|
1
|
Lumish MA, Kohn EC and Tew WP: Top
advances of the year: Ovarian cancer. Cancer. 130:837–845.
2024.
|
|
2
|
Tang H, Kulkarni S, Peters C, Eddison J,
Al-Ani M and Madhusudan S: The current status of
DNA-repair-directed precision oncology strategies in epithelial
ovarian cancers. Int J Mol Sci. 24:72932023.
|
|
3
|
Feng J, Xu L, Chen Y, Lin R, Li H and He
H: Trends in incidence and mortality for ovarian cancer in China
from 1990 to 2019 and its forecasted levels in 30 years. J Ovarian
Res. 16:1392023.
|
|
4
|
Bodelon C, Killian JK, Sampson JN,
Anderson WF, Matsuno R, Brinton LA, Lissowska J, Anglesio MS,
Bowtell DDL, Doherty JA, et al: Molecular classification of
epithelial ovarian cancer based on methylation profiling: Evidence
for survival heterogeneity. Clin Cancer Res. 25:5937–5946.
2019.
|
|
5
|
Zhang M, Cheng S, Jin Y, Zhao Y and Wang
Y: Roles of CA125 in diagnosis, prediction, and oncogenesis of
ovarian cancer. Biochim Biophys Acta Rev Cancer.
1875:1885032021.
|
|
6
|
Lheureux S, Braunstein M and Oza AM:
Epithelial ovarian cancer: Evolution of management in the era of
precision medicine. CA Cancer J Clin. 69:280–304. 2019.
|
|
7
|
Natanzon Y, Goode EL and Cunningham JM:
Epigenetics in ovarian cancer. Semin Cancer Biol. 51:160–169.
2018.
|
|
8
|
Moufarrij S, Dandapani M, Arthofer E,
Gomez S, Srivastava A, Lopez-Acevedo M, Villagra A and Chiappinelli
KB: Epigenetic therapy for ovarian cancer: Promise and progress.
Clin Epigenetics. 11:72019.
|
|
9
|
Peng S, Zhang X and Wu Y: Potential
applications of DNA methylation testing technology in female tumors
and screening methods. Biochim Biophys Acta Rev Cancer.
1878:1889412023.
|
|
10
|
Antonino M, Nicolò M, Jerome Renee L,
Federico M, Chiara V, Stefano S, Maria S, Salvatore C, Antonio B,
Calvo-Henriquez C, et al: Single-nucleotide polymorphism in chronic
rhinosinusitis: A systematic review. Clin Otolaryngol. 47:14–23.
2022.
|
|
11
|
Matei D and Nephew KP: Epigenetic attire
in ovarian cancer: The emperor's new clothes. Cancer Res.
80:3775–3785. 2020.
|
|
12
|
Matei D, Fang F, Shen C, Schilder J,
Arnold A, Zeng Y, Berry WA, Huang T and Nephew KP: Epigenetic
resensitization to platinum in ovarian cancer. Cancer Res.
72:2197–2205. 2012.
|
|
13
|
Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi
Y and Cao L: DNA methylation, its mediators and genome integrity.
Int J Biol Sci. 11:604–617. 2015.
|
|
14
|
Singh A, Gupta S and Sachan M: Epigenetic
biomarkers in the management of ovarian cancer: Current
prospectives. Front Cell Dev Biol. 7:1822019.
|
|
15
|
Ma L, Li C, Yin H, Huang J, Yu S, Zhao J,
Tang Y, Yu M, Lin J, Ding L and Cui Q: The mechanism of DNA
methylation and miRNA in breast cancer. Int J Mol Sci.
24:93602023.
|
|
16
|
Coughlan AY and Testa G: Exploiting
epigenetic dependencies in ovarian cancer therapy. Int J Cancer.
149:1732–1743. 2021.
|
|
17
|
Xie W, Sun H, Li X, Lin F, Wang Z and Wang
X: Ovarian cancer: Epigenetics, drug resistance, and progression.
Cancer Cell Int. 21:4342021.
|
|
18
|
Klymenko Y and Nephew KP: Epigenetic
crosstalk between the tumor microenvironment and ovarian cancer
cells: A therapeutic road less traveled. Cancers (Basel).
10:2952018.
|
|
19
|
Yang Y, Wu L, Shu X, Lu Y, Shu XO, Cai Q,
Beeghly-Fadiel A, Li B, Ye F, Berchuck A, et al: Genetic data from
nearly 63,000 women of european descent predicts DNA methylation
biomarkers and epithelial ovarian cancer risk. Cancer Res.
79:505–517. 2019.
|
|
20
|
Lo Riso P, Villa CE, Gasparoni G, Vingiani
A, Luongo R, Manfredi A, Jungmann A, Bertolotti A, Borgo F, Garbi
A, et al: A cell-of-origin epigenetic tracer reveals clinically
distinct subtypes of high-grade serous ovarian cancer. Genome Med.
12:942020.
|
|
21
|
Tomar T, Alkema NG, Schreuder L, Meersma
GJ, de Meyer T, van Criekinge W, Klip HG, Fiegl H, van
Nieuwenhuysen E, Vergote I, et al: Methylome analysis of extreme
chemoresponsive patients identifies novel markers of platinum
sensitivity in high-grade serous ovarian cancer. BMC Med.
15:1162017.
|
|
22
|
Feng LY, Yan BB, Huang YZ and Li L:
Abnormal methylation characteristics predict chemoresistance and
poor prognosis in advanced high-grade serous ovarian cancer. Clin
Epigenetics. 13:1412021.
|
|
23
|
Yang D, Khan S, Sun Y, Hess K, Shmulevich
I, Sood AK and Zhang W: Association of BRCA1 and BRCA2 mutations
with survival, chemotherapy sensitivity, and gene mutator phenotype
in patients with ovarian cancer. JAMA. 306:1557–1565. 2011.
|
|
24
|
Chan KY, Ozçelik H, Cheung AN, Ngan HY and
Khoo US: Epigenetic factors controlling the BRCA1 and BRCA2 genes
in sporadic ovarian cancer. Cancer Res. 62:4151–4156. 2002.
|
|
25
|
Stewart C, Ralyea C and Lockwood S:
Ovarian cancer: An integrated review. Semin Oncol Nurs. 35:151–156.
2019.
|
|
26
|
Moschetta M, George A, Kaye SB and
Banerjee S: BRCA somatic mutations and epigenetic BRCA
modifications in serous ovarian cancer. Ann Oncol. 27:1449–1455.
2016.
|
|
27
|
Glajzer J, Castillo-Tong DC, Richter R,
Vergote I, Kulbe H, Vanderstichele A, Ruscito I, Trillsch F, Mustea
A, Kreuzinger C, et al: Impact of BRCA mutation status on tumor
dissemination pattern, surgical outcome and patient survival in
primary and recurrent high-grade serous ovarian cancer: A
multicenter retrospective study by the ovarian cancer
therapy-innovative models prolong survival (OCTIPS) consortium. Ann
Surg Oncol. 30:35–45. 2023.
|
|
28
|
Jung Y, Hur S, Liu J, Lee S, Kang BS, Kim
M and Choi YJ: Peripheral blood BRCA1 methylation profiling to
predict familial ovarian cancer. J Gynecol Oncol. 32:e232021.
|
|
29
|
Barrett JE, Jones A, Evans I, Reisel D,
Herzog C, Chindera K, Kristiansen M, Leavy OC, Manchanda R, Bjørge
L, et al: The DNA methylome of cervical cells can predict the
presence of ovarian cancer. Nat Commun. 13:4482022.
|
|
30
|
Wu TI, Huang RL, Su PH, Mao SP, Wu CH and
Lai HC: Ovarian cancer detection by DNA methylation in cervical
scrapings. Clin Epigenetics. 11:1662019.
|
|
31
|
Bartlett TE, Chindera K, McDermott J,
Breeze CE, Cooke WR, Jones A, Reisel D, Karegodar ST, Arora R, Beck
S, et al: Epigenetic reprogramming of fallopian tube fimbriae in
BRCA mutation carriers defines early ovarian cancer evolution. Nat
Commun. 7:116202016.
|
|
32
|
Ibragimova I and Cairns P: Assays for
hypermethylation of the BRCA1 gene promoter in tumor cells to
predict sensitivity to PARP-inhibitor therapy. Methods Mol Biol.
780:277–291. 2011.
|
|
33
|
Sahnane N, Carnevali I, Formenti G,
Casarin J, Facchi S, Bombelli R, Di Lauro E, Memoli D, Salvati A,
Rizzo F, et al: BRCA methylation testing identifies a subset of
ovarian carcinomas without germline variants that can benefit from
PARP inhibitor. Int J Mol Sci. 21:97082020.
|
|
34
|
Taniguchi T, Tischkowitz M, Ameziane N,
Hodgson SV, Mathew CG, Joenje H, Mok SC and D'Andrea AD: Disruption
of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian
tumors. Nat Med. 9:568–574. 2003.
|
|
35
|
Cancer Genome Atlas Research Network:
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011.
|
|
36
|
Alsop K, Fereday S, Meldrum C, deFazio A,
Emmanuel C, George J, Dobrovic A, Birrer MJ, Webb PM, Stewart C, et
al: BRCA mutation frequency and patterns of treatment response in
BRCA mutation-positive women with ovarian cancer: A report from the
Australian ovarian cancer study group. J Clin Oncol. 30:2654–2663.
2012.
|
|
37
|
Mikeska T, Alsop K; Australian Ovarian
Cancer Study Group; Mitchell G, Bowtell DD and Dobrovic A: No
evidence for PALB2 methylation in high-grade serous ovarian cancer.
J Ovarian Res. 6:262013.
|
|
38
|
McAlpine JN, Porter H, Köbel M, Nelson BH,
Prentice LM, Kalloger SE, Senz J, Milne K, Ding J, Shah SP, et al:
BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and
presence of immune cell infiltrates in ovarian high-grade serous
carcinoma. Mod Pathol. 25:740–750. 2012.
|
|
39
|
Shariati-Kohbanani M, Zare-Bidaki M,
Taghavi MM, Taghipour Z, Shabanizadeh A, Kennedy D, Dahim H,
Salahshoor MR, Jalili C and Kazemi Arababadi M: DNA methylation and
microRNA patterns are in association with the expression of BRCA1
in ovarian cancer. Cell Mol Biol (Noisy-le-grand). 62:16–23.
2016.
|
|
40
|
Soslow RA, Han G, Park KJ, Garg K, Olvera
N, Spriggs DR, Kauff ND and Levine DA: Morphologic patterns
associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod
Pathol. 25:625–536. 2012.
|
|
41
|
Kraya AA, Maxwell KN, Eiva MA, Wubbenhorst
B, Pluta J, Feldman M, Nayak A, Powell DJ Jr, Domchek SM,
Vonderheide RH and Nathanson KL: PTEN loss and BRCA1 promoter
hypermethylation negatively predict for immunogenicity in
BRCA-deficient ovarian cancer. JCO Precis Oncol.
6:e21001592022.
|
|
42
|
Ebata T, Yamashita S, Takeshima H, Yoshida
H, Kawata Y, Kino N, Yasugi T, Terao Y, Yonemori K, Kato T and
Ushijima T: DNA methylation of the immediate upstream region of
BRCA1 major transcription start sites is an independent favorable
prognostic factor in patients with high-grade serous ovarian
cancer. Gynecol Oncol. 167:513–518. 2022.
|
|
43
|
Bai X, Fu Y, Xue H, Guo K, Song Z, Yu Z,
Jia T, Yan Y, Zhao L, Mi X, et al: BRCA1 promoter hypermethylation
in sporadic epithelial ovarian carcinoma: Association with low
expression of BRCA1, improved survival and co-expression of DNA
methyltransferases. Oncol Lett. 7:1088–1096. 2014.
|
|
44
|
Pradjatmo H: Methylation status and
expression of BRCA2 in epithelial ovarian cancers in Indonesia.
Asian Pac J Cancer Prev. 16:8599–8604. 2015.
|
|
45
|
Lane DP: p53 and human cancers. Br Med
Bull. 50:582–599. 1994.
|
|
46
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012.
|
|
47
|
Chen YC, Young MJ, Chang HP, Liu CY, Lee
CC, Tseng YL, Wang YC, Chang WC and Hung JJ: Estradiol-mediated
inhibition of DNMT1 decreases p53 expression to induce
M2-macrophage polarization in lung cancer progression. Oncogenesis.
11:252022.
|
|
48
|
Suh SO, Chen Y, Zaman MS, Hirata H,
Yamamura S, Shahryari V, Liu J, Tabatabai ZL, Kakar S, Deng G, et
al: MicroRNA-145 is regulated by DNA methylation and p53 gene
mutation in prostate cancer. Carcinogenesis. 32:772–778. 2011.
|
|
49
|
Cunningham JM, Winham SJ, Wang C, Weiglt
B, Fu Z, Armasu SM, McCauley BM, Brand AH, Chiew YE, Elishaev E, et
al: DNA methylation profiles of ovarian clear cell carcinoma.
Cancer Epidemiol Biomarkers Prev. 31:132–141. 2022.
|
|
50
|
Chmelarova M, Krepinska E, Spacek J, Laco
J, Beranek M and Palicka V: Methylation in the p53 promoter in
epithelial ovarian cancer. Clin Transl Oncol. 15:160–163. 2013.
|
|
51
|
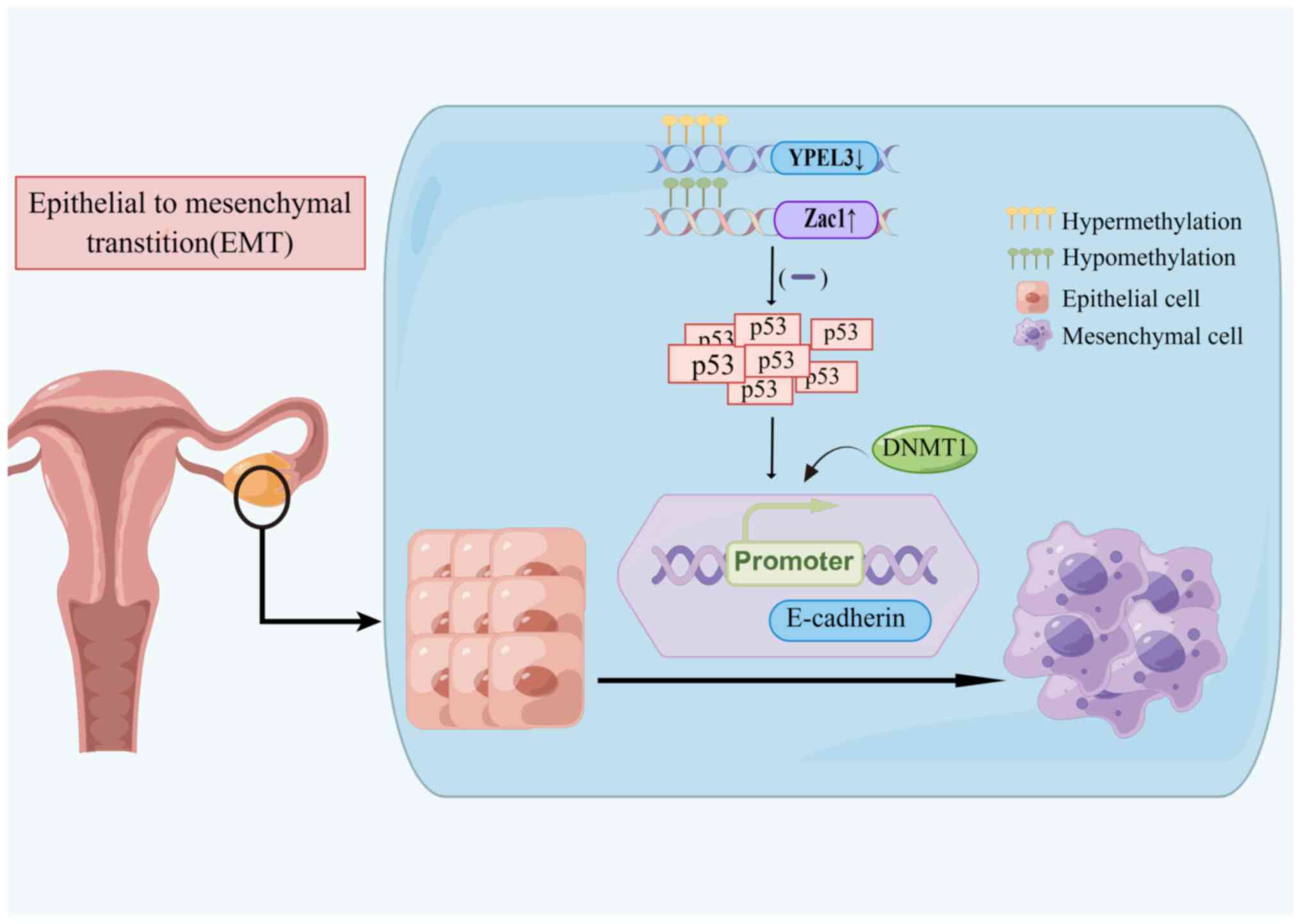
Kelley KD, Miller KR, Todd A, Kelley AR,
Tuttle R and Berberich SJ: YPEL3, a p53-regulated gene that induces
cellular senescence. Cancer Res. 70:3566–3575. 2010.
|
|
52
|
Abdollahi A: LOT1 (ZAC1/PLAGL1) and its
family members: Mechanisms and functions. J Cell Physiol.
210:16–25. 2007.
|
|
53
|
Su HC, Wu SC, Yen LC, Chiao LK, Wang JK,
Chiu YL, Ho CL and Huang SM: Gene expression profiling identifies
the role of Zac1 in cervical cancer metastasis. Sci Rep.
10:118372020.
|
|
54
|
Cheng JC, Auersperg N and Leung PC:
Inhibition of p53 represses E-cadherin expression by increasing DNA
methyltransferase-1 and promoter methylation in serous borderline
ovarian tumor cells. Oncogene. 30:3930–3942. 2011.
|
|
55
|
Bin Y, Ding Y, Xiao W and Liao A: RASSF1A:
A promising target for the diagnosis and treatment of cancer. Clin
Chim Acta. 504:98–108. 2020.
|
|
56
|
Wei B, Wu F, Xing W, Sun H, Yan C, Zhao C,
Wang D, Chen X, Chen Y, Li M and Ma J: A panel of DNA methylation
biomarkers for detection and improving diagnostic efficiency of
lung cancer. Sci Rep. 11:167822021.
|
|
57
|
Tang Q, Cheng J, Cao X, Surowy H and
Burwinkel B: Blood-based DNA methylation as biomarker for breast
cancer: A systematic review. Clin Epigenetics. 8:1152016.
|
|
58
|
Shi H, Li Y, Wang X, Lu C, Yang L, Gu C,
Xiong J, Huang Y, Wang S and Lu M: Association between RASSF1A
promoter methylation and ovarian cancer: A meta-analysis. PLoS One.
8:e767872013.
|
|
59
|
Dammann R, Schagdarsurengin U, Strunnikova
M, Rastetter M, Seidel C, Liu L, Tommasi S and Pfeifer GP:
Epigenetic inactivation of the Ras-association domain family 1
(RASSF1A) gene and its function in human carcinogenesis. Histol
Histopathol. 18:665–677. 2003.
|
|
60
|
Terp SK, Stoico MP, Dybkaer K and Pedersen
IS: Early diagnosis of ovarian cancer based on methylation profiles
in peripheral blood cell-free DNA: A systematic review. Clin
Epigenetics. 15:242023.
|
|
61
|
Rezk NA, Mohamed RH, Alnemr AA and Harira
M: Promoter methylation of RASSF1A gene in egyptian patients with
ovarian cancer. Appl Biochem Biotechnol. 185:153–162. 2018.
|
|
62
|
Bhagat R, Chadaga S, Premalata CS, Ramesh
G, Ramesh C, Pallavi VR and Krishnamoorthy L: Aberrant promoter
methylation of the RASSF1A and APC genes in epithelial ovarian
carcinoma development. Cell Oncol (Dordr). 35:473–479. 2012.
|
|
63
|
Xing BL, Li T, Tang ZH, Jiao L, Ge SM,
Qiang X and OuYang J: Cumulative methylation alternations of gene
promoters and protein markers for diagnosis of epithelial ovarian
cancer. Genet Mol Res. 14:4532–4540. 2015.
|
|
64
|
Vo LT, Thuan TB, Thu DM, Uyen NQ, Ha NT
and To TV: Methylation profile of BRCA1, RASSF1A and ER in
Vietnamese women with ovarian cancer. Asian Pac J Cancer Prev.
14:7713–7718. 2013.
|
|
65
|
Wang H, Cui M, Zhang S, He J, Song L and
Chen Y: Relationship between RAS association domain family protein
1A promoter methylation and the clinicopathological characteristics
in patients with ovarian cancer: A systematic meta-analysis.
Gynecol Obstet Invest. 83:349–357. 2018.
|
|
66
|
Ho CM, Yen TL, Chien TY and Huang SH:
Distinct promotor methylation at tumor suppressive genes in ovarian
cancer stromal progenitor cells and ovarian cancer and its clinical
implication. Am J Cancer Res. 12:5325–5341. 2022.
|
|
67
|
Ho CM, Shih DTB, Hsiao CC, Huang SH, Chang
SF and Cheng WF: Gene methylation of human ovarian carcinoma
stromal progenitor cells promotes tumorigenesis. J Transl Med.
13:3672015.
|
|
68
|
Reyes HD, Devor EJ, Warrier A, Newtson AM,
Mattson J, Wagner V, Duncan GN, Leslie KK and Gonzalez-Bosquet J:
Differential DNA methylation in high-grade serous ovarian cancer
(HGSOC) is associated with tumor behavior. Sci Rep.
9:179962019.
|
|
69
|
Feng Q, Deftereos G, Hawes SE, Stern JE,
Willner JB, Swisher EM, Xi L, Drescher C, Urban N and Kiviat N: DNA
hypermethylation, Her-2/neu overexpression and p53 mutations in
ovarian carcinoma. Gynecol Oncol. 111:320–329. 2008.
|
|
70
|
Xie G, Hu C and Huang M: Methylation
status of RASSF1A and clinical efficacy of neoadjuvant therapy in
patients with advanced epithelial ovarian cancer. Zhong Nan Da Xue
Xue Bao Yi Xue Ban. 36:631–633. 2011.In Chinese.
|
|
71
|
Giannopoulou L, Chebouti I, Pavlakis K,
Kasimir-Bauer S and Lianidou ES: RASSF1A promoter methylation in
high-grade serous ovarian cancer: A direct comparison study in
primary tumors, adjacent morphologically tumor cell-free tissues
and paired circulating tumor DNA. Oncotarget. 8:21429–21443.
2017.
|
|
72
|
S SK, Swamy SN, Premalatha CS, Pallavi VR
and Gawari R: Aberrant promoter hypermethylation of RASSF1a and
BRCA1 in circulating cell-free tumor DNA serves as a biomarker of
ovarian carcinoma. Asian Pac J Cancer Prev. 20:3001–3005. 2019.
|
|
73
|
Marfella CG and Imbalzano AN: The Chd
family of chromatin remodelers. Mutat Res. 618:30–40. 2007.
|
|
74
|
Zhao R, Yan Q, Lv J, Huang H, Zheng W,
Zhang B and Ma W: CHD5, a tumor suppressor that is epigenetically
silenced in lung cancer. Lung Cancer. 76:324–231. 2012.
|
|
75
|
Xie CR, Li Z, Sun HG, Wang FQ, Sun Y, Zhao
WX, Zhang S, Zhao WX, Wang XM and Yin ZY: Mutual regulation between
CHD5 and EZH2 in hepatocellular carcinoma. Oncotarget.
6:40940–40952. 2015.
|
|
76
|
Qu Y, Dang S and Hou P: Gene methylation
in gastric cancer. Clin Chim Acta. 424:53–65. 2013.
|
|
77
|
Du Z, Li L, Huang X, Jin J, Huang S, Zhang
Q and Tao Q: The epigenetic modifier CHD5 functions as a novel
tumor suppressor for renal cell carcinoma and is predominantly
inactivated by promoter CpG methylation. Oncotarget. 7:21618–21630.
2016.
|
|
78
|
Ma Z, Song J, Liu S, Han L, Chen Y, Wang
Y, Yu C and Hou L: Decreased expression of the CHD5 gene and its
clinicopathological significance in breast cancer: Correlation with
aberrant DNA methylation. Oncol Lett. 12:4021–4026. 2016.
|
|
79
|
Dong C, Yuan T, Wu Y, Wang Y, Fan TW,
Miriyala S, Lin Y, Yao J, Shi J, Kang T, et al: Loss of FBP1 by
Snail-mediated repression provides metabolic advantages in
basal-like breast cancer. Cancer Cell. 23:316–331. 2013.
|
|
80
|
Li H, Qi Z, Niu Y, Yang Y, Li M, Pang Y,
Liu M, Cheng X, Xu M and Wang Z: FBP1 regulates proliferation,
metastasis, and chemoresistance by participating in C-MYC/STAT3
signaling axis in ovarian cancer. Oncogene. 40:5938–5949. 2021.
|
|
81
|
Wang Y, Shao F and Chen L: ALDH1A2
suppresses epithelial ovarian cancer cell proliferation and
migration by downregulating STAT3. Onco Targets Ther. 11:599–608.
2018.
|
|
82
|
Choi JA, Kwon H, Cho H, Chung JY, Hewitt
SM and Kim JH: ALDH1A2 is a candidate tumor suppressor gene in
ovarian cancer. Cancers (Basel). 11:15532019.
|
|
83
|
Zhu L, Zhang S and Jin Y: Foxd3 suppresses
NFAT-mediated differentiation to maintain self-renewal of embryonic
stem cells. EMBO Rep. 15:1286–1296. 2014.
|
|
84
|
He GY, Hu JL, Zhou L, Zhu XH, Xin SN,
Zhang D, Lu GF, Liao WT, Ding YQ and Liang L: The
FOXD3/miR-214/MED19 axis suppresses tumour growth and metastasis in
human colorectal cancer. Br J Cancer. 115:1367–1378. 2016.
|
|
85
|
Cheng AS, Li MS, Kang W, Cheng VY, Chou
JL, Lau SS, Go MY, Lee CC, Ling TK, Ng EK, et al: Helicobacter
pylori causes epigenetic dysregulation of FOXD3 to promote gastric
carcinogenesis. Gastroenterology. 144:122–133.e9. 2013.
|
|
86
|
Luo GF, Chen CY, Wang J, Yue HY, Tian Y,
Yang P, Li YK and Li Y: FOXD3 may be a new cellular target
biomarker as a hypermethylation gene in human ovarian cancer.
Cancer Cell Int. 19:442019.
|
|
87
|
Katsaros D, Yu H, Levesque MA, Danese S,
Genta F, Richiardi G, Fracchioli S, Khosravi MJ, Diamandi A,
Gordini G, et al: IGFBP-3 in epithelial ovarian carcinoma and its
association with clinico-pathological features and patient
survival. Eur J Cancer. 37:478–485. 2001.
|
|
88
|
Wiley A, Katsaros D, Fracchioli S and Yu
H: Methylation of the insulin-like growth factor binding protein-3
gene and prognosis of epithelial ovarian cancer. Int J Gynecol
Cancer. 16:210–218. 2006.
|
|
89
|
Torng PL, Lin CW, Chan MW, Yang HW, Huang
SC and Lin CT: Promoter methylation of IGFBP-3 and p53 expression
in ovarian endometrioid carcinoma. Mol Cancer. 8:1202009.
|
|
90
|
Urrutia R: KRAB-containing zinc-finger
repressor proteins. Genome Biol. 4:2312003.
|
|
91
|
Cheng Y, Geng H, Cheng SH, Liang P, Bai Y,
Li J, Srivastava G, Ng MH, Fukagawa T, Wu X, et al: KRAB zinc
finger protein ZNF382 is a proapoptotic tumor suppressor that
represses multiple oncogenes and is commonly silenced in multiple
carcinomas. Cancer Res. 70:6516–6526. 2010.
|
|
92
|
Mase S, Shinjo K, Totani H, Katsushima K,
Arakawa A, Takahashi S, Lai HC, Lin RI, Chan MWY, Sugiura-Ogasawara
M and Kondo Y: ZNF671 DNA methylation as a molecular predictor for
the early recurrence of serous ovarian cancer. Cancer Sci.
110:1105–1116. 2019.
|
|
93
|
Yan J, Zhang J, Zhang X, Li X, Li L, Li Z,
Chen R, Zhang L, Wu J, Wang X, et al: SPARC is down-regulated by
DNA methylation and functions as a tumor suppressor in T-cell
lymphoma. Exp Cell Res. 364:125–132. 2018.
|
|
94
|
Nagaraju GP and El-Rayes BF: SPARC and DNA
methylation: Possible diagnostic and therapeutic implications in
gastrointestinal cancers. Cancer Lett. 328:10–17. 2013.
|
|
95
|
Singh A, Gupta S and Sachan M: Evaluation
of the diagnostic potential of candidate hypermethylated genes in
epithelial ovarian cancer in North Indian population. Front Mol
Biosci. 8:7190562021.
|
|
96
|
Niskakoski A, Kaur S, Staff S,
Renkonen-Sinisalo L, Lassus H, Järvinen HJ, Mecklin JP, Bützow R
and Peltomäki P: Epigenetic analysis of sporadic and
Lynch-associated ovarian cancers reveals histology-specific
patterns of DNA methylation. Epigenetics. 9:1577–1587. 2014.
|
|
97
|
Socha MJ, Said N, Dai Y, Kwong J,
Ramalingam P, Trieu V, Desai N, Mok SC and Motamed K: Aberrant
promoter methylation of SPARC in ovarian cancer. Neoplasia.
11:126–135. 2009.
|
|
98
|
Gusyatiner O and Hegi ME: Glioma
epigenetics: From subclassification to novel treatment options.
Semin Cancer Biol. 51:50–58. 2018.
|
|
99
|
Shilpa V, Bhagat R, Premalata CS, Pallavi
VR, Ramesh G and Krishnamoorthy L: Relationship between promoter
methylation & tissue expression of MGMT gene in ovarian cancer.
Indian J Med Res. 140:616–623. 2014.
|
|
100
|
Martínez-Jiménez F, Muiños F, Sentís I,
Deu-Pons J, Reyes-Salazar I, Arnedo-Pac C, Mularoni L, Pich O,
Bonet J, Kranas H, et al: A compendium of mutational cancer driver
genes. Nat Rev Cancer. 20:555–572. 2020.
|
|
101
|
Eun Kwon H and Taylor HS: The role of HOX
genes in human implantation. Ann N Y Acad Sci. 1034:1–18. 2004.
|
|
102
|
Widschwendter M, Apostolidou S, Jones AA,
Fourkala EO, Arora R, Pearce CL, Frasco MA, Ayhan A, Zikan M,
Cibula D, et al: HOXA methylation in normal endometrium from
premenopausal women is associated with the presence of ovarian
cancer: A proof of principle study. Int J Cancer. 125:2214–2218.
2009.
|
|
103
|
Wu Q, Lothe RA, Ahlquist T, Silins I,
Tropé CG, Micci F, Nesland JM, Suo Z and Lind GE: DNA methylation
profiling of ovarian carcinomas and their in vitro models
identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol
Cancer. 6:452007.
|
|
104
|
Montavon C, Gloss BS, Warton K, Barton CA,
Statham AL, Scurry JP, Tabor B, Nguyen TV, Qu W, Samimi G, et al:
Prognostic and diagnostic significance of DNA methylation patterns
in high grade serous ovarian cancer. Gynecol Oncol. 124:582–588.
2012.
|
|
105
|
Singh A, Gupta S, Badarukhiya JA and
Sachan M: Detection of aberrant methylation of HOXA9 and HIC1
through multiplex MethyLight assay in serum DNA for the early
detection of epithelial ovarian cancer. Int J Cancer.
147:1740–1752. 2020.
|
|
106
|
Faaborg L, Fredslund Andersen R, Waldstrøm
M, Høgdall E, Høgdall C, Adimi P, Jakobsen A and Dahl Steffensen K:
Analysis of HOXA9 methylated ctDNA in ovarian cancer using
sense-antisense measurement. Clin Chim Acta. 522:152–157. 2021.
|
|
107
|
Rusan M, Andersen RF, Jakobsen A and
Steffensen KD: Circulating HOXA9-methylated tumour DNA: A novel
biomarker of response to poly (ADP-ribose) polymerase inhibition in
BRCA-mutated epithelial ovarian cancer. Eur J Cancer. 125:121–129.
2020.
|
|
108
|
van Wijnen AJ, Bagheri L, Badreldin AA,
Larson AN, Dudakovic A, Thaler R, Paradise CR and Wu Z: Biological
functions of chromobox (CBX) proteins in stem cell self-renewal,
lineage-commitment, cancer and development. Bone.
143:1156592021.
|
|
109
|
Lin J, Chen L, Wu D, Lin J, Liu B and Guo
C: Potential diagnostic and prognostic values of CBX8 Expression in
liver hepatocellular carcinoma, kidney renal clear cell carcinoma,
and ovarian cancer: A study based on TCGA data mining. Comput Math
Methods Med. 2022:13728792022.
|
|
110
|
Li Q, Pan Y, Cao Z and Zhao S:
Comprehensive analysis of prognostic value and immune infiltration
of chromobox family members in colorectal cancer. Front Oncol.
10:5826672020.
|
|
111
|
Sung HY, Yang SD, Park AK, Ju W and Ahn
JH: Aberrant hypomethylation of solute carrier family 6 member 12
promoter induces metastasis of ovarian cancer. Yonsei Med J.
58:27–34. 2017.
|
|
112
|
Higa A, Mulot A, Delom F, Bouchecareilh M,
Nguyên DT, Boismenu D, Wise MJ and Chevet E: Role of pro-oncogenic
protein disulfide isomerase (PDI) family member anterior gradient 2
(AGR2) in the control of endoplasmic reticulum homeostasis. J Biol
Chem. 286:44855–44868. 2011.
|
|
113
|
Zhang S, Liu Q, Wei Y, Xiong Y, Gu Y,
Huang Y, Tang F and Ouyang Y: Anterior gradient-2 regulates cell
communication by coordinating cytokine-chemokine signaling and
immune infiltration in breast cancer. Cancer Sci. 114:2238–2253.
2023.
|
|
114
|
He J, Fu Y, Hu J, Chen J and Lou G:
Hypomethylation-mediated AGR2 overexpression facilitates cell
proliferation, migration, and invasion of lung adenocarcinoma.
Cancer Manag Res. 13:5177–5185. 2021.
|
|
115
|
Sung HY, Choi EN, Lyu D, Park AK, Ju W and
Ahn JH: Aberrant hypomethylation-mediated AGR2 overexpression
induces an aggressive phenotype in ovarian cancer cells. Oncol Rep.
32:815–820. 2014.
|
|
116
|
Chan DW, Lam WY, Chen F, Yung MMH, Chan
YS, Chan WS, He F, Liu SS, Chan KKL, Li B and Ngan HYS: Genome-wide
DNA methylome analysis identifies methylation signatures associated
with survival and drug resistance of ovarian cancers. Clin
Epigenetics. 13:1422021.
|
|
117
|
Zhan T, Rindtorff N and Boutros M: Wnt
signaling in cancer. Oncogene. 36:1461–1473. 2017.
|
|
118
|
Li HJ, Ke FY, Lin CC, Lu MY, Kuo YH, Wang
YP, Liang KH, Lin SC, Chang YH, Chen HY, et al: ENO1 promotes lung
cancer metastasis via HGFR and WNT signaling-driven
epithelial-to-mesenchymal transition. Cancer Res. 81:4094–4109.
2021.
|
|
119
|
Xu X, Zhang M, Xu F and Jiang S: Wnt
signaling in breast cancer: biological mechanisms, challenges and
opportunities. Mol Cancer. 19:1652020.
|
|
120
|
Hu W, Li M, Chen Y and Gu X: UBE2S
promotes the progression and Olaparib resistance of ovarian cancer
through Wnt/β-catenin signaling pathway. J Ovarian Res.
14:1212021.
|
|
121
|
Deshmukh A, Arfuso F, Newsholme P and
Dharmarajan A: Epigenetic demethylation of sFRPs, with emphasis on
sFRP4 activation, leading to Wnt signalling suppression and histone
modifications in breast, prostate, and ovary cancer stem cells. Int
J Biochem Cell Biol. 109:23–32. 2019.
|
|
122
|
Takada T, Yagi Y, Maekita T, Imura M,
Nakagawa S, Tsao SW, Miyamoto K, Yoshino O, Yasugi T, Taketani Y
and Ushijima T: Methylation-associated silencing of the Wnt
antagonist SFRP1 gene in human ovarian cancers. Cancer Sci.
95:741–744. 2004.
|
|
123
|
Yen HY, Tsao CW, Lin YW, Kuo CC, Tsao CH
and Liu CY: Regulation of carcinogenesis and modulation through
Wnt/β-catenin signaling by curcumin in an ovarian cancer cell line.
Sci Rep. 9:172672019.
|
|
124
|
Ho CM, Lai HC, Huang SH, Chien TY, Lin MC
and Chang SF: Promoter methylation of sFRP5 in patients with
ovarian clear cell adenocarcinoma. Eur J Clin Invest. 40:310–318.
2010.
|
|
125
|
Ho CM, Huang CJ, Huang CY, Wu YY, Chang SF
and Cheng WF: Promoter methylation status of HIN-1 associated with
outcomes of ovarian clear cell adenocarcinoma. Mol Cancer.
11:532012.
|
|
126
|
Lin HW, Fu CF, Chang MC, Lu TP, Lin HP,
Chiang YC, Chen CA and Cheng WF: CDH1, DLEC1 and SFRP5 methylation
panel as a prognostic marker for advanced epithelial ovarian
cancer. Epigenomics. 10:1397–1413. 2018.
|
|
127
|
Su HY, Lai HC, Lin YW, Liu CY, Chen CK,
Chou YC, Lin SP, Lin WC, Lee HY and Yu MH: Epigenetic silencing of
SFRP5 is related to malignant phenotype and chemoresistance of
ovarian cancer through Wnt signaling pathway. Int J Cancer.
127:555–567. 2010.
|
|
128
|
Kumar D, Patel SA, Hassan MK, Mohapatra N,
Pattanaik N and Dixit M: Reduced IQGAP2 expression promotes EMT and
inhibits apoptosis by modulating the MEK-ERK and p38 signaling in
breast cancer irrespective of ER status. Cell Death Dis.
12:3892021.
|
|
129
|
Deng Z, Wang L, Hou H, Zhou J and Li X:
Epigenetic regulation of IQGAP2 promotes ovarian cancer progression
via activating Wnt/β-catenin signaling. Int J Oncol. 48:153–160.
2016.
|
|
130
|
Ge YX, Wang CH, Hu FY, Pan LX, Min J, Niu
KY, Zhang L, Li J and Xu T: New advances of TMEM88 in cancer
initiation and progression, with special emphasis on Wnt signaling
pathway. J Cell Physiol. 233:79–87. 2018.
|
|
131
|
de Leon M, Cardenas H, Vieth E, Emerson R,
Segar M, Liu Y, Nephew K and Matei D: Transmembrane protein 88
(TMEM88) promoter hypomethylation is associated with platinum
resistance in ovarian cancer. Gynecol Oncol. 142:539–547. 2016.
|
|
132
|
Yang L, Pang Y and Moses HL: TGF-beta and
immune cells: An important regulatory axis in the tumor
microenvironment and progression. Trends Immunol. 31:220–227.
2010.
|
|
133
|
Morikawa M, Derynck R and Miyazono K:
TGF-β and the TGF-β family: Context-dependent roles in cell and
tissue physiology. Cold Spring Harb Perspect Biol.
8:a0218732016.
|
|
134
|
Matsumura N, Huang Z, Mori S, Baba T,
Fujii S, Konishi I, Iversen ES, Berchuck A and Murphy SK:
Epigenetic suppression of the TGF-beta pathway revealed by
transcriptome profiling in ovarian cancer. Genome Res. 21:74–82.
2011.
|
|
135
|
Cardenas H, Vieth E, Lee J, Segar M, Liu
Y, Nephew KP and Matei D: TGF-β induces global changes in DNA
methylation during the epithelial-to-mesenchymal transition in
ovarian cancer cells. Epigenetics. 9:1461–1472. 2014.
|
|
136
|
Chou JL, Su HY, Chen LY, Liao YP,
Hartman-Frey C, Lai YH, Yang HW, Deatherage DE, Kuo CT, Huang YW,
et al: Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4
target gene and tumor suppressor, is associated with poor prognosis
in human ovarian cancer. Lab Invest. 90:414–425. 2010.
|
|
137
|
Chou JL, Huang RL, Shay J, Chen LY, Lin
SJ, Yan PS, Chao WT, Lai YH, Lai YL, Chao TK, et al:
Hypermethylation of the TGF-β target, ABCA1 is associated with poor
prognosis in ovarian cancer patients. Clin Epigenetics.
7:12015.
|
|
138
|
Wegner M: All purpose Sox: The many roles
of Sox proteins in gene expression. Int J Biochem Cell Biol.
42:381–390. 2010.
|
|
139
|
Shonibare Z, Monavarian M, O'Connell K,
Altomare D, Shelton A, Mehta S, Jaskula-Sztul R, Phaeton R, Starr
MD, Whitaker R, et al: Reciprocal SOX2 regulation by SMAD1-SMAD3 is
critical for anoikis resistance and metastasis in cancer. Cell Rep.
40:1110662022.
|
|
140
|
Ween MP, Oehler MK and Ricciardelli C:
Transforming growth factor-beta-induced protein (TGFBI)/(βig-H3): A
matrix protein with dual functions in ovarian cancer. Int J Mol
Sci. 13:10461–10477. 2012.
|
|
141
|
Wang N, Zhang H, Yao Q, Wang Y, Dai S and
Yang X: TGFBI promoter hypermethylation correlating with paclitaxel
chemoresistance in ovarian cancer. J Exp Clin Cancer Res.
31:62012.
|
|
142
|
Ho CM, Lin MC, Huang SH, Huang CJ, Lai HC,
Chien TY and Chang SF: PTEN promoter methylation and LOH of
10q22-23 locus in PTEN expression of ovarian clear cell
adenocarcinomas. Gynecol Oncol. 112:307–313. 2009.
|
|
143
|
Ediriweera MK, Tennekoon KH and Samarakoon
SR: Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer:
Biological and therapeutic significance. Semin Cancer Biol.
59:147–160. 2019.
|
|
144
|
Li M, Balch C, Montgomery JS, Jeong M,
Chung JH, Yan P, Huang TH, Kim S and Nephew KP: Integrated analysis
of DNA methylation and gene expression reveals specific signaling
pathways associated with platinum resistance in ovarian cancer. BMC
Med Genomics. 2:342009.
|
|
145
|
Li GN, Zhao XJ, Wang Z, Luo MS, Shi SN,
Yan DM, Li HY, Liu JH, Yang Y, Tan JH, et al: Elaiophylin triggers
paraptosis and preferentially kills ovarian cancer drug-resistant
cells by inducing MAPK hyperactivation. Signal Transduct Target
Ther. 7:3172022.
|
|
146
|
Walch A, Specht K, Braselmann H, Stein H,
Siewert JR, Hopt U, Höfler H and Werner M: Coamplification and
coexpression of GRB7 and ERBB2 is found in high grade
intraepithelial neoplasia and in invasive Barrett's carcinoma. Int
J Cancer. 112:747–753. 2004.
|
|
147
|
Chen K, Liu MX, Mak CS, Yung MM, Leung TH,
Xu D, Ngu SF, Chan KK, Yang H, Ngan HY and Chan DW:
Methylation-associated silencing of miR-193a-3p promotes ovarian
cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways.
Theranostics. 8:423–436. 2018.
|
|
148
|
Sung HY, Yang SD, Ju W and Ahn JH:
Aberrant epigenetic regulation of GABRP associates with aggressive
phenotype of ovarian cancer. Exp Mol Med. 49:e3352017.
|
|
149
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531.
2004.
|
|
150
|
Kuhlmann JD, Rasch J, Wimberger P and
Kasimir-Bauer S: microRNA and the pathogenesis of ovarian cancer-a
new horizon for molecular diagnostics and treatment? Clin Chem Lab
Med. 50:601–615. 2012.
|
|
151
|
Cairns RA: Drivers of the Warburg
phenotype. Cancer J. 21:56–61. 2015.
|
|
152
|
Zhou Y, Zheng X, Lu J, Chen W, Li X and
Zhao L: Ginsenoside 20(S)-Rg3 inhibits the Warburg effect via
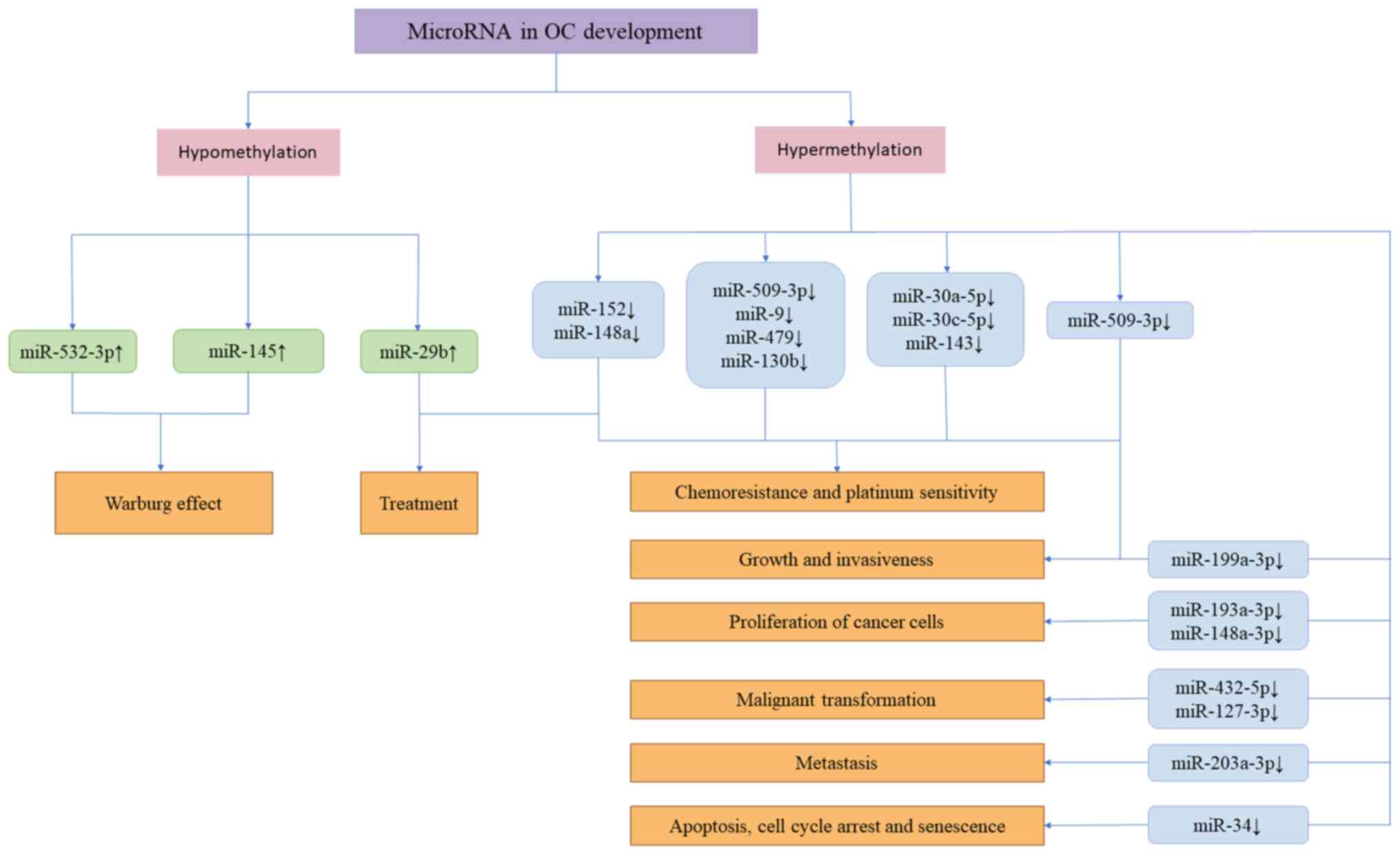
modulating DNMT3A/MiR-532-3p/HK2 pathway in ovarian cancer cells.
Cell Physiol Biochem. 45:2548–2559. 2018.
|
|
153
|
Zhang S, Pei M, Li Z, Li H, Liu Y and Li
J: Double-negative feedback interaction between DNA
methyltransferase 3A and microRNA-145 in the Warburg effect of
ovarian cancer cells. Cancer Sci. 109:2734–2745. 2018.
|
|
154
|
Li J, Zhang S, Zou Y, Wu L, Pei M and
Jiang Y: miR-145 promotes miR-133b expression through c-myc and
DNMT3A-mediated methylation in ovarian cancer cells. J Cell
Physiol. 235:4291–4301. 2020.
|
|
155
|
Teng Y, Zuo X, Hou M, Zhang Y, Li C, Luo W
and Li X: A double-negative feedback interaction between
MicroRNA-29b and DNMT3A/3B contributes to ovarian cancer
progression. Cell Physiol Biochem. 39:2341–2352. 2016.
|
|
156
|
Chhabra R, Rockfield S, Guergues J, Nadeau
OW, Hill R, Stevens SM Jr and Nanjundan M: Global miRNA/proteomic
analyses identify miRNAs at 14q32 and 3p21, which contribute to
features of chronic iron-exposed fallopian tube epithelial cells.
Sci Rep. 11:62702021.
|
|
157
|
Chen Q, Wang Y, Dang H and Wu X:
MicroRNA-148a-3p inhibits the proliferation of cervical cancer
cells by regulating the expression levels of DNMT1 and UTF1. Oncol
Lett. 22:6172021.
|
|
158
|
Wu YH, Huang YF, Wu PY, Chang TH, Huang SC
and Chou CY: The downregulation of miR-509-3p expression by
collagen type XI alpha 1-regulated hypermethylation facilitates
cancer progression and chemoresistance via the DNA
methyltransferase 1/Small ubiquitin-like modifier-3 axis in ovarian
cancer cells. Res Sq. rs.3.rs–2592453. 2023.
|
|
159
|
Han X, Zhen S, Ye Z, Lu J, Wang L, Li P,
Li J, Zheng X, Li H, Chen W, et al: A feedback loop between
miR-30a/c-5p and DNMT1 mediates cisplatin resistance in ovarian
cancer cells. Cell Physiol Biochem. 41:973–986. 2017.
|
|
160
|
Vera O, Jimenez J, Pernia O,
Rodriguez-Antolin C, Rodriguez C, Sanchez Cabo F, Soto J, Rosas R,
Lopez-Magallon S, Esteban Rodriguez I, et al: DNA methylation of
miR-7 is a mechanism involved in platinum response through MAFG
overexpression in cancer cells. Theranostics. 7:4118–4134.
2017.
|
|
161
|
Li X, Pan Q, Wan X, Mao Y, Lu W, Xie X and
Cheng X: Methylation-associated Has-miR-9 deregulation in
paclitaxel-resistant epithelial ovarian carcinoma. BMC Cancer.
15:5092015.
|
|
162
|
Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L
and Wang Z: Epigenetic silencing of miR-130b in ovarian cancer
promotes the development of multidrug resistance by targeting
colony-stimulating factor 1. Gynecol Oncol. 124:325–334. 2012.
|
|
163
|
Xu S, Fu GB, Tao Z, OuYang J, Kong F,
Jiang BH, Wan X and Chen K: MiR-497 decreases cisplatin resistance
in ovarian cancer cells by targeting mTOR/P70S6K1. Oncotarget.
6:26457–26471. 2015.
|
|
164
|
Han X, Liu D, Zhou Y, Wang L, Hou H, Chen
H, Zhang L, Chen W, Li X and Zhao L: The negative feedback between
miR-143 and DNMT3A regulates cisplatin resistance in ovarian
cancer. Cell Biol Int. 45:227–237. 2021.
|
|
165
|
He L, Zhu W, Chen Q, Yuan Y, Wang Y, Wang
J and Wu X: Ovarian cancer cell-secreted exosomal miR-205 promotes
metastasis by inducing angiogenesis. Theranostics. 9:8206–8220.
2019.
|
|
166
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007.
|
|
167
|
Loginov VI, Pronina IV, Filippova EA,
Burdennyy AM, Lukina SS, Kazubskaya TP, Uroshlev LA, Fridman MV,
Brovkina OI, Apanovich NV, et al: Aberrant methylation of 20 miRNA
genes specifically involved in various steps of ovarian carcinoma
spread: From primary tumors to peritoneal macroscopic metastases.
Int J Mol Sci. 23:13002022.
|
|
168
|
Deng Y, Zhao F, Hui L, Li X, Zhang D, Lin
W, Chen Z and Ning Y: Suppressing miR-199a-3p by promoter
methylation contributes to tumor aggressiveness and cisplatin
resistance of ovarian cancer through promoting DDR1 expression. J
Ovarian Res. 10:502017.
|
|
169
|
Schmid G, Notaro S, Reimer D, Abdel-Azim
S, Duggan-Peer M, Holly J, Fiegl H, Rössler J, Wiedemair A, Concin
N, et al: Expression and promotor hypermethylation of miR-34a in
the various histological subtypes of ovarian cancer. BMC Cancer.
16:1022016.
|
|
170
|
Zuberi M, Khan I, Mir R, Gandhi G, Ray PC
and Saxena A: Utility of serum miR-125b as a diagnostic and
prognostic indicator and its alliance with a panel of tumor
suppressor genes in epithelial ovarian cancer. PLoS One.
11:e01539022016.
|
|
171
|
He J, Xu Q, Jing Y, Agani F, Qian X,
Carpenter R, Li Q, Wang XR, Peiper SS, Lu Z, et al: Reactive oxygen
species regulate ERBB2 and ERBB3 expression via miR-199a/125b and
DNA methylation. EMBO Rep. 13:1116–1122. 2012.
|
|
172
|
Ye Z, Li J, Han X, Hou H, Chen H, Zheng X,
Lu J, Wang L, Chen W, Li X and Zhao L: TET3 inhibits TGF-β1-induced
epithelial-mesenchymal transition by demethylating miR-30d
precursor gene in ovarian cancer cells. J Exp Clin Cancer Res.
35:722016.
|
|
173
|
Vogt M, Munding J, Grüner M, Liffers ST,
Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A and Hermeking H:
Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG
methylation in colorectal, pancreatic, mammary, ovarian,
urothelial, and renal cell carcinomas and soft tissue sarcomas.
Virchows Arch. 458:313–322. 2011.
|
|
174
|
Cai Y, Tsai HC, Yen RC, Zhang YW, Kong X,
Wang W, Xia L and Baylin SB: Critical threshold levels of DNA
methyltransferase 1 are required to maintain DNA methylation across
the genome in human cancer cells. Genome Res. 27:533–544. 2017.
|
|
175
|
Li H, Lei Y, Li S, Li F and Lei J:
MicroRNA-20a-5p inhibits the autophagy and cisplatin resistance in
ovarian cancer via regulating DNMT3B-mediated DNA methylation of
RBP1. Reprod Toxicol. 109:93–100. 2022.
|
|
176
|
Ye Z, Jiang Y and Wu J: DNMT3B attenuated
the inhibition of TET3 on epithelial-mesenchymal transition in
TGF-β1-induced ovarian cancer by methylating the TET3 promoter.
Reprod Biol. 22:1007012022.
|
|
177
|
Del Castillo Falconi VM, Díaz-Chávez J,
Torres-Arciga K, Luna-Maldonado F, Gudiño-Gomez AA, Pedroza-Torres
A, Castro-Hernández C, Cantú de León D and Herrera LA: Expression
of DNA methyltransferase 3B isoforms is associated with DNA
satellite 2 hypomethylation and clinical prognosis in advanced
high-grade serous ovarian carcinoma. Int J Mol Sci.
23:127592022.
|
|
178
|
Lyko F and Brown R: DNA methyltransferase
inhibitors and the development of epigenetic cancer therapies. J
Natl Cancer Inst. 97:1498–1506. 2005.
|
|
179
|
Natoli M, Gallon J, Lu H, Amgheib A,
Pinato DJ, Mauri FA, Marafioti T, Akarca AU, Ullmo I, Ip J, et al:
Transcriptional analysis of multiple ovarian cancer cohorts reveals
prognostic and immunomodulatory consequences of ERV expression. J
Immunother Cancer. 9:e0015192021.
|
|
180
|
Ma G, Li Y, Meng F, Sui C, Wang Y and
Cheng D: Hsa_ circ_0000119 promoted ovarian cancer development via
enhancing the methylation of CDH13 by sponging miR-142-5p. J
Biochem Mol Toxicol. 37:e232642023.
|
|
181
|
Wong-Brown MW, van der Westhuizen A and
Bowden NA: Sequential azacitidine and carboplatin induces immune
activation in platinum-resistant high-grade serous ovarian cancer
cell lines and primes for checkpoint inhibitor immunotherapy. BMC
Cancer. 22:1002022.
|
|
182
|
Liu M, Thomas SL, DeWitt AK, Zhou W, Madaj
ZB, Ohtani H, Baylin SB, Liang G and Jones PA: Dual inhibition of
DNA and histone methyltransferases increases viral mimicry in
ovarian cancer cells. Cancer Res. 78:5754–5766. 2018.
|
|
183
|
Shim JI, Ryu JY, Jeong SY, Cho YJ, Choi
JJ, Hwang JR, Choi JY, Sa JK and Lee JW: Combination effect of poly
(ADP-ribose) polymerase inhibitor and DNA demethylating agents for
treatment of epithelial ovarian cancer. Gynecol Oncol. 165:270–280.
2022.
|
|
184
|
McDonald JI, Diab N, Arthofer E, Hadley M,
Kanholm T, Rentia U, Gomez S, Yu A, Grundy EE, Cox O, et al:
Epigenetic therapies in ovarian cancer alter repetitive element
expression in a TP53-dependent manner. Cancer Res. 81:5176–5189.
2021.
|
|
185
|
Steele N, Finn P, Brown R and Plumb JA:
Combined inhibition of DNA methylation and histone acetylation
enhances gene re-expression and drug sensitivity in vivo. Br J
Cancer. 100:758–763. 2009.
|
|
186
|
Fang F, Balch C, Schilder J, Breen T,
Zhang S, Shen C, Li L, Kulesavage C, Snyder AJ, Nephew KP and Matei
DE: A phase 1 and pharmacodynamic study of decitabine in
combination with carboplatin in patients with recurrent,
platinum-resistant, epithelial ovarian cancer. Cancer.
116:4043–4053. 2010.
|
|
187
|
Yin B, Ding J, Hu H, Yang M, Huang B, Dong
W, Li F and Han L: Overexpressed CMTM6 improves prognosis and
associated with immune infiltrates of ovarian cancer. Front Mol
Biosci. 9:7690322022.
|
|
188
|
Gomez S, Cox OL, Walker RR III, Rentia U,
Hadley M, Arthofer E, Diab N, Grundy EE, Kanholm T, McDonald JI, et
al: Inhibiting DNA methylation and RNA editing upregulates
immunogenic RNA to transform the tumor microenvironment and prolong
survival in ovarian cancer. J Immunother Cancer.
10:e0049742022.
|
|
189
|
Giri AK and Aittokallio T: DNMT inhibitors
increase methylation in the cancer genome. Front Pharmacol.
10:3852019.
|
|
190
|
Bauerschlag DO, Ammerpohl O, Bräutigam K,
Schem C, Lin Q, Weigel MT, Hilpert F, Arnold N, Maass N,
Meinhold-Heerlein I and Wagner W: Progression-free survival in
ovarian cancer is reflected in epigenetic DNA methylation profiles.
Oncology. 80:12–20. 2011.
|
|
191
|
Khajehnoori S, Zarei F, Mazaheri M and
Dehghani-Firoozabadi A: Epidrug modulated expression of MiR-152 and
MiR-148a reverse cisplatin resistance in ovarian cancer cells: An
experimental in-vitro study. Iran J Pharm Res. 19:509–519.
2020.
|
|
192
|
Belsky DW, Caspi A, Corcoran DL, Sugden K,
Poulton R, Arseneault L, Baccarelli A, Chamarti K, Gao X, Hannon E,
et al: DunedinPACE, a DNA methylation biomarker of the pace of
aging. Elife. 11:e734202022.
|