Introduction
Aldehyde dehydrogenases (ALDHs) comprise a family of
19 NAD(P+)-dependent enzymes that metabolize endogenously and
exogenously produced aldehydes, by irreversibly catalyzing their
oxidation to their respective carboxylic acids (1,2).
ALDHs have a broad spectrum of biological activities, including but
not limited to biosynthesis of retinoic acid and alcohol
metabolism.
ALDHs are expressed in stem cells in general; ALDHs
to some extent are also expressed in cancer cells that resemble
normal stem cells in terms of cell cycle dormancy and metabolic
adjustments for decreased generation of reactive oxygen species
(3). These cancer cells with
slower proliferation most often have tumor-initiating properties
and tend to be resistant to chemotherapy and cytotoxic agents.
Their capacity to withstand oxidative stress is limited, but it is
noteworthy that they may give rise to aggressive cancer cell clones
with a high pace of proliferation and growth (4-5).
Some of these effects are often attributed to polyploid cancer
cells (6), or to cancer cells
that have been fused to macrophages or other non-tumor cells
(7,8); however the cancer 'stem-like' cells
are not exclusively polyploid. Furthermore, the degree of metabolic
dormancy and the precise phase of cell cycle arrest vary, as well
as the readiness to re-enter the cell cycle (9). The shared aspect among these cell
phenotypes is that they all exhibit an obligatory state of arrested
tumor growth, which confers cancer resistance to adverse
conditions.
Quiescent cells are in a metabolic state that
generates a lower level of oxidative stress, thus resulting in
decreased expression of enzymes that protect from reactive oxygen
species. However, the cancer stem-like cells have alterations in
chromatin in key genes that encode enzymes operating as components
of essential antioxidant systems. These chromatin alterations allow
them to express those genes rapidly and therefore adapt and survive
acute exposure to oxidant stress (5,10).
During chemotherapy or inflammation, in the critical phase of
cytotoxic exposure that causes cell stress and growth suppression,
ALDH enzymes may protect cancer stem cells (CSCs), before favorable
conditions and appropriate stimuli permit the generation of
daughter clones with different phenotypes. A key member of the ALDH
family that possesses properties that are well-suited to its
central role in the initial cellular recovery, occurring prior to
the acute expression of key rapid-response genes, is aldehyde
dehydrogenase 1 family member A1 (ALDH1A1) (11).
ALDH1A1 is a member of the aldehyde dehydrogenase
gene subfamily that encodes enzymes with the ability to oxidize
retinaldehyde, owing to a larger substrate entry channel (1,12).
The protein ALDH1A1, which localizes to the cytosol and the
nucleus, is overexpressed in a number of diverse cancer types;
however it is not consistently associated with a negative disease
prognosis: ALDH1A1 plays the role of a tumor suppressor under
certain conditions that can be attributed to the maintenance of an
optimal intracellular milieu. Its precise funtion in normal stem
cells, such as hematopoietic stem cells (HSCs), is indicated by
research findings that suggest a degree of redundancy between
certain similar ALDH enzymes (13,14). Nevertheless, ALDH1A1 is an enzyme
with critical functions in CSCs (2). In contrast to normal HSCs, in some
leukemia cells ALDH1A1 may posses non-redundant functions.
Acute myeloid leukemia (AML), is a hematopoietic
malignancy associated with high morbidity and mortality rates
(15). Understanding the
molecular mechanisms underlying AML is crucial for developing
effective therapies. The expression of ALDH1A1 specifically
protects leukemia-initiating cells (LSCs) from a number of
antineoplastic agents; i) protection from cyclophosphamide by
ALDH1A1 gene transfer in cultured cells (16), and ii) conversely, ALDH1A1 gene
knockout sensitizes LSCs to cyclophosphamide (17), while the enzymatic activity of
ALDH in AML blast cells, has been proven to be essential for the
establishment of human AML xenografts in mice (18-20). ALDH(+) cells from samples of
patients with AML with ≥1.9% ALDH(+) cells were quiescent,
refractory to cytarabine treatment, and capable of leukemic
engraftment in a xenogenic mouse transplantation model (21).
Conversely, AML cells null for ALDH1A1 RNA
expression were obtained from patients with a favorable prognosis,
and were sensitive to chemotherapeutic agents (22).
It is important to emphasize that even after
generation of a multi-omic profile of samples of patients with AML,
the integrated classification continues to categorize
ALDH1A1-overexpressing samples to the worst AML prognosis group:
This analysis indicates the significant impact of
ALDH1A1-expressing AML cells in an unfavorable disease course
(23).
Biological links between AML, miRNAs and
ALDH1A1
It was recently shown that ALDH1A1 RNA abundance is
correlated with the outcome of AML; especially when compared to the
other members of the ALDH family, ALDH1A1 had the greatest
statistical capacity to differentiate between patients with AML
with a favorable and an unfavorable prognosis (24,25). A number of agents are known to
inhibit ALDH activity, with at least one, DIMATE, demonstrating the
ability to selectively kill LSCs while leaving normal HSCs intact
(26). However, there is always
room for improvement, especially in respect to the development of
methodologies for genetic interference.
The need to develop alternatives arises from the
plasticity of leukemia cell populations, that allows the emergence
of altered phenotypes. This is due to the capacity of leukemia
stem-like cells to undergo phenotype changes in response to the
metabolite content of their microenvironment, and most notably, in
response to changes in the oxidative state (4,27).
Over a decade ago, miRNAs, a class of noncoding
RNAs, emerged as key regulators of gene expression in AML, making
them one of several potential avenues for genetic intervention in
AML cells (28). Especially
relevant in AML biology, are the mutual interactions between
miRNAs, including miR-146 for example, with NF-κB, a transcription
factor that regulates a substantial proportion of inflammatory
genes and miRNAs involved in malignant progression (29-31). In addition, recent data suggest a
strong association of miRNA expression with macrophage
polarization, which regulates immune responses against AML
(32). The list of miRNAs and
their mRNA targets that are relevant in AML disease progression
continues to grow, rendering therapeutic manipulation of miRNAs an
increasingly relevant aim, especially in light of interesting
preclinical data that emerge from a recent study (33).
As examined in the present review, the interactions
between miRNAs and ALDH1A1 can be complex and not ubiquitous
between different cell phenotypes. In other words, the mutual
effects between a given miRNA and ALDH1A1 can be enhancing or
suppressing, but in different cells this may change. Furthermore,
it cannot be excluded that ALDH1A1 induces the expression of a
given miRNA, which then acts as a negative feedback trigger and
leads to repression of ALDH1A1. For this reason, in the present
review, the miRNAs that have exhibited potential to act directly on
ALDH1A1 expression are focused on, since this type of interaction
can be expected to have the least variability.
miRNAs that may be included in the list of
miRNAs with the potential to target ALDH1A1
There is a substantial number of miRNAs that may
target the gene ALDH1A1. A few of them have already been recognized
as tumor suppressors in AML, making their preclinical assay the
next step forward in elucidating their application potential. As is
reviewed next, experiments on cultured cells provide direct
evidence, while bioinformatic analysis also suggests that there are
numerous miRNAs that interfere with ALDH1A1 expression.
A number of the prospective ALDH1A1-interacting
miRNAs have been identified via high-throughput sequencing of RNA
isolated by cross-linking immunoprecipitation (HITS-CLIP), by
photoactivatable ribonucleoside-enhanced CLIP [PAR-CLIP], and
similar methods, aimed to determine the Argonaut: miRNA binding
sites in the transcriptome, as a means for localizing the RNA bound
by each relevant species, since Argonaute proteins use small RNA
guides to identify complementary sites in transcripts targeted for
silencing or repression (34).
As will be discussed further, both activating as
well as repressing miRNAs have been identified. For some miRNAs,
experimental evidence has directly demonstrated their capacity to
interfere with ALDH1A1 expression negatively, making them thereby
strong candidates for further research.
In vitro assays of miRNAs interfering
with ALDH1A1 expression
In leukemia study models, there are no published
studies that examine direct interference of miRNAs with ALDH1A1
gene expression. However, there are a few studies on solid tumor
model systems that describe ALDH1A1 RNA-interacting miRNAs.
The human papillomavirus HPV16 caused an increase
both in ALDH1A1 mRNA as well as ALDH1 enzymatic activity in
oropharyngeal squamous cell carcinoma cells, which was mediated by
repressing miR-181a/d, two miRNAs, that otherwise suppressed
anchorage independent growth and CSC phenotype (35). However, in AML research, miR-181a
has shown both favorable as well as unfavorable prognostic
associations and molecular mechanistic effects, rendering this
miRNA a challenging candidate for developing ALDH1A1 inhibitors for
AML (36-40). One potential use for miR-181a,
based on both favorable and unfavorable associations, is the
trigger of cell proliferation to render AML cells sensitive to both
pharmacological, as well as immunological intervention. Preclinical
studies have shown encouraging results, making miR-181a, a
candidate for context-dependent development of interventions
(40).
In gastric cancer cells, miR-625 reversed multidrug
resistance by repressing ALDH1A1; miR-625 silencing increased the
IC50 values of four chemotherapeutic agents (ADR, VCR,
5FU and CDDP). Depletion of ALDH1A1 by siRNA reversed those effects
(41). In AML, miR-625 has shown
the potential to suppress metastatic and proliferative functions
(42), cell viability (43,44) and invasiveness (45). miR-625 is therefore a noteworthy
candidate for repression of ALDH1A1 in AML.
In breast cancer, it was revealed that miR-140 was
significantly downregulated in stem-like cells from ductal
carcinoma in situ tumor cells in comparison to normal
mammary stem cells. miR-140 directly targeted the 3′ untranslated
region of ALDH1A1, to inhibit protein expression (46). miR-140 has shown the ability to
function as a tumor suppressor in AML study models (47,48), and a previous study demonstrated
the same effect specifically for miR-140-3p (49). miR-140 is therefore a plausible
candidate for inhibition of ALDH1A1 in AML model systems, where it
can be examined to verify whether it functions via the same
mechanism as that in breast cancer cells. To underscore the
importance of the evidence provided for miR-140 regulation of
ALDH1A1, the widely recognized curated miRNA platform, miRTarBase,
only selected miR-140 as a candidate regulator for ALDH1A1
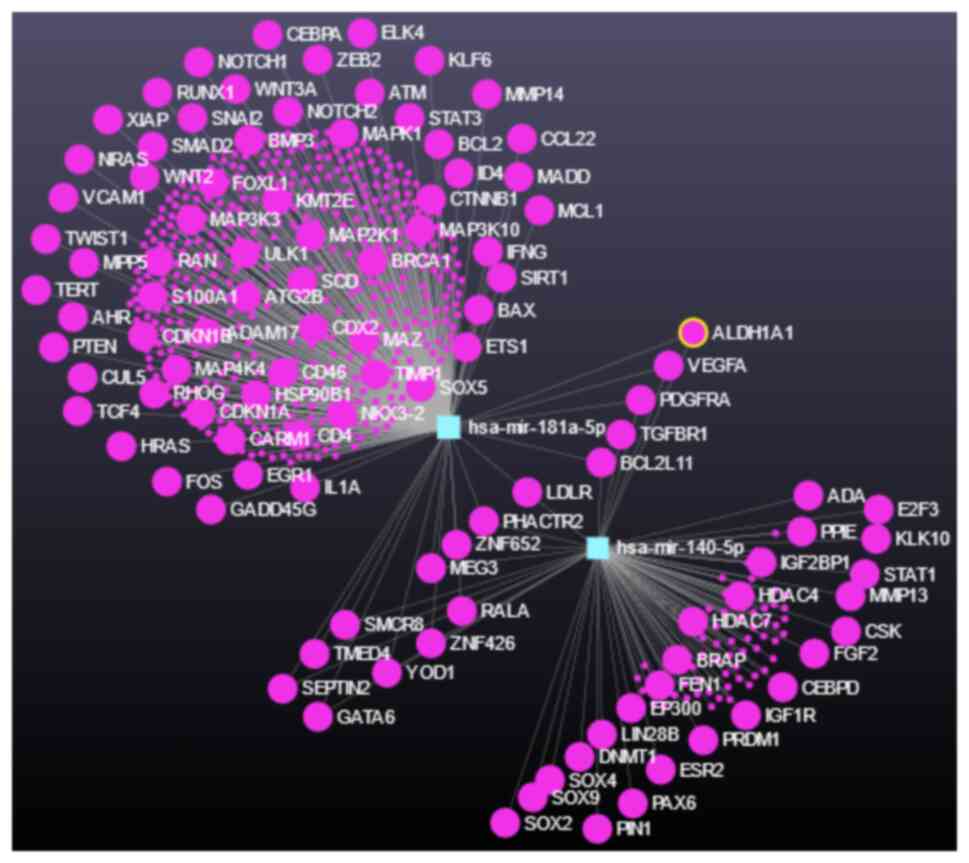
(50,51). In addition, the database,
mirtargetlink 2.0, confirmed this assessment (miR-140-5p, as
supported by the experimental evidence), with the additional
listing of miR-181a-5p as weakly supported, due to the lack of
experimental evidence (52)
(Fig. 1).
In cervical CSCs derived from tumorspheres of the
cell lines, Hela and CaSki, miR-23b reduced ALDH1A1 protein levels,
by specifically binding to the 3′UTR of ALDH1A1 mRNA.
Overexpression of miR-23b substantially reduced the size and number
of tumorspheres, and rendered cells sensitive to cisplatin
(53). miR-23b appears to
decreased in AML (54).
Re-expression in leukemia cells can increase oxidative stress, by
repressing peroxiredoxin III (55). However, miR-23b has been
correlated with the Warburg effect and with a poor prognosis,
making its utility in AML uncertain (56).
It can therefore be concluded that from the miRNAs
that regulate ALDH1A1 in solid tumor study systems, miR-181,
miR-625, miR-140, and even miR-23b can be further investigated to
determine their effects on ALDH1A1 in AML. These investigations
however, must employ a rigorous approach in respect to the precise
time course, dose response, and dynamic distribution in model
systems that resemble human tissue as close as possible, to address
the key issue of context-dependent effects that is pervasive in
miRNA biology, and which is also evident especially in the case of
miR-23b as aforementioned.
miRNAs predicted to regulate ALDH1A1
expression by bioinfomatic analysis platforms
There are a number of miRNAs predicted to target
ALDH1A1 as revealed using the miRNA database, TarBase (57), accessed through miRNet2.0
(58). These are summarized in
Table I. A similar result was
obtained by directly using the database Tarbase (Table II).
 | Table IList of microRNAs predicted to
interfere with ALDH1A1 gene expression. |
Table I
List of microRNAs predicted to
interfere with ALDH1A1 gene expression.
| miRNet | miR_id | miR_acc | Experiment | PMID or
database |
|---|
|
mirnet-hsa-29920 |
hsa-mir-181a-5p | MIMAT0000256 | qRT-PCR | 26693182 |
|
mirnet-hsa-44218 | hsa-mir-140-5p | MIMAT0000431 | Luc/Wblot | 23752191 |
|
mirnet-hsa-647281 | hsa-mir-185-5p | MIMAT0000455 | HITS-CLIP | TarBase |
|
mirnet-hsa-647282 |
hsa-mir-200c-3p | MIMAT0000617 | PAR-CLIP | TarBase |
|
mirnet-hsa-647283 | hsa-mir-21-5p | MIMAT0000076 | PAR-CLIP | TarBase |
|
mirnet-hsa-647284 | hsa-mir-221-3p | MIMAT0000278 | HITS-CLIP | TarBase |
|
mirnet-hsa-647285 | hsa-mir-221-5p | MIMAT0004568 | HITS-CLIP | TarBase |
|
mirnet-hsa-647286 | hsa-mir-222-3p | MIMAT0000279 | HITS-CLIP | TarBase |
|
mirnet-hsa-647287 | hsa-mir-222-5p | MIMAT0004569 | HITS-CLIP | TarBase |
|
mirnet-hsa-647288 | hsa-mir-22-5p | MIMAT0004495 | HITS-CLIP | TarBase |
|
mirnet-hsa-647289 | hsa-mir-362-3p | MIMAT0004683 | HITS-CLIP | TarBase |
|
mirnet-hsa-647290 |
hsa-mir-374a-3p | MIMAT0004688 | HITS-CLIP | TarBase |
|
mirnet-hsa-647291 | hsa-mir-4517 | MIMAT0019054 | PAR-CLIP | TarBase |
|
mirnet-hsa-647292 | hsa-let-7b-5p | MIMAT0000063 | Microarrays | TarBase |
|
mirnet-hsa-647293 |
hsa-mir-103a-3p | MIMAT0000101 | Microarrays | TarBase |
|
mirnet-hsa-647294 | hsa-mir-107 | MIMAT0000104 | Microarrays | TarBase |
|
mirnet-hsa-647295 | hsa-mir-16-5p | MIMAT0000069 | Microarrays | TarBase |
|
mirnet-hsa-647296 | hsa-mir-191-5p | MIMAT0000440 | Microarrays | TarBase |
|
mirnet-hsa-647297 | hsa-mir-195-5p | MIMAT0000461 | Microarrays | TarBase |
|
mirnet-hsa-647298 | hsa-mir-21-3p | MIMAT0004494 | Microarrays | TarBase |
|
mirnet-hsa-647299 | hsa-mir-210-3p | MIMAT0000267 | Microarrays | TarBase |
|
mirnet-hsa-647300 | hsa-mir-26a-5p | MIMAT0000082 | Microarrays | TarBase |
|
mirnet-hsa-647301 | hsa-mir-27a-5p | MIMAT0004501 | Microarrays | TarBase |
|
mirnet-hsa-647302 | hsa-mir-30d-5p | MIMAT0000245 | Microarrays | TarBase |
|
mirnet-hsa-647303 | hsa-mir-34b-5p | MIMAT0000685 | Microarrays | TarBase |
|
mirnet-hsa-647304 | hsa-mir-34c-5p | MIMAT0000686 | Microarrays | TarBase |
|
mirnet-hsa-647305 | hsa-mir-941 | MIMAT0004984 | Microarrays | TarBase |
 | Table IIList of microRNA species that
interfere with ALDH1A1 expression. |
Table II
List of microRNA species that
interfere with ALDH1A1 expression.
| miRNA_name | miRNA_id | Experiments | Publications | Cell_lines | micro_tscore |
|---|
|
hsa-miR-103a-3p | MIMAT0000101 | 2 | 2 | 2 | −0.27 |
| hsa-miR-15a-5p | MIMAT0000068 | 2 | 2 | 1 | 0.37 |
| hsa-miR-16-5p | MIMAT0000069 | 2 | 2 | 2 | 0.46 |
| hsa-miR-210-3p | MIMAT0000267 | 2 | 1 | 2 | 0.42 |
| hsa-miR-26b-5p | MIMAT0000083 | 2 | 2 | 2 | 0.62 |
| hsa-miR-34a-5p | MIMAT0000255 | 2 | 2 | 2 | −1 |
| hsa-let-7g-3p | MIMAT0004584 | 1 | 1 | 1 | −1 |
| hsa-miR-101-3p | MIMAT0000099 | 1 | 1 | 1 | −1 |
| hsa-miR-107 | MIMAT0000104 | 1 | 1 | 1 | −1 |
|
hsa-miR-1271-3p | MIMAT0022712 | 1 | 1 | 1 | −1 |
|
hsa-miR-130a-3p | MIMAT0000425 | 1 | 1 | 1 | −1 |
|
hsa-miR-130b-3p | MIMAT0000691 | 1 | 1 | 1 | −1 |
| hsa-miR-195-5p | MIMAT0000461 | 1 | 1 | 1 | −1 |
|
hsa-miR-199a-3p | MIMAT0000232 | 1 | 1 | 1 | −1 |
|
hsa-miR-199b-3p | MIMAT0004563 | 1 | 1 | 1 | −1 |
| hsa-miR-21-5p | MIMAT0000076 | 1 | 1 | 1 | 0.74 |
| hsa-miR-221-5p | MIMAT0004568 | 1 | 1 | 1 | 0.41 |
| hsa-miR-29b-3p | MIMAT0000100 | 1 | 1 | 1 | −1 |
|
hsa-miR-301a-3p | MIMAT0000688 | 1 | 1 | 1 | −1 |
|
hsa-miR-301b-3p | MIMAT0004958 | 1 | 1 | 1 | −1 |
| hsa-miR-30d-5p | MIMAT0000245 | 1 | 1 | 1 | −1 |
| hsa-miR-454-3p | MIMAT0003885 | 1 | 1 | 1 | −1 |
| hsa-miR-542-5p | MIMAT0003340 | 1 | 1 | 1 | 0.38 |
| hsa-miR-941 | MIMAT0004984 | 1 | 1 | 1 | −1 |
miR-16 has been revealed to be typically
downregulated in leukemia, an event which contributes to the
uncontrolled growth and survival of leukemic cells (59,60). It has been shown to be increased
in patients with AML in remission (61). In murine myeloid cells expressing
internal tandem duplications of the juxtamembrane region of the
gene FLT3 (FLT3/ITD) that constitutes a marker for poor prognosis
for AML, miR-16 was significantly down-regulated; and conversely,
it was upregulated upon FLT3 inhibition (62). Its reduced expression was revealed
to be associated with the dysregulation of several target genes
involved in cell cycle control and apoptosis (63). miR-16 was demonstrated to target
multiple oncogenes and regulators of apoptosis, such as BCL2 (an
anti-apoptotic protein) and cyclins (involved in cell cycle
progression) (60). By targeting
these genes, miR-16 inhibited cell proliferation and promoted
programmed cell death. Thus, miR-16 may be a prospective candidate
for ALDH1A1 inhibition in AML model systems, due to the established
anti-leukemic effects of this miRNA.
Another miRNA, miR-34a has been revealed to be
associated with prognosis in AML (64), and experiments on epithelial
cancer cells indicate that miR-34a has the potential to repress
ALDH1A1, without findings revealing whether repression is direct or
indirect (65-67). Research is required to elucidate
the mechanism of miR-34a interference with ALDH1A1 gene expression,
and specifically whether or not miR-34a can directly target the 3′
untranslated region of ALDH1A1 in AML cells.
For the miR-30 family, members 30a, 30b and 30c,
were repressed in AML bone marrow samples, while miR-30d was found
underexpressed in serum samples of patients with chronic
lymphocytic leukemia, but an association with AML has yet to be
shown (68,69). However, in oral squamous cell
carcinoma specimens, miR-30a was shown to promote expression of
ALDH1 member ALDH1A2 (70),
making miR-30a an unlikely candidate for development as an ALDH1A1
inhibitor.
Lastly, miR-200c has exhibited the potential to
regulate ALDH1A1 expression (71,72) even if this effect can be
indirectly linked to miR-200c. This miRNA, has shown relevance in
blocking oncogenic signaling in AML; in particular, miR-200c
repression was identified as a key molecular mechanism of oncogene
MUC1 induction of PD-L1 expression, which has a critical function
in the progression of AML (73).
miR-200c, therefore is a noteworthy candidate to assess its
potential as an ALDH1A1 inhibitor in AML model systems.
miRNAs with a potential to regulate
ALDH1A1 expression
Integrating miRNA and mRNA expression profiling in
AML revealed that miR-155 has a critical association with immunity
(74). miR-155 was revealed to
suppress ALDH1A1 in a solid tumor model. In a metastatic cell line
model that allows investigation of extravasation and colonization
of circulating cancer cells to lungs of mice, miR-155
overexpression in tumors suppressed ALDH1A1, PIR and PDCD4
(75). However, in AML, miR-155
has an association with poor disease outcome; in cytogenetically
normal patients, overexpression of miR-155 was associated with a
shorter disease-free and overall survival (76). miR-155 was also revealed to be
associated with a 'core enriched' stem cell gene expression score;
other miRNAs that were associated with this score were miRNAs known
to be highly expressed and functionally relevant in embryonic
(miR-20a) (77) or HSCs (miR-99,
miR-125a/b and miR-126) (78).
For some miRs in that signature (the 'core enriched' stem cell gene
expression score), there are functional studies showing that their
overexpression causes leukemia in model systems [miR-92a (79) and miR-125b (80)]. Furthermore, primary AML blast
cells harboring the FLT3-ITD mutation had high expression of
miR-155; 8-chloro-adenosine killed LSCs and supressed miR-155
without killing normal stem cells (81). Other miRNAs that regulate the
maintenance of stemness of primitive hematopoietic progenitor
cells, include miR-22 and miR-29 (82).
A notable observation was made with another miRNA,
namely miR-143; overexpression of a miR-143-3p mimic repressed
viability and proliferation of AML cells and overexpression of
lysine acetyltransferase 6A (KAT6A) partially reversed the
inhibitory effects of the miR-143-3p mimic on viability and
proliferation of AML cells. A miR-143-3p mimic decreased the
expression levels of IL-1β, TNF-α and IL-6, and increased the
expression levels of TGF-β and IL-10 (83). The induction of TGF-β and IL-10
may be potentially detrimental in AML, if these two cytokines are
secreted by AML cells in the microenviroment, since they can have a
negative effect on the antitumor immune response by inhibiting the
function of T cells (84).
Nevertheless their effects require extensive characterization in
more clinically-relevant research models (85,86).
It is extremely important to emphasize that the role
of individual miRNAs is highly context-dependent; overexpression of
miR-125b in osteoblasts, for example, leads to increased bone mass
and strength, while preserving bone formation and quality (87). Thus, it is crucial to determine
the characteristics of any given miRNA before selecting it for
intervention. Furthermore, any such intervention can be expected to
have complex pathological consequences, which necessitates a
precise understanding of the effects of any given miRNA.
Impact of the ALDH1A1-targeting miRNAs on
the cellular phenotype
As aforementioned, the interaction between miRNAs
and ALDH1A1 may not have ubiquitous effects for all cell types, due
to the complexity of their interacting pathways. In this context,
it is not yet known whether the interactions between the
aforementioned miRNAs and ALDH1A1 occur in all cell types, and
especially in AML cells. However, there are also indications that
miRNA-mediated control of ALDH1A1 levels in cells may function as
part of a general adaptation mechanism and should be further
investigated. For example, the expression of most of the miRNAs
aforementioned has been revealed to be regulated by TGF-β, and it
was shown that they are involved in the process of
epithelial-mesenchymal transition (EMT); in particular, miR-140
suppressed the TGF-β pathway in mouse embryonic fibroblasts and
conversely, TGF-β suppressed the accumulation of miR-140 forming a
double negative feedback loop (88). EMT is a phenotype adaptation that
is triggered in cells to adjust to new conditions, which is not
limited for epithelial cells as the name suggests, but it is also
used by various types of acute leukemia cells (89). In this sense, after being
stimulated by various factors in the local microenvironment,
including TGF-β, transcriptional reprogramming is activated to
transform cells into a mesenchymal phenotype (90). As regards CSCs, numerous studies
have reported that cells undergoing EMT exhibit CSC or CSC-like
properties (91,92) under the influence of TGF-β
(93). On the other hand, core
pathways operating in CSCs may also induce EMT. For example,
ALDH1A1 and ALDH1A3 may induce TGF-β expression by activating
retinoic acid receptor, RARA, and androgen receptor in prostate
cancer (94). In concordance,
retinoic acid was shown to increase TGF-β2 expression and secretion
of active and latent forms of TGF-β2 in pancreatic cancer cells
(95).
Prospects of targeting miRNAs that
regulate ALDH1A1 expression
Although the field of RNA therapeutics has made
substantial progress over the last decade, there are currently only
a few miRNAs that are clinically targeted in intervention studies
(96), due to the observation of
off-target effects and toxicity (97). This is to be expected given the
complex manner of miRNA function.
Of the miRNAs reviewed herein, only two are
currently targets of intervention in clinical trials, namely miR-29
and miR-155.
Cobomarsen (MRG-106) is a miR-155 inhibitor
developed by Viridian Therapeutics, and has demonstrated efficacy
in the treatment of cutaneous T-cell lymphoma (98).
In addition, Remlarsen and MRG-229 also developed by
the same manufacturer, are miR-29 mimics. Remlarsen repressed
collagen expression and the development of fibroplasia in
incisional skin wounds (99).
MRG-229, developed for idiopathic pulmonary fibrosis, reduces
experimentally induced fibrotic activity in both in vitro
and ex vivo (lung slices derived from donors without a
history of lung disease) human disease models in non-human
primates, and was reportedly well tolerated at clinically relevant
doses with no adverse findings observed (100).
By contrast, numerous studies focus on developing
miRNA-based biomarker applications, such as the study NCT05809050,
'Study of miRNA-155 in Acute Leukemia'.
miRNAs that can be studied further in model
systems for AML preclinical drug development, based on database
output
In conclusion, in AML research, ALDH1A1 repression
by miRNAs is a rather under-studied topic. From the miRNAs
identified through bioinformatic analysis, it is suggested that
miR-16 and possibly also miR-200, are potential candidates for
further analyses. To underscore this assessment, miR-16-5p was
implicated by miRNet in ALDH1A1 regulation in chronic myeloid
leukemia (101). Another
potential incentive for considering miR-16 development, is the
rather acceptable safety profile observed in a phase 1 clinical
trial for patients with recurrent malignant pleural mesothelioma.
The approach undertaken was to use 'bacterial minicells', which
were anucleate nanoparticles produced by inactivating the genes
that control normal bacterial cell division, allowing efficient
packaging of cytotoxic drugs and internalization into cancer cells
(102). In acute lymphoblastic
leukemia (ALL), a distinct type of leukemia from AML, miR-16-5p was
proposed to enhance sensitivity to the p53-Mdm2 inhibitor, RG7388,
which was evaluated in a clinical trial (NCT04029688) (103), making an application of
miR-16-5p in ALL at least theoretically conceivable. To support
this additional prospective application research, when examining
RNA samples from pediatric patients with either AML or ALL using
the program 'Therapeutically Applicable Research to Generate
Effective Treatments (TARGET)' (https://www.cancer.gov/ccg/research/genome-sequencing/target/about)
(104,105), it becomes apparent that high
ALDH1A1 RNA expression is associated with a decreased patient
survival in both types of acute leukemia (Fig. 2). In general however, caution
should be exercised when translating the data for the miR-16-5p
regulation of ALDH1A1, for the development of prospective
preclinical treatment schemes, either for AML or for ALL.
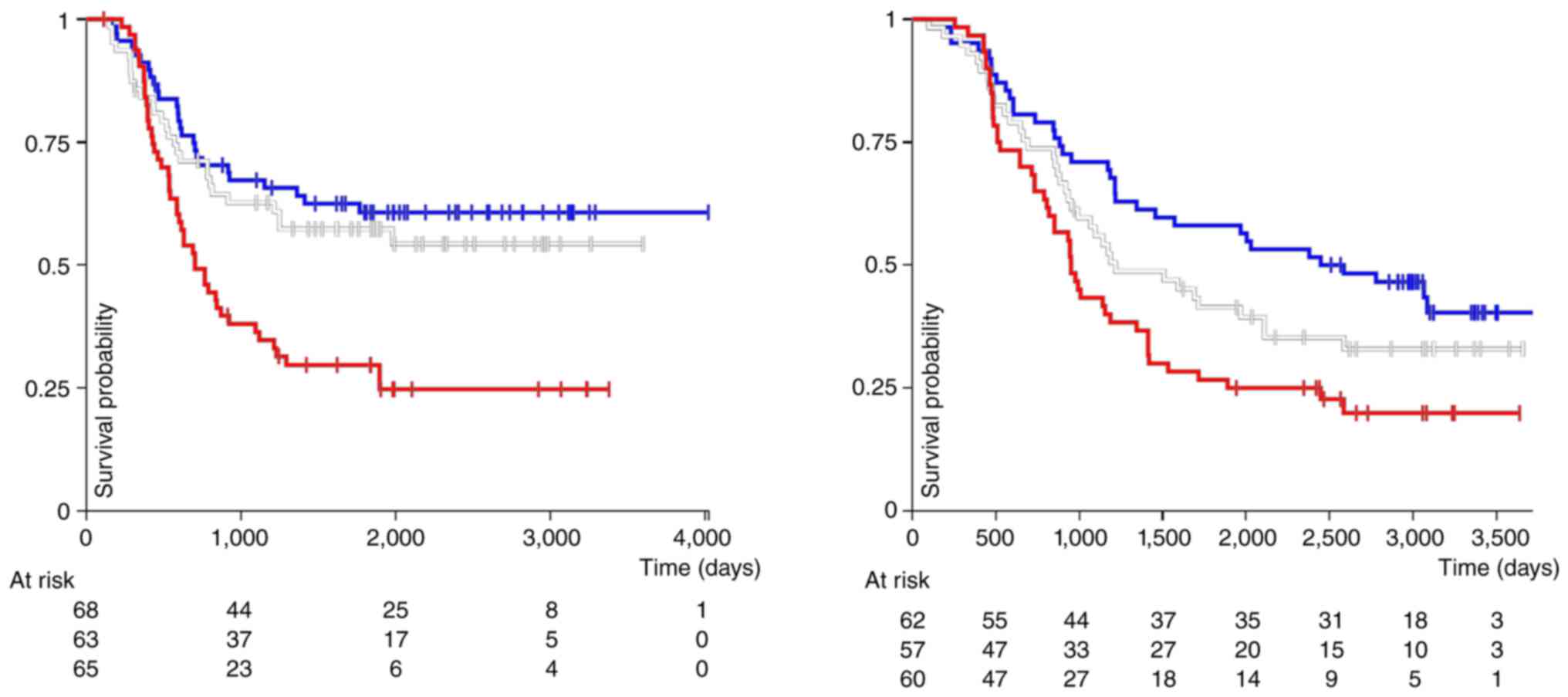 | Figure 2Analysis of the association of RNA
expression from the gene ALDH1A1 with overall survival of pediatric
patients with either acute myeloid leukemia (left panel, P=0.00014;
log-rank test=17.68) or acute lymphoblastic leukemia (right panel,
P=0.005; log-rank test=10.25), performed using the online software
platform UCSC Xena (https://xenabrowser.net/, accessed on March 10, 2024)
(123). The x-axis corresponds
to the time passed in days, and the y-axis corresponds to patient
overall survival. Units used are log2 (normalized counts
+1). The red line corresponds to samples with the highest
expression (>8.12, n=65 for AML; and >8.7, n=60 for ALL). The
white line shows samples with intermediate ALDH1A1 mRNA expression
(>4.61 and <8.12, n=63 for AML; >6.0 and <8.7, n=57 for
ALL). Shown by the blue line are samples with the lowest ALDH1A1
RNA expression (<4.61, n=68 for AML; and <6.0, n=62 for ALL).
The results published in this figure are in whole based upon data
generated by the study 'Therapeutically Applicable Research to
Generate Effective Treatments' (https://www.cancer.gov/ccg/research/genome-sequencing/target)
initiative, phs000218. The data used for this analysis are
available at the Genomic Data Commons (https://portal.gdc.cancer.gov). |
Nevertheless, the miRNAs identified as direct
regulators of ALDH1A1 in solid tumor cell studies, namely
miR-181a/b, miR-625, miR-140 and miR-23b, can be studied in
preclinical AML model systems with an anticipated beneficial
outcome. This assumption is based on the implication of their
repression in mechanisms of leukemia progression, which suggests
that their exogenous re-introduction could inhibit at least a
portion of the leukemic clones, prompting the question of whether
these clones comprise cells expressing high levels of ALDH1A1
RNA.
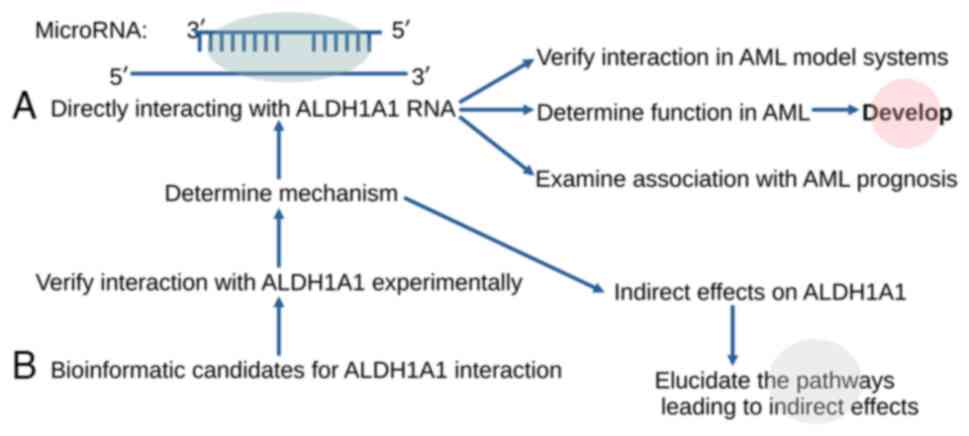
The available miRNA database information and the
existing experimental evidence render it possible to implement a
strategy for the development of candidate inhibitors of ALDH1A1
expression (Fig. 3), taking into
account the impact of the miRNA candidates on the metabolic status
of the cells, where the inhibition is aimed to take place.
Although several miRNAs, especially miR-181, have
been identified as prospective candidates for the development of
AML therapy, in clinical trials, miRNAs are mostly evaluated as
prospective biomarkers (40).
Prospective delivery methods and study
systems for targeting miRNAs that regulate ALDH1A1 expression
miRNAs can be delivered to the bone marrow through a
number of methodological developments that include exosomal
delivery, activated hydrogel, cell-specific ligand-decorated
nanocarriers, and encapsulating co-polymers (60,106-109). The advances that have been made
during the last 10 years in RNA therapeutic applications, and in
particular in small interfering RNAs, can help accelerate progress
of research in miRNA delivery (96). Strategies explored in miRNA
delivery research include lipid-based nanoparticles, polymeric
vectors, dendrimer-based vectors, cell-derived membrane vesicles,
three-dimensional scaffolds, as well as biodegradable and
biocompatible nanoparticles derived from polymers and metals
(110). Antagonists of miRNAs
may be clinically evaluated using antisence oligonucleotides, an
approach that currently appears most feasible (111,112).
Recently, a novel approach that was based on
programmable editing of primary miRNA, switched stem cell
differentiation and improved tissue regeneration, promoting in
vitro cartilage formation and calvarial bone healing in rats
(113). The bone, and especially
the bone marrow, are targets for potential anti-osteoporosis
treatments in experimental research (114). Furthermore, treatments for bone
metastasis for solid tumors may affect not only tumor cells but
also the balance between osteoclasts and osteoblasts, and thereby
modulate the properties of the bone as a niche (115-117). While the development of such
applications is not directly related to AML, it is a field that may
provide effective methods for delivery of miRNAs into the bone
marrow for treatment of AML, also including ex vivo
manipulation of selected marrow cell types that can be used as
vehicles with anti-leukemia activity. Another significant
development to anticipate are bone marrow organoids, which can help
bridge in vitro research and clinical applications, while
limiting the use of animal models (118). The organoids can help with the
accurate selection of the cell types that are targeted with the
experimental miRNA-based intervention, enabling improved assessment
of the outcomes in a cell-specific manner.
Although miRNAs are intensively studied, the
complexity of their regulation has limited their clinical
application mostly to a biomarker-oriented field. However, there
are a few studies that continue to explore interventions based on
direct regulation of miRNA function (96-99). In this sense, it is urgent to
overcome two fundamental problems that may be encountered in
miRNA-based therapy. The first is the development of a treatment
strategy that targets only specific types of cells and tissues.
Since miRNA target all cells in a systemic application using miRNA
mimics without a specific tropism, side effects are inevitable.
Therefore, the design of target-selective constructs (such as a
modified viral vector) that will express a specific miRNA based on
genetic engineering appears to be a more relevant approach
(119). In such a case, using a
promoter of a gene that has limited expression only in target cells
(or tissues) and placing the miRNA in the construct under the
control of this promoter may provide possible success in terms of
ensuring expression only in the intended target cells. The second
issue that may be encountered in miRNA therapy is the off-target
effects caused by miRNAs generally targeting more than one mRNA. In
fact, overcoming the off-target effects is challenging in the
native miRNA-involving applications when compared with the
synthetic modified versions of miRNAs. Although there have been
attempts to increase the selectivity and specificity of
experimental interventions, significant progress is still required
in order to develop approaches that permit a rigorous selection of
target genes for the artificial miRNA constructs (120-122).
Conclusions
Despite extensive research on miRNAs, the intricacy
of their regulation has limited their clinical application mostly
to a biomarker-focused field. However, there are a few studies that
continue to explore interventions based on miRNA regulation. Due to
the certainty of off-target effects, it is imperative to accurately
ascertain the clusters of candidate target genes in relevant model
systems. In the case of ALDH1A1, while there are miRNAs, such as
miR-155 with varying effects in different systems, there are other
miRNAs that may qualify for preclinical development of
interventions, such as miR-181.
It is enticing to consider including miRNA-targeted
interventions in standard or experimental AML treatments. To
combine two novel approaches is extremely risky from the point of
view of drug development, but may be fruitful as an experimental
approach for the aim of enriching our understanding of AML biology.
The next more immediate step in drug development would be to
consider combining miRNA-targeted interventions with standard AML
treatments. Although combination of miRNA-based approaches with
approved anti-neoplastic agents is an appealing aim, at this stage
the main challenge that needs to be overcome before moving forward,
is to determine the methodological approach that will permit a
greater investment of resources in the field of preclinical
development of miRNA-based interventions. The reason for
recommending this caution is due to the inherent complexity of
miRNA-interacting pathways, which inevitably exert numerous
effects. The primary concern is therefore to determine and manage
the substantial biomedical impact of a given miRNA, before the
drugs that are pharmacologically compatible with that miRNA can be
included into a testing protocol.
ALDH1A1 has critical roles in LSC biology and
thereby in therapy resistance. miRNAs are directly involved in the
regulation of ALDH1A1 in cells. Although miRNAs directly targeting
ALDH1A1 have been mostly demonstrated in solid tissues, there is a
strong possibility that they also target ALDH1A1 in LSCs. Given
this perspective, it is understandable that these studies are
somewhat overlooked, despite the critical roles of ALDH1A1 in LSCs
and its impact on therapy resistance. It is crucial to
comprehensively identify miRNAs that target ALDH1A1 in both HSCs
and LSCs. Once the miRNA networks targeting ALDH1A1 in HSCs and
LSCs are revealed, any differences between the two should be
identified and the molecular mechanisms that cause these
differences can then be rigorously investigated.
Availability of data and materials
Not applicable.
Authors' contributions
All authors (SAV, LV, PZ, DAS and VZ) contributed to
the conceptualization of the project, to the interpretation and
analysis of the data to be included in the review, and wrote and
prepared the draft of the manuscript. All authors read and approved
the final manuscript. Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
No funding was received.
References
|
1
|
Smith C, Gasparetto M, Jordan C, Pollyea
DA and Vasiliou V: The effects of alcohol and aldehyde
dehydrogenases on disorders of hematopoiesis. Adv Exp Med Biol.
815:349–359. 2015. View Article : Google Scholar
|
|
2
|
Duan X, Hu H, Wang L and Chen L: Aldehyde
dehydrogenase 1 family: A potential molecule target for diseases.
Cell Biol Int. May 27–2024.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lavudi K, Nuguri SM, Pandey P, Kokkanti RR
and Wang QE: ALDH and cancer stem cells: Pathways, challenges, and
future directions in targeted therapy. Life Sci. 356:1230332024.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vlahopoulos S, Pan L, Varisli L, Dancik
GM, Karantanos T and Boldogh I: OGG1 as an epigenetic reader
affects NFκB: What this means for cancer. Cancers (Basel).
16:1482023. View Article : Google Scholar
|
|
5
|
Vlahopoulos SA: Divergent processing of
cell stress signals as the basis of cancer progression: Licensing
NFκB on Chromatin. Int J Mol Sci. 25:86212024. View Article : Google Scholar
|
|
6
|
Carroll C, Manaprasertsak A, Boffelli
Castro A, van den Bos H, Spierings DCJ, Wardenaar R, Bukkuri A,
Engström N, Baratchart E, Yang M, et al: Drug-resilient Cancer Cell
Phenotype Is Acquired via Polyploidization Associated with Early
Stress Response Coupled to HIF2α Transcriptional Regulation. Cancer
Res Commun. 4:691–705. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fredebohm J, Boettcher M, Eisen C, Gaida
MM, Heller A, Keleg S, Tost J, Greulich-Bode KM, Hotz-Wagenblatt A,
Lathrop M, et al: Establishment and characterization of a highly
tumourigenic and cancer stem cell enriched pancreatic cancer cell
line as a well defined model system. PLoS One. 7:e485032012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kaigorodova EV, Kozik AV and Grishchenko
MY: Decoding Metastasis: From cell death to fusion in cancer
progression. Curr Cancer Drug Targets. Jul 15–2024.Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Truskowski K, Amend SR and Pienta KJ:
Dormant cancer cells: Programmed quiescence, senescence, or both?
Cancer Metastasis Rev. 42:37–47. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Park MN: The therapeutic potential of a
strategy to prevent acute myeloid leukemia stem cell reprogramming
in older patients. Int J Mol Sci. 24:120372023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dancik GM, Varisli L and Vlahopoulos SA:
The molecular context of oxidant stress response in cancer
establishes ALDH1A1 as a Critical Target: What this means for acute
myeloid leukemia. Int J Mol Sci. 24:93722023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shortall K, Djeghader A, Magner E and
Soulimane T: Insights into aldehyde dehydrogenase enzymes: A
structural perspective. Front Mol Biosci. 8:6595502021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gasparetto M and Smith CA: ALDHs in normal
and malignant hematopoietic cells: Potential new avenues for
treatment of AML and other blood cancers. Chem Biol Interact.
276:46–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yue H, Hu Z, Hu R, Guo Z, Zheng Y, Wang Y
and Zhou Y: ALDH1A1 in Cancers: Bidirectional function, drug
resistance, and regulatory mechanism. Front Oncol. 12:9187782022.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou Y, Huang G, Cai X, Liu Y, Qian B and
Li D: Global, regional, and national burden of acute myeloid
leukemia, 1990-2021: a systematic analysis for the global burden of
disease study 2021. Biomark Res. 12:1012024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Magni M, Shammah S, Schiró R, Mellado W,
Dalla-Favera R and Gianni AM: Induction of
cyclophosphamide-resistance by aldehyde-dehydrogenase gene
transfer. Blood. 87:1097–1103. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moreb JS, Maccow C, Schweder M and
Hecomovich J: Expression of antisense RNA to aldehyde dehydrogenase
class-1 sensitizes tumor cells to 4-hydroperoxycyclophosphamide in
vitro. J Pharmacol Exp Ther. 293:390–396. 2000.PubMed/NCBI
|
|
18
|
Smith C, Gasparetto M, Humphries K,
Pollyea DA, Vasiliou V and Jordan CT: Aldehyde dehydrogenases in
acute myeloid leukemia. Ann N Y Acad Sci. 1310:58–68. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cheung AM, Wan TS, Leung JC, Chan LY,
Huang H, Kwong YL, Liang R and Leung AY: Aldehyde dehydrogenase
activity in leukemic blasts defines a subgroup of acute myeloid
leukemia with adverse prognosis and superior NOD/SCID engrafting
potential. Leukemia. 21:1423–1430. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dancik GM, Voutsas IF and Vlahopoulos S:
Aldehyde dehydrogenase enzyme functions in acute leukemia stem
cells. Front Biosci (Sch Ed). 14:82022. View Article : Google Scholar
|
|
21
|
Hoang VT, Buss EC, Wang W, Hoffmann I,
Raffel S, Zepeda-Moreno A, Baran N, Wuchter P, Eckstein V, Trumpp
A, et al: The rarity of ALDH(+) cells is the key to separation of
normal versus leukemia stem cells by ALDH activity in AML patients.
Int J Cancer. 137:525–536. 2015. View Article : Google Scholar
|
|
22
|
Gasparetto M, Pei S, Minhajuddin M, Khan
N, Pollyea DA, Myers JR, Ashton JM, Becker MW, Vasiliou V,
Humphries KR, et al: Targeted therapy for a subset of acute myeloid
leukemias that lack expression of aldehyde dehydrogenase 1A1.
Haematologica. 102:1054–1065. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Batten DJ, Crofts JJ and Chuzhanova N:
Towards In Silico identification of genes contributing to
similarity of patients' multi-omics profiles: A case study of acute
myeloid leukemia. Genes (Basel). 14:17952023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dancik GM, Voutsas IF and Vlahopoulos S:
Lower RNA expression of ALDH1A1 distinguishes the favorable risk
group in acute myeloid leukemia. Mol Biol Rep. 49:3321–3331. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dancik GM, Varisli L, Tolan V and
Vlahopoulos S: Aldehyde dehydrogenase genes as prospective
actionable targets in acute myeloid leukemia. Genes (Basel).
14:18072023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Venton G, Pérez-Alea M, Baier C, Fournet
G, Quash G, Labiad Y, Martin G, Sanderson F, Poullin P, Suchon P,
et al: Aldehyde dehydrogenases inhibition eradicates leukemia stem
cells while sparing normal progenitors. Blood Cancer J. 6:e4692016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pei S, Minhajuddin M, Adane B, Khan N,
Stevens BM, Mack SC, Lai S, Rich JN, Inguva A, Shannon KM, et al:
AMPK/FIS1-Mediated mitophagy is required for self-renewal of human
AML stem cells. Cell Stem Cell. 23:86–100.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Marcucci G, Mrózek K, Radmacher MD, Garzon
R and Bloomfield CD: The prognostic and functional role of
microRNAs in acute myeloid leukemia. Blood. 117:1121–1129. 2011.
View Article : Google Scholar :
|
|
29
|
Xiang M, Birkbak NJ, Vafaizadeh V, Walker
SR, Yeh JE, Liu S, Kroll Y, Boldin M, Taganov K, Groner B, et al:
STAT3 induction of miR-146b forms a feedback loop to inhibit the
NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes.
Sci Signal. 7:ra112014. View Article : Google Scholar
|
|
30
|
Karin M: NF-kappaB as a critical link
between inflammation and cancer. Cold Spring Harb Perspect Biol.
1:a0001412009. View Article : Google Scholar
|
|
31
|
Vlahopoulos SA, Cen O, Hengen N, Agan J,
Moschovi M, Critselis E, Adamaki M, Bacopoulou F, Copland JA,
Boldogh I, et al: Dynamic aberrant NF-κB spurs tumorigenesis: a new
model encompassing the microenvironment. Cytokine Growth Factor
Rev. 26:389–403. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jimbu L, Mesaros O, Joldes C, Neaga A,
Zaharie L and Zdrenghea M: MicroRNAs associated with a bad
prognosis in acute myeloid leukemia and their impact on macrophage
polarization. Biomedicines. 12:1212024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wallace JA and O'Connell RM: MicroRNAs and
acute myeloid leukemia: Therapeutic implications and emerging
concepts. Blood. 130:1290–1301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Boudreau RL, Jiang P, Gilmore BL, Spengler
RM, Tirabassi R, Nelson JA, Ross CA, Xing Y and Davidson BL:
Transcriptome-wide discovery of microRNA binding sites in human
brain. Neuron. 81:294–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee SH, Lee CR, Rigas NK, Kim RH, Kang MK,
Park NH and Shin KH: Human papillomavirus 16 (HPV16) enhances tumor
growth and cancer stemness of HPV-negative oral/oropharyngeal
squamous cell carcinoma cells via miR-181 regulation.
Papillomavirus Res. 1:116–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu X, Liao W, Peng H, Luo X, Luo Z, Jiang
H and Xu L: miR-181a promotes G1/S transition and cell
proliferation in pediatric acute myeloid leukemia by targeting ATM.
J Cancer Res Clin Oncol. 142:77–87. 2016. View Article : Google Scholar
|
|
37
|
Nanbakhsh A, Visentin G, Olive D, Janji B,
Mussard E, Dessen P, Meurice G, Zhang Y, Louache F, Bourhis JH and
Chouaib S: miR-181a modulates acute myeloid leukemia susceptibility
to natural killer cells. Oncoimmunology. 4:e9964752015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huang X, Schwind S, Santhanam R, Eisfeld
AK, Chiang CL, Lankenau M, Yu B, Hoellerbauer P, Jin Y, Tarighat
SS, et al: Targeting the RAS/MAPK pathway with miR-181a in acute
myeloid leukemia. Oncotarget. 7:59273–59286. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Seipel K, Messerli C, Wiedemann G, Bacher
U and Pabst T: MN1, FOXP1 and hsa-miR-181a-5p as prognostic markers
in acute myeloid leukemia patients treated with intensive induction
chemotherapy and autologous stem cell transplantation. Leuk Res.
89:1062962020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fletcher D, Brown E, Javadala J,
Uysal-Onganer P and Guinn BA: microRNA expression in acute myeloid
leukaemia: New targets for therapy? EJHaem. 3:596–608. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gong X, Xu B, Zi L and Chen X: miR-625
reverses multidrug resistance in gastric cancer cells by directly
targeting ALDH1A1. Cancer Manag Res. 11:6615–6624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ma L, Wang YY and Jiang P: LncRNA
LINC00909 promotes cell proliferation and metastasis in pediatric
acute myeloid leukemia via miR-625-mediated modulation of
Wnt/β-catenin signaling. Biochem Biophys Res Commun. 527:654–661.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shang Z, Ming X, Wu J and Xiao Y:
Downregulation of circ_0012152 inhibits proliferation and induces
apoptosis in acute myeloid leukemia cells through the
miR-625-5p/SOX12 axis. Hematol Oncol. 39:539–548. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Aliabedi B, Mousavi SH, Ebrahimi M,
Alizadeh S, Hedayati Asl AA, Mohammad M and Samieyan Dehkordi S:
Hsa-miR-625 Upregulation promotes apoptosis in acute myeloid
leukemia cell line by targeting integrin-linked kinase pathway.
Asian Pac J Cancer Prev. 23:1159–1167. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Samieyan Dehkordi S, Mousavi SH, Ebrahimi
M, Alizadeh SH, Hedayati Asl AA, Mohammad M and Aliabedi B:
Upregulation of hsa-miR-625-5p inhibits invasion of acute myeloid
leukemia cancer cells through ILK/AKT Pathway. Cell J. 24:76–84.
2022.PubMed/NCBI
|
|
46
|
Li Q, Yao Y, Eades G, Liu Z, Zhang Y and
Zhou Q: Downregulation of miR-140 promotes cancer stem cell
formation in basal-like early stage breast cancer. Oncogene.
33:2589–2600. 2014. View Article : Google Scholar :
|
|
47
|
Li H, Bi K, Feng S, Wang Y and Zhu C:
MiR-140 Targets lncRNA DNAJC3-AS1 to Suppress Cell Proliferation in
Acute Myeloid Leukemia. Mediterr J Hematol Infect Dis.
14:e20220052022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y, Wang F, Lu Y, Li Y, Ran H, Yan F
and Tian Y: MiR-140 targets lncRNA FAM230B to suppress cell
proliferation in acute myeloid leukemia running title: MiR-140
targets FAM230B in AML. Hematology. 27:700–705. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huang J, Jin S, Guo R, Wu W, Yang C, Qin
Y, Chen Q, He X, Qu J and Yang Z: Histone lysine demethylase KDM5B
facilitates proliferation and suppresses apoptosis in human acute
myeloid leukemia cells through the miR-140-3p/BCL2 axis. RNA.
30:435–447. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang HY, Lin YC, Cui S, Huang Y, Tang Y,
Xu J, Bao J, Li Y, Wen J, Zuo H, et al: miRTarBase update 2022: an
informative resource for experimentally validated miRNA-target
interactions. Nucleic Acids Res. 50(D1): D222–D230. 2022.
View Article : Google Scholar
|
|
51
|
Kariuki D, Asam K, Aouizerat BE, Lewis KA,
Florez JC and Flowers E: Review of databases for experimentally
validated human microRNA-mRNA interactions. Database (Oxford).
2023:baad0142023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kern F, Aparicio-Puerta E, Li Y, Fehlmann
T, Kehl T, Wagner V, Ray K, Ludwig N, Lenhof HP, Meese E and Keller
A: miRTargetLink 2.0-interactive miRNA target gene and target
pathway networks. Nucleic Acids Res. 49(W1): W409–W416. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang W, Li Y, Liu N, Gao Y and Li L:
MiR-23b controls ALDH1A1 expression in cervical cancer stem cells.
BMC Cancer. 17:2922017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Barrera-Ramirez J, Lavoie JR, Maganti HB,
Stanford WL, Ito C, Sabloff M, Brand M, Rosu-Myles M, Le Y and
Allan DS: Micro-RNA profiling of exosomes from marrow-derived
mesenchymal stromal cells in patients with acute myeloid leukemia:
Implications in Leukemogenesis. Stem Cell Rev Rep. 13:817–825.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Jiang W, Min J, Sui X, Qian Y, Liu Y, Liu
Z, Zhou H, Li X and Gong Y: MicroRNA-26a-5p and microRNA-23b-3p
up-regulate peroxiredoxin III in acute myeloid leukemia. Leuk
Lymphoma. 56:460–471. 2015. View Article : Google Scholar :
|
|
56
|
Gaál Z, Oláh É, Rejtő L, Bálint BL and
Csernoch L: Expression Levels of Warburg-Effect Related microRNAs
Correlate with each Other and that of Histone Deacetylase Enzymes
in Adult Hematological Malignancies with Emphasis on Acute Myeloid
Leukemia. Pathol Oncol Res. 23:207–216. 2017. View Article : Google Scholar
|
|
57
|
Sethupathy P, Corda B and Hatzigeorgiou
AG: TarBase: A comprehensive database of experimentally supported
animal microRNA targets. RNA. 12:192–197. 2006. View Article : Google Scholar :
|
|
58
|
Chang L, Zhou G, Soufan O and Xia J:
miRNet 2.0: Network-based visual analytics for miRNA functional
analysis and systems biology. Nucleic Acids Res. 48(W1): W244–W251.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar
|
|
60
|
Liberati FR, Di Russo S, Barolo L, Peruzzi
G, Farina MV, Spizzichino S, Di Fonzo F, Quaglio D, Pisano L, Botta
B, et al: Combined Delivery of miR-15/16 through Humanized ferritin
nanocages for the treatment of chronic lymphocytic leukemia.
Pharmaceutics. 16:4022024. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gao SM, Yang J, Chen C, Zhang S, Xing CY,
Li H, Wu J and Jiang L: miR-15a/16-1 enhances retinoic
acid-mediated differentiation of leukemic cells and is up-regulated
by retinoic acid. Leuk Lymphoma. 52:2365–2371. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kim KT, Carroll AP, Mashkani B, Cairns MJ,
Small D and Scott RJ: MicroRNA-16 is down-regulated in mutated FLT3
expressing murine myeloid FDC-P1 cells and interacts with Pim-1.
PLoS One. 7:e445462012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Abraham M, Klein S, Bulvik B, Wald H,
Weiss ID, Olam D, Weiss L, Beider K, Eizenberg O and Wald O, et al:
The CXCR4 inhibitor BL-8040 induces the apoptosis of AML blasts by
downregulating ERK, BCL-2, MCL-1 and cyclin-D1 via altered
miR-15a/16-1 expression. Leukemia. 31:2336–2346. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Abdellateif MS, Hassan NM, Kamel MM and
El-Meligui YM: Bone marrow microRNA-34a is a good indicator for
response to treatment in acute myeloid leukemia. Oncol Res.
32:577–584. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ma W, Xiao GG, Mao J, Lu Y, Song B, Wang
L, Fan S, Fan P, Hou Z, Li J, et al: Dysregulation of the
miR-34a-SIRT1 axis inhibits breast cancer stemness. Oncotarget.
6:10432–10444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hsieh PL, Liao YW, Hsieh CW, Chen PN and
Yu CC: Soy isoflavone genistein impedes cancer stemness and
mesenchymal transition in head and neck cancer through activating
miR-34a/RTCB Axis. Nutrients. 12:19242020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Xu C, Cao X, Cao X, Liu L, Qiu Y, Li X,
Zhou L, Ning Y, Ren K and Cao J: Isovitexin Inhibits Stemness and
Induces Apoptosis in Hepatocellular Carcinoma SK-Hep-1 Spheroids by
Upregulating miR-34a Expression. Anticancer Agents Med Chem.
20:1654–1663. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Fuster O, Llop M, Dolz S, García P, Such
E, Ibáñez M, Luna I, Gómez I, López M, Cervera J, et al: Adverse
prognostic value of MYBL2 overexpression and association with
microRNA-30 family in acute myeloid leukemia patients. Leuk Res.
37:1690–1696. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Farzadfard E, Kalantari T and Tamaddon G:
Serum Expression of Seven MicroRNAs in Chronic Lymphocytic Leukemia
Patients. J Blood Med. 11:97–102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Shiah SG, Hsiao JR, Chang HJ, Hsu YM, Wu
GH, Peng HY, Chou ST, Kuo CC and Chang JY: MiR-30a and miR-379
modulate retinoic acid pathway by targeting DNA methyltransferase
3B in oral cancer. J Biomed Sci. 27:462020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Nurwidya F, Takahashi F, Winardi W, Tajima
K, Mitsuishi Y, Murakami A, Kobayashi I, Nara T, Hashimoto M, Kato
M, et al: Zinc-finger E-box-binding homeobox 1 (ZEB1) plays a
crucial role in the maintenance of lung cancer stem cells resistant
to gefitinib. Thorac Cancer. 12:1536–1548. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hashida S, Yamamoto H, Shien K, Miyoshi Y,
Ohtsuka T, Suzawa K, Watanabe M, Maki Y, Soh J, Asano H, et al:
Acquisition of cancer stem cell-like properties in non-small cell
lung cancer with acquired resistance to afatinib. Cancer Sci.
106:1377–1384. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Pyzer AR, Stroopinsky D, Rosenblatt J,
Anastasiadou E, Rajabi H, Washington A, Tagde A, Chu JH, Coll M,
Jiao AL, et al: MUC1 inhibition leads to decrease in PD-L1 levels
via upregulation of miRNAs. Leukemia. 31:2780–2790. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Havelange V, Stauffer N, Heaphy CC,
Volinia S, Andreeff M, Marcucci G, Croce CM and Garzon R:
Functional implications of microRNAs in acute myeloid leukemia by
integrating microRNA and messenger RNA expression profiling.
Cancer. 117:4696–4706. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Thomsen KG, Terp MG, Lund RR, Søkilde R,
Elias D, Bak M, Litman T, Beck HC, Lyng MB and Ditzel HJ: miR-155,
identified as anti-metastatic by global miRNA profiling of a
metastasis model, inhibits cancer cell extravasation and
colonization in vivo and causes significant signaling alterations.
Oncotarget. 6:29224–29239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Metzeler KH, Maharry K, Kohlschmidt J,
Volinia S, Mrózek K, Becker H, Nicolet D, Whitman SP, Mendler JH,
Schwind S, et al: A stem cell-like gene expression signature
associates with inferior outcomes and a distinct microRNA
expression profile in adults with primary cytogenetically normal
acute myeloid leukemia. Leukemia. 27:2023–2031. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Rizzo M, Mariani L, Pitto L, Rainaldi G
and Simili M: miR-20a and miR-290, multi-faceted players with a
role in tumourigenesis and senescence. J Cell Mol Med.
14:2633–2640. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gerrits A, Walasek MA, Olthof S, Weersing
E, Ritsema M, Zwart E, van Os R, Bystrykh LV and de Haan G: Genetic
screen identifies microRNA cluster 99b/let-7e/125a as a regulator
of primitive hematopoietic cells. Blood. 119:377–387. 2012.
View Article : Google Scholar
|
|
79
|
Li Y, Vecchiarelli-Federico LM, Li YJ,
Egan SE, Spaner D, Hough MR and Ben-David Y: The miR-17-92 cluster
expands multipotent hematopoietic progenitors whereas imbalanced
expression of its individual oncogenic miRNAs promotes leukemia in
mice. Blood. 119:4486–4498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Bousquet M, Harris MH, Zhou B and Lodish
HF: MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA.
107:21558–21563. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Buettner R, Nguyen LXT, Kumar B, Morales
C, Liu C, Chen LS, Pemovska T, Synold TW, Palmer J, Thompson R, et
al: 8-chloro-adenosine activity in FLT3-ITD acute myeloid leukemia.
J Cell Physiol. 234:16295–16303. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Testa U and Pelosi E: MicroRNAs expressed
in hematopoietic stem/progenitor cells are deregulated in acute
myeloid leukemias. Leuk Lymphoma. 56:1466–1474. 2015. View Article : Google Scholar
|
|
83
|
Xu D, Jiang J, He G, Zhou H and Ji C:
miR-143-3p represses leukemia cell proliferation by inhibiting
KAT6A expression. Anticancer Drugs. 33:e662–e669. 2022. View Article : Google Scholar
|
|
84
|
Buggins AG, Milojkovic D, Arno MJ, Lea NC,
Mufti GJ, Thomas NS and Hirst WJ: Microenvironment produced by
acute myeloid leukemia cells prevents T cell activation and
proliferation by inhibition of NF-kappaB, c-Myc, and pRb pathways.
J Immunol. 167:6021–6030. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sun YX, Kong HL, Liu CF, Yu S, Tian T, Ma
DX and Ji CY: The imbalanced profile and clinical significance of T
helper associated cytokines in bone marrow microenvironment of the
patients with acute myeloid leukemia. Hum Immunol. 75:113–118.
2014. View Article : Google Scholar
|
|
86
|
Alhattab DM, Isaioglou I, Alshehri S, Khan
ZN, Susapto HH, Li Y, Marghani Y, Alghuneim AA, Díaz-Rúa R,
Abdelrahman S, et al: Fabrication of a three-dimensional bone
marrow niche-like acute myeloid Leukemia disease model by an
automated and controlled process using a robotic multicellular
bioprinting system. Biomater Res. 27:1112023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ito S, Minamizaki T, Kohno S, Sotomaru Y,
Kitaura Y, Ohba S, Sugiyama T, Aubin JE, Tanimoto K and Yoshiko Y:
Overexpression of miR-125b in osteoblasts improves age-related
changes in bone mass and quality through suppression of osteoclast
formation. Int J Mol Sci. 22:67452021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Pais H, Nicolas FE, Soond SM, Swingler TE,
Clark IM, Chantry A, Moulton V and Dalmay T: Analyzing mRNA
expression identifies Smad3 as a microRNA-140 target regulated only
at protein level. RNA. 16:489–494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Varisli L and Vlahopoulos S:
Epithelial-Mesenchymal transition in acute leukemias. Int J Mol
Sci. 25:21732024. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Imodoye SO, Adedokun KA, Muhammed AO,
Bello IO, Muhibi MA, Oduola T and Oyenike MA: Understanding the
complex milieu of epithelial-mesenchymal transition in cancer
metastasis: New insight into the roles of transcription factors.
Front Oncol. 11:7628172021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kong D, Banerjee S, Ahmad A, Li Y, Wang Z,
Sethi S and Sarkar FH: Epithelial to mesenchymal transition is
mechanistically linked with stem cell signatures in prostate cancer
cells. PLoS One. 5:e124452010. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Muraoka-Cook RS, Shin I, Yi JY, Easterly
E, Barcellos-Hoff MH, Yingling JM, Zent R and Arteaga CL: Activated
type I TGFbeta receptor kinase enhances the survival of mammary
epithelial cells and accelerates tumor progression. Oncogene.
25:3408–3423. 2006. View Article : Google Scholar
|
|
94
|
Gorodetska I, Lukiyanchuk V, Gawin M,
Sliusar M, Linge A, Lohaus F, Hölscher T, Kati Erdmann, Fuessel S,
Borkowetz A, et al: Blood-based detection of MMP11 as a marker of
prostate cancer progression regulated by the ALDH1A1-TGF-β1
signaling mechanism. bioRxiv: https://doi.org/10.1101/2024.07.16.603771.
|
|
95
|
Singh B, Murphy RF, Ding XZ, Roginsky AB,
Bell RH and Adrian TE: On the role of transforming growth
factor-beta in the growth inhibitory effects of retinoic acid in
human pancreatic cancer cells. Mol Cancer. 6:822007. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Seyhan AA: Trials and Tribulations of
MicroRNA Therapeutics. Int J Mol Sci. 25:14692024. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hong DS, Kang YK, Borad M, Sachdev J,
Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, et al: Phase 1
study of MRX34, a liposomal miR-34a mimic, in patients with
advanced solid tumours. Br J Cancer. 122:1630–1637. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Witten L and Slack FJ: miR-155 as a novel
clinical target for hematological malignancies. Carcinogenesis.
41:2–7. 2020. View Article : Google Scholar
|
|
99
|
Gallant-Behm CL, Piper J, Lynch JM, Seto
AG, Hong SJ, Mustoe TA, Maari C, Pestano LA, Dalby CM, Jackson AL,
et al: A MicroRNA-29 Mimic (Remlarsen) Represses Extracellular
Matrix Expression and Fibroplasia in the Skin. J Invest Dermatol.
139:1073–1081. 2019. View Article : Google Scholar
|
|
100
|
Chioccioli M, Roy S, Newell R, Sauler M,
Ahangari F, Ding S, DeIuliis J, Aurelien N, Montgomery RL and
Kaminski N: A lung targeted miR-29 mimic as a therapy for pulmonary
fibrosis. EBioMedicine. 85:1043042022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Narendra G, Raju B, Verma H and Silakari
O: Identification of potential genes associated with ALDH1A1
overexpression and cyclophosphamide resistance in chronic
myelogenous leukemia using network analysis. Med Oncol. 38:1232021.
View Article : Google Scholar : PubMed/NCBI
|
|
102
|
van Zandwijk N, Pavlakis N, Kao SC, Linton
A, Boyer MJ, Clarke S, Huynh Y, Chrzanowska A, Fulham MJ, Bailey
DL, et al: Safety and activity of microRNA-loaded minicells in
patients with recurrent malignant pleural mesothelioma: A
first-in-man, phase 1, open-label, dose-escalation study. Lancet
Oncol. 18:1386–1396. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zanjirband M, Rahgozar S and Aberuyi N:
miR-16-5p enhances sensitivity to RG7388 through targeting PPM1D
expression (WIP1) in childhood acute lymphoblastic leukemia. Cancer
Drug Resist. 6:242–256. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhang J, Mullighan CG, Harvey RC, Wu G,
Chen X, Edmonson M, Buetow KH, Carroll WL, Chen IM, Devidas M, et
al: Key pathways are frequently mutated in high-risk childhood
acute lymphoblastic leukemia: a report from the Children's Oncology
Group. Blood. 118:3080–3087. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Huang BJ, Smith JL, Farrar JE, Wang YC,
Umeda M, Ries RE, Leonti AR, Crowgey E, Furlan SN, Tarlock K, et
al: Integrated stem cell signature and cytomolecular risk
determination in pediatric acute myeloid leukemia. Nat Commun.
13:54872022. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Won Lee G, Thangavelu M, Joung Choi M,
Yeong Shin E, Sol Kim H, Seon Baek J, Woon Jeong Y, Eun Song J,
Carlomagno C, Miguel Oliveira J, et al: Exosome mediated transfer
of miRNA-140 promotes enhanced chondrogenic differentiation of bone
marrow stem cells for enhanced cartilage repair and regeneration. J
Cell Biochem. 121:3642–3652. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Wang N, Liu X, Tang Z, Wei X, Dong H, Liu
Y, Wu H, Wu Z, Li X, Ma X and Guo Z: Increased BMSC exosomal
miR-140-3p alleviates bone degradation and promotes bone
restoration by targeting Plxnb1 in diabetic rats. J
Nanobiotechnology. 20:972022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Rajagopal K, Arjunan P, Marepally S and
Madhuri V: Controlled differentiation of mesenchymal stem cells
into Hyaline Cartilage in miR-140-Activated Collagen Hydrogel.
Cartilage. 13(2_suppl): 571S–581S. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhou Y, Jia H, Hu A, Liu R, Zeng X and
Wang H: Nanoparticles targeting delivery antagomir-483-5p to bone
marrow mesenchymal stem cells treat osteoporosis by increasing bone
formation. Curr Stem Cell Res Ther. 18:115–126. 2023. View Article : Google Scholar
|
|
110
|
Diener C, Keller A and Meese E: Emerging
concepts of miRNA therapeutics: From cells to clinic. Trends Genet.
38:613–626. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Kim T and Croce CM: MicroRNA: Trends in
clinical trials of cancer diagnosis and therapy strategies. Exp Mol
Med. 55:1314–1321. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Grillone K, Caridà G, Luciano F, Cordua A,
Di Martino MT, Tagliaferri P and Tassone P: A systematic review of
non-coding RNA therapeutics in early clinical trials: A new
perspective against cancer. J Transl Med. 22:7312024. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Truong VA, Chang YH, Dang TQ, Tu Y, Tu J,
Chang CW, Chang YH, Liu GS and Hu YC: Programmable editing of
primary MicroRNA switches stem cell differentiation and improves
tissue regeneration. Nat Commun. 15:83582024. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Wen C, Xu X, Zhang Y, Xia J, Liang Y and
Xu L: Bone targeting nanoparticles for the treatment of
osteoporosis. Int J Nanomedicine. 19:1363–1383. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Gu J, Jiang L, Chen Z and Qi J: A simple
nanoplatform of thermo-sensitive liposomes and gold nanorods to
treat bone metastasis through improved chemotherapy combined with
photothermal therapy. Int J Pharm X. 8:1002822024.PubMed/NCBI
|
|
116
|
Li S, Kang Y and Zeng Y: Targeting tumor
and bone microenvironment: Novel therapeutic opportunities for
castration-resistant prostate cancer patients with bone metastasis.
Biochim Biophys Acta Rev Cancer. 1879:1890332024. View Article : Google Scholar
|
|
117
|
Xu M and Li S: Nano-drug delivery system
targeting tumor microenvironment: A prospective strategy for
melanoma treatment. Cancer Lett. 574:2163972023. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
de Janon A, Mantalaris A and Panoskaltsis
N: Three-Dimensional Human Bone Marrow Organoids for the Study and
Application of Normal and Abnormal Hematoimmunopoiesis. J Immunol.
210:895–904. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Herrera-Carrillo E, Liu YP and Berkhout B:
Improving miRNA Delivery by Optimizing miRNA expression cassettes
in diverse virus vectors. Hum Gene Ther Methods. 28:177–190. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Calloni R and Bonatto D: Scaffolds for
Artificial miRNA expression in animal cells. Hum Gene Ther Methods.
26:162–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Lundstrom K: Trans-amplifying RNA hitting
new grounds: Gene regulation by microRNA. Mol Ther Nucleic Acids.
35:1021912024. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Yıldız A, Hasani A, Hempel T, Köhl N,
Beicht A, Becker R, Hubich-Rau S, Suchan M, Poleganov MA, Sahin U
and Beissert T: Trans-amplifying RNA expressing functional miRNA
mediates target gene suppression and simultaneous transgene
expression. Mol Ther Nucleic Acids. 35:1021622024. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Goldman MJ, Craft B, Hastie M, Repečka K,
McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN, et al:
Visualizing and interpreting cancer genomics data via the Xena
platform. Nat Biotechnol. 38:675–678. 2020. View Article : Google Scholar : PubMed/NCBI
|