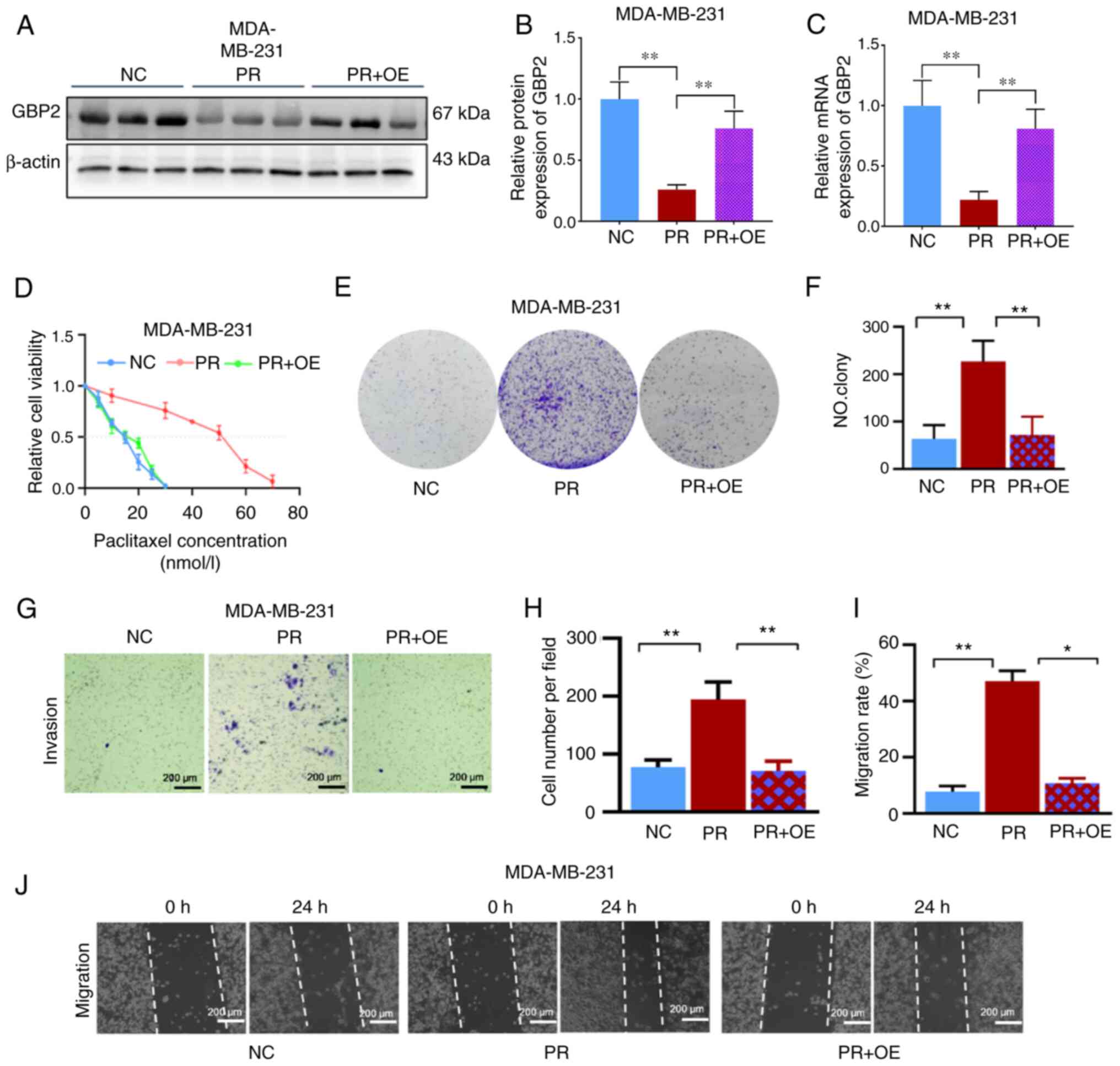
|
1
|
International Agency for Research on
Cancer (IARC): Global Cancer Observatory. IARC; Lyon: 2020
|
|
2
|
Smolarz B, Nowak AZ and Romanowicz H:
Breast cancer-epidemiology, classification, pathogenesis and
treatment (review of literature). Cancers (Basel). 14:25692022.
|
|
3
|
da Silva JL, Cardoso Nunes NC, Izetti P,
de Mesquita GG and de Melo AC: Triple negative breast cancer: A
thorough review of biomarkers. Crit Rev Oncol Hematol.
145:1028552020.
|
|
4
|
M Braden A, V Stankowski R, M Engel J and
A Onitilo A: Breast cancer biomarkers: risk assessment, diagnosis,
prognosis, prediction of treatment efficacy and toxicity, and
recurrence. Curr Pharm Des. 20:4879–4898. 2014.
|
|
5
|
Vestal DJ and Jeyaratnam JA: The
guanylate-binding proteins: emerging insights into the biochemical
properties and functions of this family of large interferon-induced
guanosine triphosphatase. J Interferon Cytokine Res. 31:89–97.
2011.
|
|
6
|
Guimarães DP, Oliveira IM, de Moraes E,
Paiva GR, Souza DM, Barnas C, Olmedo DB, Pinto CE, Faria PA, De
Moura Gallo CV, et al: Interferon-inducible guanylate binding
protein (GBP)-2: A novel p53-regulated tumor marker in esophageal
squamous cell carcinomas. Int J Cancer. 124:272–279. 2009.
|
|
7
|
Yu S, Yu X, Sun L, Zheng Y, Chen L, Xu H,
Jin J, Lan Q, Chen CC and Li M: GBP2 enhances glioblastoma invasion
through Stat3/fibronectin pathway. Oncogene. 39:5042–5055.
2020.
|
|
8
|
Ye S, Li S, Qin L, Zheng W, Liu B, Li X,
Ren Z, Zhao H, Hu X, Ye N and Li G: GBP2 promotes clear cell renal
cell carcinoma progression through immune infiltration and
regulation of PD-L1 expression via STAT1 signaling. Oncol Rep.
49:492023.
|
|
9
|
Wang J, Min H, Hu B, Xue X and Liu Y:
Guanylate-binding protein-2 inhibits colorectal cancer cell growth
and increases the sensitivity to paclitaxel of paclitaxel-resistant
colorectal cancer cells by interfering Wnt signaling. J Cell
Biochem. 121:1250–1259. 2020.
|
|
10
|
Zhang S, Chen K, Zhao Z, Zhang X, Xu L,
Liu T and Yu S: Lower expression of GBP2 associated with less
immune cell infiltration and poor prognosis in skin cutaneous
melanoma (SKCM). J Immunother. 45:274–283. 2022.
|
|
11
|
Godoy P, Cadenas C, Hellwig B, Marchan R,
Stewart J, Reif R, Lohr M, Gehrmann M, Rahnenführer J, Schmidt M
and Hengstler JG: Interferon-inducible guanylate binding protein
(GBP2) is associated with better prognosis in breast cancer and
indicates an efficient T cell response. Breast Cancer. 21:491–499.
2014.
|
|
12
|
Nyabuto GO, Wilson JP, Heilman SA, Kalb
RC, Kopacz JP, Abnave AV and Vestal DJ: The large GTPase, GBP-2,
regulates Rho family GTPases to inhibit migration and invadosome
formation in breast cancer cells. Cancers (Basel). 13:56322021.
|
|
13
|
Zhang J, Zhang Y, Wu W, Wang F, Liu X,
Shui G and Nie C: Guanylate-binding protein 2 regulates
Drp1-mediated mitochondrial fission to suppress breast cancer cell
invasion. Cell Death Dis. 8:e31512017.
|
|
14
|
Škubník J, Jurášek M, Ruml T and Rimpelová
S: Mitotic poisons in research and medicine. Molecules.
25:46322020.
|
|
15
|
Alqahtani FY, Aleanizy FS, El Tahir E,
Alkahtani HM and AlQuadeib BT: Paclitaxel. Profiles Drug Subst
Excip Relat Methodol. 44:205–238. 2019.
|
|
16
|
Wang H, Vo T, Hajar A, Li S, Chen X,
Parissenti AM, Brindley DN and Wang Z: Multiple mechanisms
underlying acquired resistance to taxanes in selected
docetaxel-resistant MCF-7 breast cancer cells. BMC Cancer.
14:372014.
|
|
17
|
McGrogan BT, Gilmartin B, Carney DN and
McCann A: Taxanes, microtubules and chemoresistant breast cancer.
Biochim Biophys Acta. 1785:96–132. 2008.
|
|
18
|
Wang J, Yang M, Li Y and Han B: The role
of MicroRNAs in the chemoresistance of breast cancer. Drug Dev Res.
76:368–374. 2015.
|
|
19
|
Ho CJ and Gorski SM: Molecular mechanisms
underlying autophagy-mediated treatment resistance in cancer.
Cancers (Basel). 11:17752019.
|
|
20
|
Hashemi M, Paskeh MDA, Orouei S, Abbasi P,
Khorrami R, Dehghanpour A, Esmaeili N, Ghahremanzade A, Zandieh MA,
Peymani M, et al: Towards dual function of autophagy in breast
cancer: A potent regulator of tumor progression and therapy
response. Biomed Pharmacother. 161:1145462023.
|
|
21
|
Škubník J, Svobodová Pavlíčková V, Ruml T
and Rimpelová S: Autophagy in cancer resistance to paclitaxel:
Development of combination strategies. Biomed Pharmacother.
161:1144582023.
|
|
22
|
Dunn LA, Fury MG, Xiao H, Baxi SS, Sherman
EJ, Korte S, Pfister C, Haque S, Katabi N, Ho AL and Pfister DG: A
phase II study of temsirolimus added to low-dose weekly carboplatin
and paclitaxel for patients with recurrent and/or metastatic (R/M)
head and neck squamous cell carcinoma (HNSCC). Ann Oncol.
28:2533–2538. 2017.
|
|
23
|
Li S and De Souza P: Ras isoprenylation
and pAkt inhibition by zoledronic acid and fluvastatin enhances
paclitaxel activity in T24 bladder cancer cells. Cancers (Basel).
3:662–674. 2011.
|
|
24
|
Hama Y, Ogasawara Y and Noda NN: Autophagy
and cancer: Basic mechanisms and inhibitor development. Cancer Sci.
114:2699–2708. 2023.
|
|
25
|
Osawa T, Kotani T, Kawaoka T, Hirata E,
Suzuki K, Nakatogawa H, Ohsumi Y and Noda NN: Atg2 mediates direct
lipid transfer between membranes for autophagosome formation. Nat
Struct Mol Biol. 26:281–288. 2019.
|
|
26
|
Xiao W, Wang J, Wang X, Cai S, Guo Y, Ye
L, Li D, Hu A, Jin S, Yuan B, et al: Therapeutic targeting of the
USP2-E2F4 axis inhibits autophagic machinery essential for zinc
homeostasis in cancer progression. Autophagy. 18:2615–2635.
2022.
|
|
27
|
Liu N, Luo T, Zhang J, Han LN, Duan WQ, Lu
WX, Qiu H, Lin Y, Wu YM, Zhang H, et al: YF343, a novel histone
deacetylase inhibitor, combined with CQ to inhibit-autophagy,
contributes to increased apoptosis in triple-negative breast
cancer. Curr Med Chem. 30:4605–4621. 2023.
|
|
28
|
Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang
H, Hu Y, Qiu J, Pu L, Tang J and Wang X: HIF-1α-induced expression
of m6A reader YTHDF1 drives hypoxia-induced autophagy and
malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14
translation. Signal Transduct Target Ther. 6:762021.
|
|
29
|
Niklaus NJ, Tokarchuk I, Zbinden M,
Schläfli AM, Maycotte P and Tschan MP: The multifaceted functions
of autophagy in breast cancer development and treatment. Cells.
10:14472021.
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
31
|
Dunn KW, Kamocka MM and McDonald JH: A
practical guide to evaluating colocalization in biological
microscopy. Am J Physiol Cell Physiol. 300:C723–C742. 2011.
|
|
32
|
Abd El-Aziz YS, Gillson J, Jansson PJ and
Sahni S: Autophagy: A promising target for triple negative breast
cancers. Pharmacol Res. 175:1060062022.
|
|
33
|
Folkerts H, Hilgendorf S, Vellenga E,
Bremer E and Wiersma VR: The multifaceted role of autophagy in
cancer and the microenvironment. Med Res Rev. 39:517–560. 2019.
|
|
34
|
Chen W, Bai Y, Patel C and Geng F:
Autophagy promotes triple negative breast cancer metastasis via YAP
nuclear localization. Biochem Biophys Res Commun. 520:263–268.
2019.
|
|
35
|
Wang J, Dang MN and Day ES: Inhibition of
Wnt signaling by Frizzled7 antibody-coated nanoshells sensitizes
triple-negative breast cancer cells to the autophagy regulator
chloroquine. Nano Res. 13:1693–1703. 2020.
|
|
36
|
Wang M, Zhang J, Huang Y, Ji S, Shao G,
Feng S, Chen D, Zhao K, Wang Z and Wu A: Cancer-associated
fibroblasts autophagy enhances progression of triple-negative
breast cancer cells. Med Sci Monit. 23:3904–3912. 2017.
|
|
37
|
Li M, Liu J, Li S, Feng Y, Yi F, Wang L,
Wei S and Cao L: Autophagy-related 7 modulates tumor progression in
triple-negative breast cancer. Lab Invest. 99:1266–1274. 2019.
|
|
38
|
Qiao Z, Li X, Kang N, Yang Y, Chen C, Wu
T, Zhao M, Liu Y and Ji X: A novel specific anti-CD73 antibody
inhibits triple-negative breast cancer cell motility by regulating
autophagy. Int J Mol Sci. 20:10572019.
|
|
39
|
Chen Y, Azad MB and Gibson SB: Methods for
detecting autophagy and determining autophagy-induced cell death.
Can J Physiol Pharmacol. 88:285–295. 2010.
|
|
40
|
Chen C, Gao H and Su X: Autophagy-related
signaling pathways are involved in cancer (review). Exp Ther Med.
22:7102021.
|
|
41
|
Ciccarese F, Zulato E and Indraccolo S:
LKB1/AMPK pathway and drug response in cancer: A therapeutic
perspective. Oxid Med Cell Longev. 2019:87308162019.
|
|
42
|
Lu C, Wang W, Jia Y, Liu X, Tong Z and Li
B: Inhibition of AMPK/autophagy potentiates parthenolide-induced
apoptosis in human breast cancer cells. J Cell Biochem.
115:1458–1466. 2014.
|
|
43
|
Zhou YY, Li Y, Jiang WQ and Zhou LF:
MAPK/JNK signaling: A potential autophagy regulation pathway.
Biosci Rep. 35:e001992015.
|
|
44
|
Lorzadeh S, Kohan L, Ghavami S and
Azarpira N: Autophagy and the Wnt signaling pathway: A focus on
Wnt/β-catenin signaling. Biochim Biophys Acta Mol Cell Res.
1868:1189262021.
|
|
45
|
Ravikumar B, Sarkar S, Davies JE, Futter
M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M,
Korolchuk VI, Lichtenberg M, Luo S, et al: Regulation of mammalian
autophagy in physiology and pathophysiology. Physiol Rev.
90:1383–1435. 2010.
|
|
46
|
Wang Y and Zhang H: Regulation of
autophagy by mTOR signaling pathway. Adv Exp Med Biol. 1206:67–83.
2019.
|
|
47
|
Li L, Tong M, Fu Y, Chen F, Zhang S, Chen
H, Ma X, Li D, Liu X and Zhong Q: Lipids and membrane-associated
proteins in autophagy. Protein Cell. 12:520–544. 2021.
|
|
48
|
Cheng SW, Chen PC, Ger TR, Chiu HW and Lin
YF: GBP5 serves as a potential marker to predict a favorable
response in triple-negative breast cancer patients receiving a
taxane-based chemotherapy. J Pers Med. 11:1972021.
|
|
49
|
Peng Y, Yu H, Jin Y, Qu F, Ren H, Tang Z,
Zhang Y, Qu C, Zong B and Liu S: Construction and validation of an
immune infiltration-related gene signature for the prediction of
prognosis and therapeutic response in breast cancer. Front Immunol.
12:6661372021.
|
|
50
|
Muller R: Systemic toxicity of chloroquine
and hydroxychloroquine: Prevalence, mechanisms, risk factors,
prognostic and screening possibilities. Rheumatol Int.
41:1189–1202. 2021.
|
|
51
|
Wu Y, Yan B, Xu W, Guo L, Wang Z, Li G,
Hou N, Zhang J and Ling R: Compound C enhances the anticancer
effect of aspirin in HER-2-positive breast cancer by regulating
lipid metabolism in an AMPK-independent pathway. Int J Biol Sci.
16:583–597. 2020.
|
|
52
|
Milošević Z, Banković J, Dinić J,
Tsimplouli C, Sereti E, Dragoj M, Paunović V, Milovanović Z,
Stepanović M, Tanić N, et al: Potential of the dual mTOR kinase
inhibitor AZD2014 to overcome paclitaxel resistance in anaplastic
thyroid carcinoma. Cell Oncol (Dordr). 41:409–426. 2018.
|
|
53
|
Park GB, Jeong JY and Kim D: Gliotoxin
enhances autophagic cell death via the DAPK1-TAp63 signaling
pathway in paclitaxel-resistant ovarian cancer cells. Mar Drugs.
17:4122019.
|
|
54
|
Guo Y, Yuan J, Yin S, Wang X, Shuai R and
Kang J: MAP2K6-FP enhances the sensitiveness of paclitaxel for
ovarian cancer via inducing autophagy. Int J Gynecol Cancer.
27:1082–1087. 2017.
|
|
55
|
Song H, Pan B, Yi J and Chen L: Featured
article: Autophagic activation with nimotuzumab enhanced
chemosensitivity and radiosensitivity of esophageal squamous cell
carcinoma. Exp Biol Med (Maywood). 239:529–541. 2014.
|
|
56
|
Lian B, Pei YC, Jiang YZ, Xue MZ, Li DQ,
Li XG, Zheng YZ, Liu XY, Qiao F, Sun WL, et al: Truncated HDAC9
identified by integrated genome-wide screen as the key modulator
for paclitaxel resistance in triple-negative breast cancer.
Theranostics. 10:11092–11109. 2020.
|
|
57
|
Oudin MJ, Barbier L, Schäfer C, Kosciuk T,
Miller MA, Han S, Jonas O, Lauffenburger DA and Gertler FB: MENA
confers resistance to paclitaxel in triple-negative breast cancer.
Mol Cancer Ther. 16:143–155. 2017.
|
|
58
|
Wang Y, Sui Y and Tao Y: Gambogic acid
increases the sensitivity to paclitaxel in drug-resistant
triple-negative breast cancer via the SHH signaling pathway. Mol
Med Rep. 20:4515–4522. 2019.
|