Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common malignant tumor worldwide and the second leading cause of
cancer-related deaths globally (1,2).
HCC accounts for 85-90% of all primary liver cancers (3). In 2022, there were over 430,000
newly reported cases of liver cancer in China, ranking fourth among
all cancers, with over 410,000 deaths, ranking second among all
cancer-related deaths (4).
Despite significant advances in liver resection, liver
transplantation, radiotherapy, chemotherapy and immunotherapy in
recent years, the prognosis for patients with HCC remains poor due
to high recurrence and metastasis rates, as well as resistance to
chemotherapy and targeted therapy (5,6).
Therefore, it is urgent to clarify the mechanisms of HCC metastasis
and identify new therapeutic targets.
Insulin-like growth factor binding proteins (IGFBPs)
are a group of secreted proteins that can bind to insulin-like
growth factors (IGFs). They mainly include IGFBP1-7, which can
affect the binding strength between IGF and IGF receptor (IGFR) and
subsequently regulate the activation of downstream signaling
pathways, thus controlling the growth and proliferation of target
cells (7). According to the
significant differences in expression levels of all IGFBPs between
tumors and normal tissues, combining survival analysis, IGFBP4 has
drawn our attention. Previous studies have showed that IGFBP4 is
involved in the development of multiple diseases such as lung
cancer (8), breast cancer
(9), bladder epithelial cancer
(10), bone metabolism (11) and endocrine metabolic diseases
(12). However, its role and
specific mechanisms in HCC have not been clearly defined.
In the present study, it was identified that IGFBP4
is downregulated in liver cancer. The expression of IGFBP4 was
associated with a favorable prognosis in patients with liver
cancer. Functionally, IGFBP4 inhibited cell migration and invasion
in vitro and suppressed tumor metastasis in vivo.
Mechanistically, the expression of IGFBP4 is regulated by MYBBP1A,
which targeted the methylation of the IGFBP4 promoter region to
suppress its transcription, thereby affecting its expression.
IGFBP4 could inhibit NOTCH pathway activation, furthermore inhibit
epithelial-mesenchymal transition (EMT) and impact tumor
metastasis. Additionally, MYBBP1A and IGFBP4 expression could be
combined to predict poor prognosis in HCC. In summary, the present
study expands the knowledge of the regulation of IGFBP4 mediated by
MYBBP1A, reveals a new mechanism in HCC tumorigenesis and
demonstrates its potential for precise diagnosis.
Materials and methods
Patients and specimens
The present study adhered to the ethical guidelines
outlined in the Declaration of Helsinki (1964), and protocols were
approved (approval no. 2020-IIT-834) by the Ethics Committee of The
First Affiliated Hospital of Zhejiang University (Hangzhou, China).
All patients included in the study provided written informed
consent. Surgical specimens of liver cancer tissue samples and
adjacent non-tumor tissue samples of 25 patients with HCC with or
without metastasis were retrospectively collected from The First
Affiliated Hospital of Zhejiang University, between January 2015
and December 2019. The patients were randomly selected based on
specific clinical and pathological criteria, including confirmed
diagnosis of HCC, stages I-III according to the Barcelona Clinic
Liver Cancer (BCLC) classification, and no previous treatment. The
age of the patients (22 men and 3 women) ranged from 37-80 years,
with a median age of 60 years. Demographic data were recorded but
did not influence selection criteria. The clinicopathological
characteristics of the 25 patients are listed in Table SI. Following surgical resection,
tissue samples were immediately snap-frozen in liquid nitrogen and
stored at −80°C to preserve RNA integrity. All samples were
processed using standardized protocols for RNA extraction to ensure
high-quality analysis.
Bioinformatics analysis
The transcript expression data and survival
information of the Liver HCC (LIHC) from The Cancer Genome Atlas
(TCGA) and the gene expression data in Genotype-Tissue Expression
(GTEx; https://gtexportal.org/home/) were
used for the present study. The Kaplan-Meier plotter website
(https://kmplot.com/analysis/), the GEPIA
website (http://gepia.cancer-pku.cn), and the
UALCAN website (https://ualcan.path.uab.edu/) were used to analyze the
clinical correlations with MYBBP1A and IGFBP4.The Methprimer
database (http://www.urogene.org/methprimer/) was used to
predict the methylation sites of IGFBP4 and it was found that there
were CpG islands in the promoter region of IGFBP4. The SMART
website (http://www.bioinfo-zs.com/smartapp/) was used to
analyze the correlations of expression and methylation of IGFBP4.
Genes were ranked based on their correlation with IGFBP4 expression
levels. Differentially expressed genes (DEGs) were identified using
standard thresholds of P<0.05 and |log2FC|>1. These DEGs were
then subjected to differential expression analysis and gene set
enrichment analysis (GSEA) to explore the potential pathways and
biological processes associated with IGFBP4.
Cell lines and cell culture
The human liver cancer cell lines HCCLM3 (cat. no.
TCHu270), Huh7 (cat. no. TCHu182) as well as HepG2 (cat. no.
TCHu72) used in the present study were obtained from the Cell Bank
of the Chinese Academy of Sciences Typical Culture Collection
Committee. Prior to use, all cell lines were validated by short
tandem repeat (STR). Cells were maintained and stored according to
the provider's instructions. The culture medium contained 10% fetal
bovine serum (FBS; BioInd; http://www.bioind.org/), 100 IU/ml penicillin and 100
IU/ml streptomycin. Cells were cultured in a humidified incubator
at 37°C with 5% CO2.
Reagents and drugs treatment
Referring to previous studies (13), different concentrations (0, 5, 10
and 40 μmol/l) of the methylation inhibitor Decitabine
(5-Aza-2′-deoxycytidine) (cat. no. HY-A0004; MedChemExpress)
dissolved in DMSO were used to inhibit the methylation process.
After 48 h of treatment, the cells were collected for western
blotting (WB) and reverse transcription-quantitative PCR
(RT-qPCR).
Lentivirus construction and cell
transfection
MYBBP1A-RNAi-lentivirus, IGFBP4-RNAi-lentivirus and
IGFBP4-overexpression (oe) lentivirus were purchased from Shanghai
GeneChem Co., Ltd. pFU-GW-016-hU6-MCS-CBh-gcGFP-IRES-puromycin was
used as the vector for shMYBBP1A and shMYBBP1A control. MYBBP1A
shRNA sequences were targeted: 5′-GCTGGTGAATGTGCTGAAGATGGCC-3′;
pFU-GW-014-hU6-MCS-Ubc-mCherry-IRES -neomycin was used as the
vector for shIGFBP4 and shIGFBP4 control, IGFBP4 shRNA sequences
were targeted: 5′-CTGCAGAAGCACTTCGCCAAA-3′; in addition, a random
sequence control shRNA, 5′-TTCTCCGAACGTGTCACGT-3′ was used as a
negative control (NC).
pGC-FU-CMVenhancer-3FLAG-EF1-ZsGreen1-T2A-puromycin was used as the
vector for oeIGFBP4 and its control was vector only control.
All lentivirus constructs were generated using
second-generation lentiviral vector packaging system provided by
Shanghai GeneChem Co., Ltd. Packaging was performed using the
helper plasmids Helper1.0 and Helper2.0 (Shanghai GeneChem Co.,
Ltd.). 293T cells (cat. no. GNHu17; Cell Bank of the Chinese
Academy of Sciences Typical Culture Collection Committee) were
transfected with the lentiviral constructs (target vector plasmid
20 μg, Helper1.0 vector plasmid 15 μg, Helper2.0
vector plasmid 10 μg) using Lipofectamine 2000 (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Cells were incubated at 37°C with 5% CO2 for 48 h
post-transfection. After 48 h, the viral supernatant was collected,
filtered through a 0.22-μm filter, and concentrated by
ultracentrifugation at 73,000 × g for 2 h at 4°C.
The viral particles were harvested and used to
transduce the target cells Huh7 and HCCLM3. The target cells were
infected at a multiplicity of infection of 10, and Polybrene
(Shanghai GeneChem Co., Ltd.) was added at a final concentration of
4 μg/ml to improve the infection efficiency. After
incubation at 37°C for 8-12 h, the complete medium was replaced. A
total of 48 h after infection, cells were selected using puromycin
or neomycin (3-4 μg/ml) (Sangon Biotech Co., Ltd.) for 72 h
and transfection efficiency was confirmed by RT-qPCR and WB.
RNA extraction and RT-Qpcr
Cell and tissue total RNA were extracted using an
RNA-Quick Purification Kit (Esunbio; http://www.esunbio.com/). Total RNA was reverse
transcribed into cDNAwith HiScript II Q Select RT SuperMix for qPCR
(cat. no. R233-01; Vazyme Biotech Co., Ltd.) and HiScript III
All-in-one RT SuperMix Perfect for qPCR (cat. no. R333-01; Vazyme
Biotech Co., Ltd.) according to the manufacturer's protocol.
RT-qPCR was performed with ChamQ SYBR Green Master Mix (Vazyme
Biotech Co., Ltd.) on QuantStudio 5 Real-Time PCR System (Thermo
Fisher Scientific, Inc.). After pre-denaturation at −95°C for 5min,
the reaction was carried out at 95°C for 10 sec and at 60°C for 30
sec, with 40 cycles. The expression of target genes was calculated
using the 2−ΔΔCq method and normalized to the expression
of GAPDH (14). The primers of
all genes were ordered from TsingKe Biological Technology and
listed in Table SII.
WB assays
Whole cells or tissues were lysed with RIPA buffer
(Wuhan Servicebio Technology Co., Ltd.) supplemented with protease
and phosphorylation inhibitor cocktail (Selleck Chemicals) and
quantified by BCA Protein Assay Kit (Thermo Fisher Scientific,
Inc.). Then a total of 30 μg of protein loaded per lane were
electrophoresed with 4-20% SDS-PAGE (GenScript) and transferred to
0.22-μm PVDF membranes (MilliporeSigma). Membranes were
blocked with 5% BSA (Beijing Solarbio Science & Technology Co.,
Ltd.) or 25% non-fat milk for 1 h at room temperature, and
incubated with primary antibodies over 8 h or overnight in 4°C.
After being washed with TBST (with 0.1% Tween) and incubated for 1
h with secondary antibodies at room temperature, the protein bands
were exposed to an ECL detection reagent (Fudebio; http://www.fudebio.com/) in the chemiluminescence
system (Bio-Rad Laboratories, Inc.). GAPDH was marked as an
internal control. Quantification of WB bands was performed using
ImageJ software (version 1.54k; National Institutes of Health). The
'Analyze Gel' tool in ImageJ was used to measure the grayscale
intensity of protein bands. Signal intensity for each band was
background-corrected and normalized to the internal control protein
for relative quantification. All information regarding antibodies
used in WB is included in Table
SIII.
Immunohistochemistry (IHC)
The metastatic tumors and liver tissues from mice
and clinical patients were fixed with 4% paraformaldehyde at room
temperature for 48 h and embedded in paraffin. The tissue sections
were cut to a thickness of 3 μm and followed by H&E and
IHC staining (incubation with the E-cadherin, N-cadherin and
Vimentin antibodies at 4°C for 8 h) respectively. The tissue
sections were stained with H&E (Wuhan Servicebio Technology
Co., Ltd.) at room temperature for 5 and 7 min, respectively. The
IHC assay was performed as previously described (15). Product information and dilution
ratios of primary antibodies is provided in Table SIII.
Immunofluorescence (IF)
Cells (10×104 cells/well) were seeded
onto glass coverslips in a 6-well plate and allowed to adhere
overnight at 37°C with 5% CO2. The cells were washed
twice with PBS and fixed with 4% paraformaldehyde at room
temperature for 20 min. After two washes with PBS, the cells were
stained with 5 μg/ml iFluor™ 488 phalloidin (cat. no.
40736ES75; Shanghai Yeasen Biotechnology Co., Ltd.) at room
temperature for 60 min, followed by two PBS washes. The cells were
incubated with 1 μg/ml DAPI (cat. no. 40728ES50; Shanghai
Yeasen Biotechnology Co., Ltd.) at room temperature for 10 min.
After two additional PBS washes, fluorescence images were captured
under a fluorescence microscope.
Transwell assays
Cells (5×104 cells/well) suspended in
serum-free medium were seeded into each upper Transwell chamber (24
well 8.0-μm pore size; Falcon; Corning Life Sciences), while
medium containing 10% FBS was added to the lower chamber. After
incubation for 72 h at 37°C with 5% CO2, the migratory
or invasive cells in the lower chambers were fixed at room
temperature for 10 min with 4% paraformaldehyde and stained at room
temperature for 15 min with 0.1% crystal violet. Migratory cells
were plotted as the average number of cells per field of view using
a light microscope (Olympus Corporation) from 3 independent
experiments.
Gap closure assays
Cells (5×104 cells/well) were seeded into
both sides of Culture-Inserts® (Ibidi GmbH) to make a
500-μm gap. After incubation for 48 h in Huh7 and 72 h in
HCCLM3 cells, inserts were removed to allow cell migration for the
indicated period of time in serum-free medium. Migration distances
were plotted as the average distance of two sides of cells views
using a light microscope (Olympus Corporation) from 3 independent
experiments.
Colony formation assay
Cells (1×103 cells/well) were seeded into
6 well plates and incubated for 2-3 weeks at 37°C with 5%
CO2. Then, the colonies on the plates were fixed at room
temperature for 10 min with 4% polyoxymethylene and stained at room
temperature for 15 min with 0.1% crystal violet. Viable colonies
(>50 cells) were observed and counted manually and are pictured.
The numbers of colonies are depicted as bar graphs. All experiments
were repeated three times under the same conditions and
methods.
DNA methylation analysis by bisulfite
sequencing
The genomic DNA of cells was extracted using the
standard phenol-chloroform technique followed by proteinase K
treatment to prevent protein contamination. Bisulfite conversion
was performed using an EpiTect Fast DNA Bisulfite Kit (cat. no.
59824; Qiagen China Co., Ltd.) according to the manufacturer's
protocol. The bisulfite-treated genomic DNA was subjected to PCR
for the amplification of the CpG islands of IGFBP4 promoter using
Hot Start Takara Taq DNA Polymerase (Takara Biotechnology Co.,
Ltd.). Amplified PCR products were cloned into the pGEMT-easy
vector using DNA Ligation Kit (cat. no. 6022; Takara Biotechnology
Co., Ltd.). A total of 10 insert-positive clones were isolated by
the QIAprep Spin Miniprep kit (cat. no. 27104; Qiagen China Co.,
Ltd.). RNA sequencing and analysis of the samples were performed by
Hangzhou Cosmos Wisdom Biotech Co. The PCR primers used for
bisulfite sequencing of CpG islands of the promoter region of
IGFBP4 gene are presented as follows: forward,
5′-TTYGGTAGAAAAGGATTTTTAGATG-3′ and reverse,
5′-CACRAAACAAAAAAACAACATAACC-3′.
Tumor xenograft model
A splenic venous liver metastasis tumor experiment
was performed with male immunodeficient nude mice aged 5 weeks and
weighing 20-25 g. A total of 5 mice in each group were purchased
from the Zhejiang Academy of Medical Sciences and raised in the SPF
barrier of the Experimental Animal Center of the First Affiliated
Hospital of Zhejiang University School of Medicine with 12/12-h
light-dark cycle, 21-23°C and a relative humidity of 60-65%, with
ad libitum access to food and water, with ad libitum
supply of food and water. Experiments were performed in strict
accordance with the NIH Guide for the Care and Use of Laboratory
Animals. A total of 10 BALB/c nude mice were randomly allocated
into two groups. After careful sterilization, 75 μl PBS
containing 5×106 Huh7 cells of IGFBP4
stabilized-knockdown or control cells were injected in the spleen
of mice. The detailed process was conducted as previously described
(16). Experimental nude mice
were anesthetized by intraperitoneal injection of pentobarbital
sodium according to their body weight (50 mg/kg). After anesthesia,
tumor cells were injected under the splenic capsule for
compression, hemostasis and suture. After injection, the mental
state, abdominal shape and weight of the mice were observed every
week to evaluate the tumor progression. In our laboratory animal
Centre, experienced animal care staff monitored the animals daily
and notified the laboratory staff if anything unusual happened. If
the mice showed obvious wasting, lethargy and significant abdominal
distension or where the ascites burden exceeded 10% of the body
weight (17), the humane
experimental endpoint was reached. Their whole liver was removed
when the mice were euthanized and immediately processed for
histological evaluation. For a total of 60 days after injection,
all mice were administered peritoneal injections of pentobarbital
(150 mg/kg) and euthanized by cervical dislocation. Cardiac arrest
is then followed by a pulsar examination to confirm death. The
liver metastasis model was constructed for previous research
proposes (18-23). The animal studies were approved
(approval no. 2020-IIT-rapid-1132) by the Clinical Research Ethics
Committee of the First Affiliated Hospital, School of Medicine,
Zhejiang University (Hangzhou, China).
Statistical analysis
All experiments were repeated three times under the
same conditions and methods to ensure the repeatability of the
experiment. All data are expressed as the mean ± standard deviation
(SD). Significant differences between the two unpaired groups were
tested by the unpaired Student's t-test, and paired groups were
tested using paired t-test. The one-way ANOVA test followed by
Tukey's post hoc test was used to analyze comparisons of
significant differences between three or more groups. Kaplan-Meier
and log-rank tests were performed to determine survival. The
correlation analysis was evaluated using Pearson's test. In all
statistical analyses, *P<0.05 was considered to indicate a
statistically significant difference. Statistical analysis of data
was performed using GraphPad Prism 8.0.2 software (Dotmatics).
Results
Expression of IGFBP4 is reduced in HCC
tissues and negatively correlated with poor prognosis in patients
with HCC
To determine which IGFBP family member may be the
key to regulating HCC progression, the IGFBPs family (IGFBP1-7)
gene expression was first analyzed using TCGA-LIHC datasets. The
results showed that the IGFBP1 (P<0.001), IGFBP3 (P<0.001),
IGFBP4 (P<0.001) and IGFBP5 (P=0.0042) have significantly
downregulated expression in liver cancer (Fig. 1A). Kaplan-Meier plotter website
was used to predict the single gene expression associated with HCC
prognosis, and low expression of IGFBP4 (P=2.1×10−6) and
IGFBP7 (P=2.2×10−5) was found; rather, other IGFBP
members were significantly correlated with poor prognosis of
patients with LIHC (Fig. 1B).
Thus, further studies were performed to investigate the role of
IGFBP4 in HCC progression. The expression of IGFBP4 was found to be
lower in the HCC population compared with the normal population,
especially in the Asian population (Fig. 1C). In addition, the expression of
IGFBP4 was further reduced in patients with HCC with poor liver
tumor grade and lymph node metastasis (Fig. 1D and E). The downregulated
expression of IGFBP4 in the clinical tumor specimens of patients
with HCC was further validated using RT-qPCR (P<0.0001) and WB
(Fig. 1F and G). IHC staining
also demonstrated that IGFBP4 expression was lower in HCC tissues
compared with that in the matched adjacent normal tissues (Fig. 1H). These data indicated that
IGFBP4 was downregulated in HCC and its low expression predicts
poor prognosis in patients with HCC.
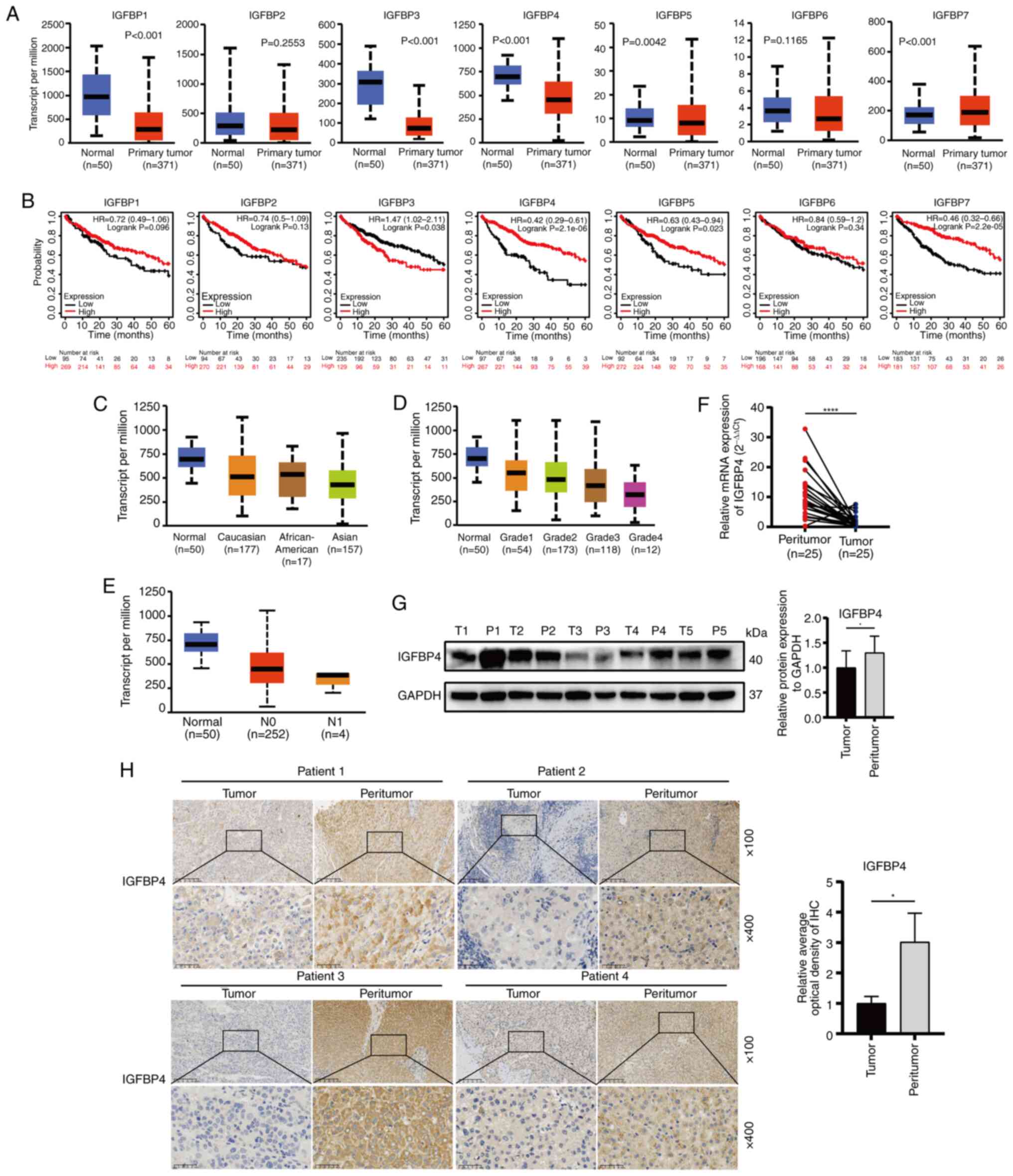 | Figure 1Expression of IGFBP4 is downregulated
in HCC. (A) Expression of IGFBP1 to IGFBP7 in normal and primary
tumor patients based on LIHC in the TCGA and Genotype-Tissue
Expression databases. (B) Kaplan-Meier survival analyzed patient
survival in different expression groups of IGFBP1 to IGFBP7 in
TCGA-LIHC [log-rank (Mantel-Cox]. (C) Expression of IGFBP4 in
patients from various ethnicities based on LIHC in the TCGA
databases. (D and E) Expression of IGFBP4 based on different tumor
grades and nodal metastasis of LIHC in TCGA databases. (F) IGFBP4
mRNA expression in paired HCC tissues (n=25) and adjacent non-tumor
tissues (n=25) evaluated by reverse transcription-quantitative PCR
(****P<0.0001, paired t-test). (G) Representative
IGFBP4 protein expression results in paired HCC tissues and
adjacent non-tumor tissues evaluated by western blotting
(*P<0.05, paired t-test). (H) Representative
immunohistochemical images of IGFBP4 staining in liver tumor or
adjacent tissues (scale bar, 100 μm; magnification, ×100,
and ×400; *P<0.05, paired t-test). IGFBP,
insulin-like growth factor binding protein; HCC, hepatocellular
carcinoma; LIHC, liver hepatocellular carcinoma; TCGA, The Cancer
Genome Atlas. |
IGFBP4 inhibits the metastatic ability of
liver cancer cells through repressing EMT
To explore the biological function of IGFBP4 in
liver cancer, its expression levels were first assessed in various
liver cancer cell lines. The results showed high IGFBP4 expression
in HCCLM3 cells, moderate expression in MHCC97H and HepG2 cells,
and low expression in Huh7 cells (Fig. 2A and B). HCCLM3 and Huh7 were
selected for further study. IGFBP4 overexpression lentivirus was
applied in Huh7 and HCCLM3 cells, and the transfection efficiency
was proven at both the mRNA and protein levels (Fig. 2C and D). The Transwell assays
revealed that IGFBP4 overexpression inhibited the migration ability
in both two cell lines in vitro (Fig. 2E). Moreover, gap closure assays
also showed that IGFBP4-overexpressing Huh7 and HCCLM3 cell lines
decreased migration ability (Fig.
2F). In addition, overexpression of IGFBP4 could also reduce
proliferation ability of liver cancer cells (Fig. S1). Phalloidin staining results
showed that NC liver cancer cells appeared more rounded and had
fewer pseudopodia compared with those with IGFBP4 overexpression
(Fig. 2G). Cancer metastasis is
closely related to the EMT of tumor cells. WB was used to verify
EMT-associated protein expression. IGFBP4 ectopic overexpression
increased the expression of E-cadherin while decreasing the
expression of N-cadherin and Vimentin in HCCLM3 and Huh7 cells
(Fig. 2H). By contrast, IGFBP4
silencing increased the expression of N-cadherin and Vimentin,
whereas it decreased the expression of E-cadherin (Fig. 2I). All these results suggested
that IGFBP4 can alter the cytoskeleton and repress EMT in liver
cancer cells.
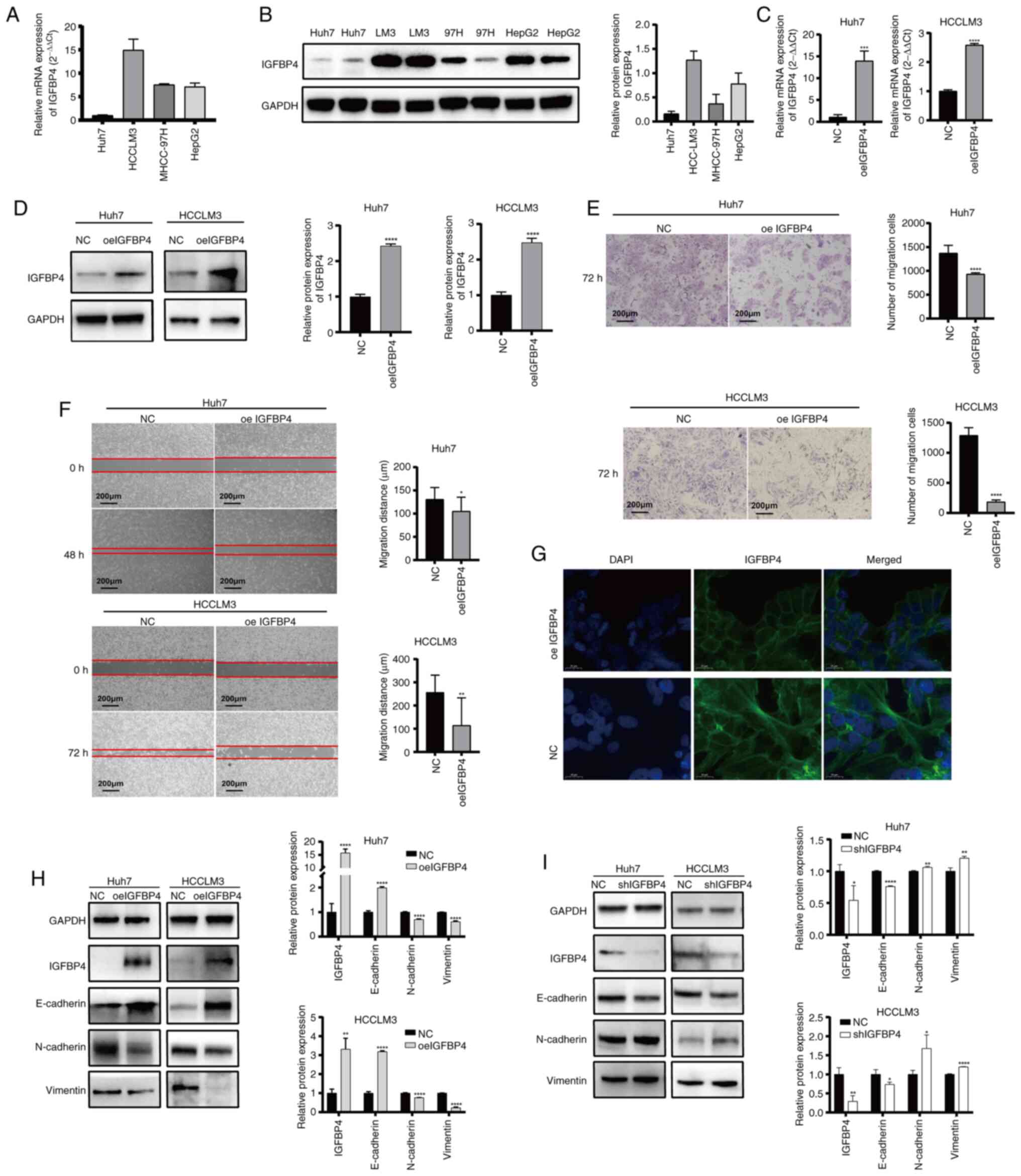 | Figure 2IGFBP4 inhibits the metastatic
ability of liver cancer cells through repressing
epithelial-mesenchymal transformation in vitro. (A and B)
Basal expression of IGFBP4 in mRNA level and protein level in Huh7,
HCCLM3, MHCC-97H and HepG2 cell lines by RT-qPCR and WB. (C) The
mRNA levels of IGFBP4 in Huh7 and HCCLM3 cell lines after
transfection with IGFBP4-oe lentivirus based on RT-qPCR
(***P<0.001 and ****P<0.0001, Student's
t-test). (D) The protein expression of IGFBP4 in Huh7 and HCCLM3
cell lines after transfection with IGFBP4-oe lentivirus based on WB
(****P<0.0001, Student's t-test). (E and F) The
migratory abilities of oeIGFBP4 Huh7 and HCCLM3 cells based on (E)
Transwell migration assays and (F) gap closure assays, respectively
(original magnification, ×100 and ×400; *P<0.05,
**P<0.01 and ****P<0.0001, Student's
t-test). (G) The cytoskeletal morphologic effects of overexpressing
IGFBP4 in liver cancer cells were detected by phalloidin staining
(phalloidin, green; DAPI, blue; scale bar, 50 μm). (H) The
protein levels of E-cadherin, N-cadherin, Vimentin and IGFBP4 in NC
and oeIGFBP4 Huh7 and HCCLM3 cells based on WB
(**P<0.01, ****P<0.0001, Student's
t-test). (I) The protein levels of E-cadherin, N-cadherin, Vimentin
and IGFBP4 in shIGFBP4 and NC groups in Huh7 and HCCLM3 cells based
on WB (*P<0.05, **P<0.01,
****P<0.0001, Student's t-test). IGFBP 4,
insulin-like growth factor binding protein 4; RT-qPCR, reverse
transcription quantitative PCR; WB, western blotting; oe-,
overexpression; sh-, short hairpin; NC, negative control. |
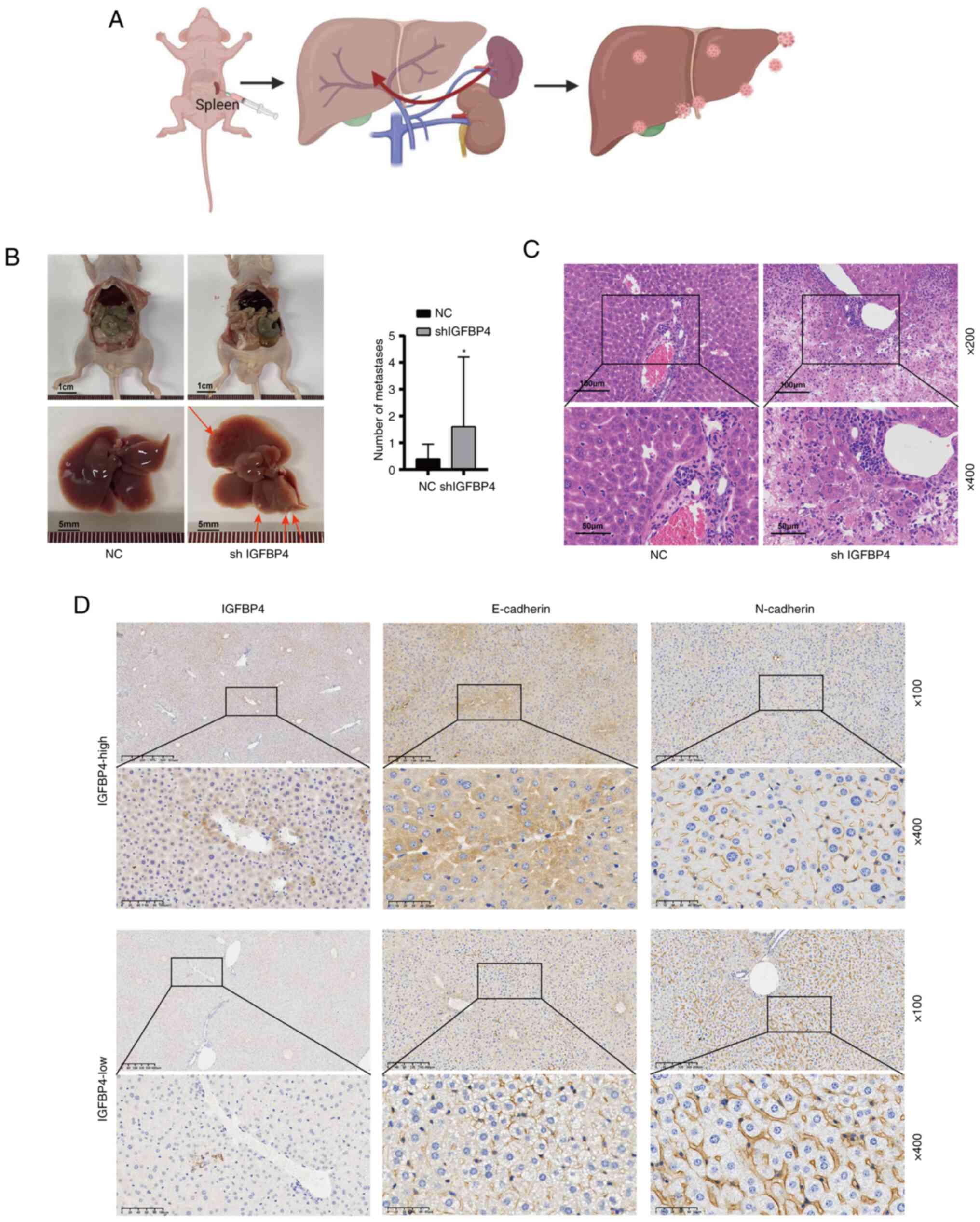
The silencing of IGFBP4 increased tumor
metastasis in vivo
To further investigate whether IGFBP4 promotes tumor
metastasis in vivo, a splenic venous liver metastasis model
was constructed by injecting lentivirus-mediated shRNA targeting
IGFBP4 transduced cells and control cells under the splenic
envelope of nude mice to mimic the liver metastasis (Fig. 3A). More liver metastatic nodules
were found in IGFBP4-knockdown group (Fig. 3B). Interestingly, intestinal
metastasis was accidentally found in one IGFBP4 knockdown mice
(Fig. S2). H&E staining
showed the microstructure of metastatic lesions (Fig. 3C). Moreover, among normal liver
tissues, it was also found that low expression of IGFBP4 in the
liver was accompanied by low E-cadherin expression and high
N-cadherin expression, and vice versa (Fig. 3D). These results demonstrated that
IGFBP4 silencing promotes cancer cell metastasis in
vivo.
IGFBP4 inhibits the activation of the
NOTCH signaling pathway
To explore the mechanism of IGFBP4 in regulating
liver cancer malignancy, differentially expressed genes (DEGs)
enrichment analysis (Fig. 4A) and
GSEA analysis were performed, and the NOTCH pathway was found
enriched (Fig. 4B). NOTCH
signaling is a strong modulator in regulating the EMT process
(24-26), thus it was hypothesized that
IGFBP4 might inhibit EMT through the NOTCH pathway. RT-qPCR was
used to verify the mRNA expression level of core genes of the NOTCH
pathway, which are involved in the present GSEA analysis. It was
found that mRNA levels of NOTCH receptor 1 (NOTCH1), hes family
bHLH transcription factor 1 (HES1) and neuralized E3 ubiquitin
protein ligase 1 (NEURL) decreased after IGFBP4 overexpression, and
increased after IGFBP4 silencing (Fig. 4C and D). WB results showed that
overexpression of IGFBP4 reduced the expression of NOTCH1 and HES1
at the protein level, while knockdown of IGFBP4 had the opposite
effect (Fig. 4E and F). Next, to
verify the relation of NOTCH pathway activation and IGFBP4
expression in the liver, IHC was performed. IHC results revealed
that NOTCH1 expression was reduced in the liver tissue with high
levels of IGFBP4, and vice versa (Fig. 4G). The Smad and β-catenin pathways
play crucial roles in EMT and tumor metastasis, which is well
established. WB assays were performed to assess the expression
levels of β-catenin, Wnt3a, Smad2/3 and phosphorylated Smad2/3.
However, no significant alterations were observed in the expression
of these proteins following IGFBP4 overexpression (Fig. S3). Additionally, the expression
changes of the transcription factor YAP1 in the NOTCH signaling
pathway were assessed. It was found that the protein expression
level of YAP1 was significantly reduced in liver cancer cells with
IGFBP4 overexpression, and the level of YAP1 was positively
correlated with the activation of the NOTCH pathway (Fig. S4). These results showed that
IGFBP4 overexpression could inhibit the NOTCH signaling pathway
activation in liver cancer cells.
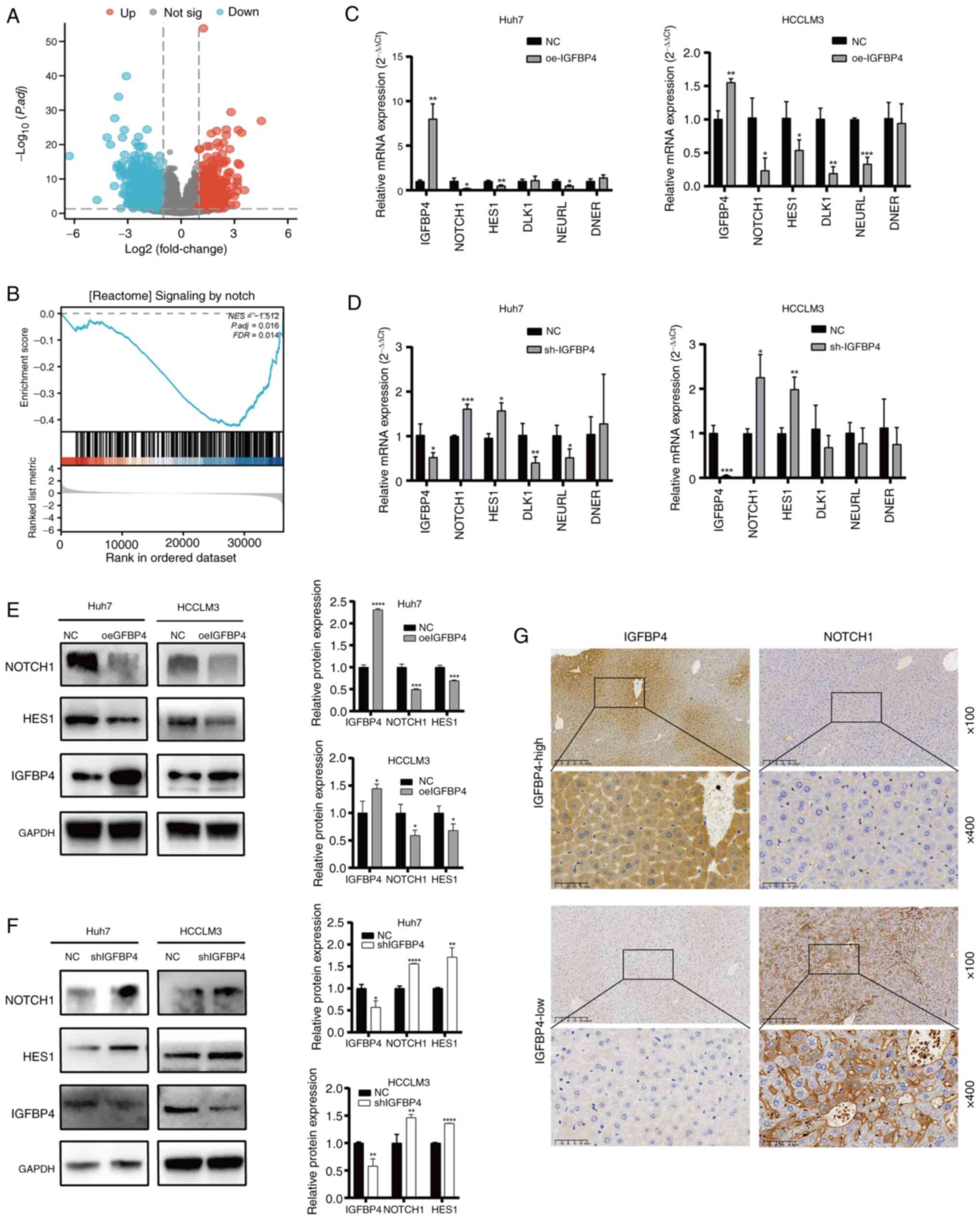 | Figure 4Overexpression of IGFBP4
downregulates NOTCH signaling pathway in liver cancer. (A) Volcano
plot of the differentially expressed genes in patients with
hepatocellular carcinoma included in The Cancer Genome Atlas
database. Red dots represent genes that were significantly
upregulated, blue dots represent genes that were significantly
downregulated, grey dots represent genes that were not
differentially expressed and the horizontal line represents a
P-value of 0.05. (B) Gene set enrichment analysis found the
differentially expressed gene of IGFBP4 was enriched in the NOTCH
signaling pathway (p adj.=0.016, FDR=0.014). (C) The mRNA levels of
the NOTCH pathway key genes NOTCH1, HES1 and related genes DLK1,
NEURL and DNER in negative control and oeIGFBP4 groups of Huh7 and
HCCLM3 cells based on RT-qPCR (*P<0.05,
**P<0.01 and ***P<0.001, Student's
t-test). (D) The mRNA levels of NOTCH1, HES1, DLK1, NEURL and DNER
in NC and shIGFBP4 groups of Huh7 and HCCLM3 based on RT-qPCR
(*P<0.05, **P<0.01 and
***P<0.001, Student's t-test). (E) The protein
expression of NOTCH1 and HES1 in oeIGFBP4 group and NC group in
Huh7 and HCCLM3 cell lines based on WB (*P<0.05,
***P<0.001, ****P<0.0001, Student's
t-test). (F) The protein expression of NOTCH1 and HES1 in NC and
shIGFBP4 group in Huh7 and HCCLM3 cell lines based on WB
(*P<0.05, **P<0.01,
****P<0.0001, Student's t-test). (G) Representative
images of immunohistochemical staining of NOTCH1 in the
high-expressing IGFBP4 and low-expressing patients with liver
tissues (original magnification, ×100 and ×400). IGFBP 4,
insulin-like growth factor binding protein 4; HES, hes family bHLH
transcription factor 1; NEURL, neuralized E3 ubiquitin protein
ligase 1; oe-, overexpression; RT-qPCR, reverse transcription
quantitative PCR; NC, negative control; WB, western blotting; sh-,
short hairpin. |
IGFBP4 is negatively regulated by MYBBP1A
via CpG island methylation-mediated degradation
Our previous study had shown that IGFBP5, another
important IGFBP family member, could be regulated by MYBBP1A and
inhibit the metastasis ability of liver cancer cells (13). Thus, it was hypothesized that
IGFBP4 may be regulated by MYBBP1A as well. In order to investigate
the relation between MYBBP1A and IGFBP4, though analyzing the data
from the TCGA database and GTEx database, it was found that the
IGFBP4 gene expression varied obviously when the expression of
MYBBP1A was changed (Fig. 5A). At
the same time, a correlation analysis between two genes was
conducted and it was identified that there was no significant
correlation between the expression of MYBBP1A and IGFBP4 in the
liver tissue of the normal liver tissue (P=0.5, R=0.098) (Fig. 5B). However, a negative correlation
of MYBBP1A and IGFBP4 expression was observed in liver tumor
tissues of patients with HCC (P=9.8×10−6, R=-0.23)
(Fig. 5C). The experimental
results demonstrated that MYBBP1A knockdown resulted in a
significant increase in the mRNA and protein expression level of
IGFBP4 (Fig. 5D and E). By
contrast, after overexpressing MYBBP1A, IGFBP4 decreased in both
mRNA and protein levels (Fig. S5A
and B). Our previous study (13) reported that MYBBP1A inhibited
IGFBP5 transcription by promoting the high methylation level of CpG
island at IGFBP5 CDS sites. Then, the Methprimer database was used
to predict the methylation sites of IGFBP4 and found that there
were CpG islands in the promoter region of IGFBP4 (Fig. 5F). The SMART database was used to
analyze the correlation between IGFBP4 methylation and expression
and it was found that the expression of IGFBP4 was significantly
negatively associated with the methylation in transcription level
(Fig. 5G). In addition, a higher
level of IGFBP4 promoter methylation was observed in patients with
higher tumor grade and stage (Fig. 5H
and I). Bisulfite sequencing PCR (BSP) technology was utilized
to compare the methylation levels of CpG islands in the IGFBP4
promoter region between the MYBBP1A knocked-down group and the NC
group (Fig. 5J). Overall
methylation in the IGFBP4 promoter region was decreased in the
MYBBP1A knockdown group compared with the control group. More
importantly, mRNA levels of IGFBP4 in HCC cells were significantly
increased after treatment with DNA methyltransferase inhibitor
Decitabine (5-Aza-2′-deoxycytidine) (0, 5, 10 and 40 μmol/l)
(Fig. 5K and L). The inhibitory
efficiency of 5Aza was verified by detecting the expression of
DNMT1 at different concentrations of 5Aza. These results all
confirmed that MYBBP1A affects the expression level of IGFBP4 by
promoting CpG island methylation of the IGFBP4 promoter region.
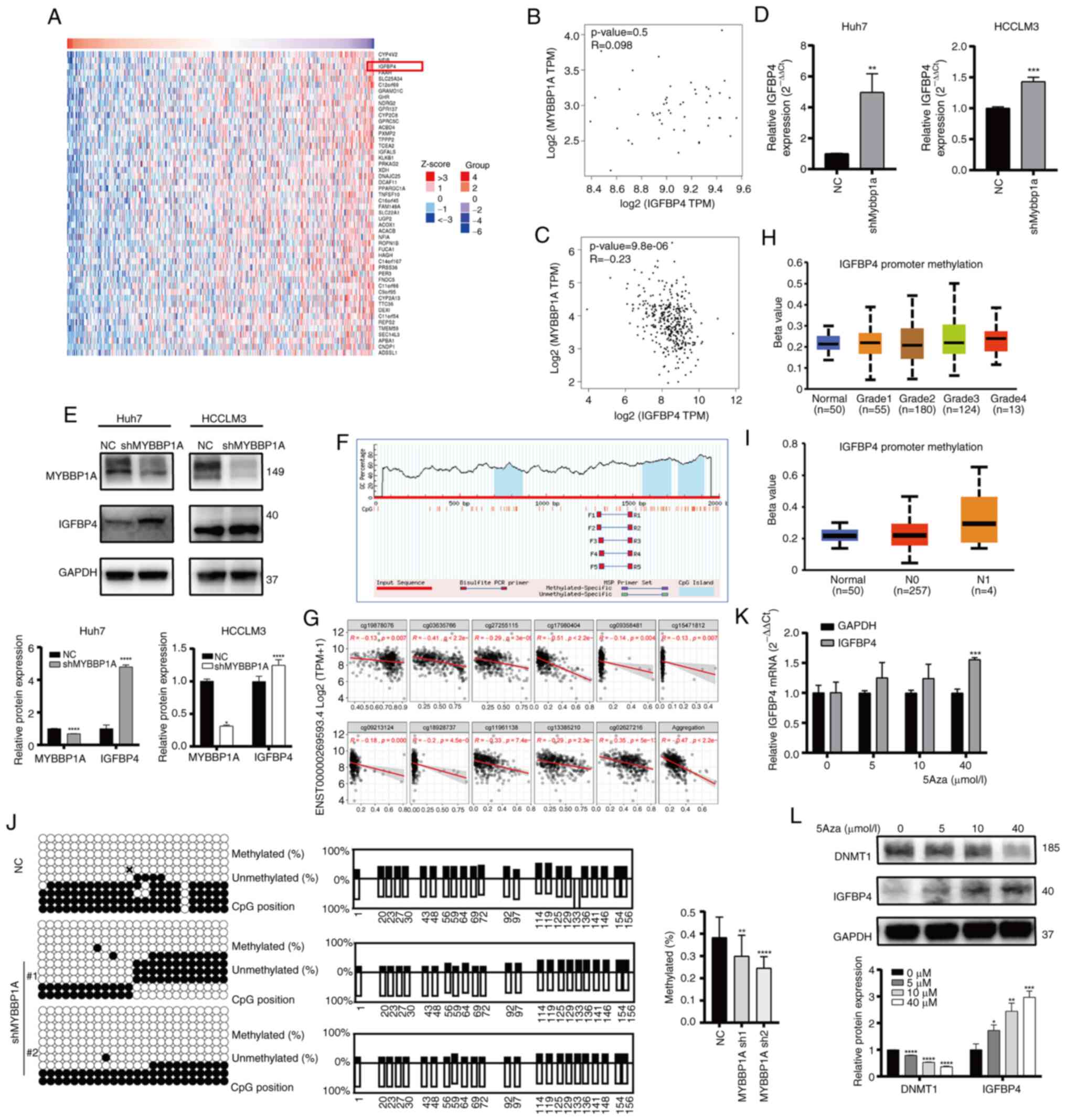 | Figure 5IGFBP4 is negatively regulated by
MYBBP1A via CpG island methylation mediated degradation. (A) Heat
map showed the top 50 genes which significantly negatively
correlated with MYBBP1A from TCGA database, IGFBP4 ranked third. (B
and C) TCGA and The Genotype-Tissue Expression databases analysis
showed that there was no significant correlation between MYBBP1A
and IGFBP4 expression in normal liver tissues (P=0.05, R=0.098),
while there was a negative correlation between MYBBP1A and IGFBP4
expression in patients with liver cancer (P=9.8×10−6,
R=-0.23). (D) The expression level of IGFBP4 mRNA in Huh7 and
HCCLM3 cell lines after MYBBP1A knockdown was detected by RT-qPCR
(**P<0.01 and ***P<0.001, Student's
t-test). (E) WB of IGFBP4 protein expression in Huh7 and HCCLM3
cell lines after knockdown of MYBBP1A (*P<0.05,
****P<0.0001, Student's t-test). (F) Using the
Methprimer website, three CpG islands were found in the IGFBP4
promoter region. (G) Transcript-level correlation showing that the
expression of IGFBP4 is negatively associated with the methylation
of cg19878076, cg03635766, cg27255115, cg17980404, cg09358481,
cg15471812, cg09213124, cg18928737, cg11961138, cg13385210 and
cg02627216 using SMART website analysis (M value, Spearman). (H and
I) IGFBP4 promoter methylation level in the liver hepatocellular
carcinoma database of TCGA under different tumor grades and lymph
node metastases. (J) The methylation level of IGFBP4 promoter
region was detected by bisulfite sequencing PCR technique. Compared
with the MYBBP1A knockdown group, the IGFBP4 promoter region was
more methylated in the control group (**P<0.01 and
****P<0.0001, Student's t-test). (K) The expression
level of IGFBP4 mRNA after stimulation with methylase inhibitor
5-Aza at different concentrations (0, 10, 20 and 40 μmol/l)
after 48 h was detected by RT-qPCR (***P<0.001,
Student's t-test). (L) WB was performed to detect the expression
levels of methylase DNMT1 and IGFBP4 protein after stimulation with
different concentrations of methylase inhibitor 5Aza (0, 10, 20, 40
μmol/l) after 48 h (*P<0.05,
**P<0.01, ***P<0.001,
****P<0.0001, Student's t-test). IGFBP 4,
insulin-like growth factor binding protein 4; TCGA, The Cancer
Genome Atlas; RT-qPCR, reverse transcription-quantitative PCR; WB,
western blotting. |
Knocking down IGFBP4 restores the
repressed metastasis ability of liver cancer caused by MYBBP1A
inhibition
To further confirm our theory, lentivirus-mediated
shRNA targeting MYBBP1A was used to construct MYBBP1A-knockdown
Huh7 and HCCLM3 cells. Transwell and gap closure assay results
demonstrated that knocking down MYBBP1A decreased the migratory
activities of liver cancer cells (Fig. 6A and B). Then, the
lentivirus-mediated shMYBBP1A and shIGFBP4 were used to
co-transfect Huh7 and HCCLM3 cells. The results showed that
knocking down MYBBP1A suppressed migration ability while knocking
down IGFBP4 restored the migration abilities of liver cancer cells
(Fig. 6C and D). Besides, colony
formation assays revealed that the proliferation ability of liver
cancer cells could also be restored (Fig. S6A and B). Moreover, IGFBP4
knockdown could restore the repressed EMT progression caused by
MYBBP1A inhibition. Specifically, inhibition of MYBBP1A increased
E-cadherin expression, while knocking down IGFBP4 restricted the
increase of E-cadherin. Similarly, MYBBP1A inhibition decreased
N-cadherin expression, and knocking down IGFBP4 restored its
expression caused by MYBBP1A inhibition (Fig. 6E). As for NOTCH signaling
regulation, IGFBP4 knockdown could restore the repressed NOTCH1 and
Hes1 expression caused by MYBBP1A inhibition (Fig. 6F). These results demonstrated that
IGFBP4 was involved in regulating EMT, cancer metastasis and EMT
and NOTCH activation mediated by MYBBP1A in liver cancer.
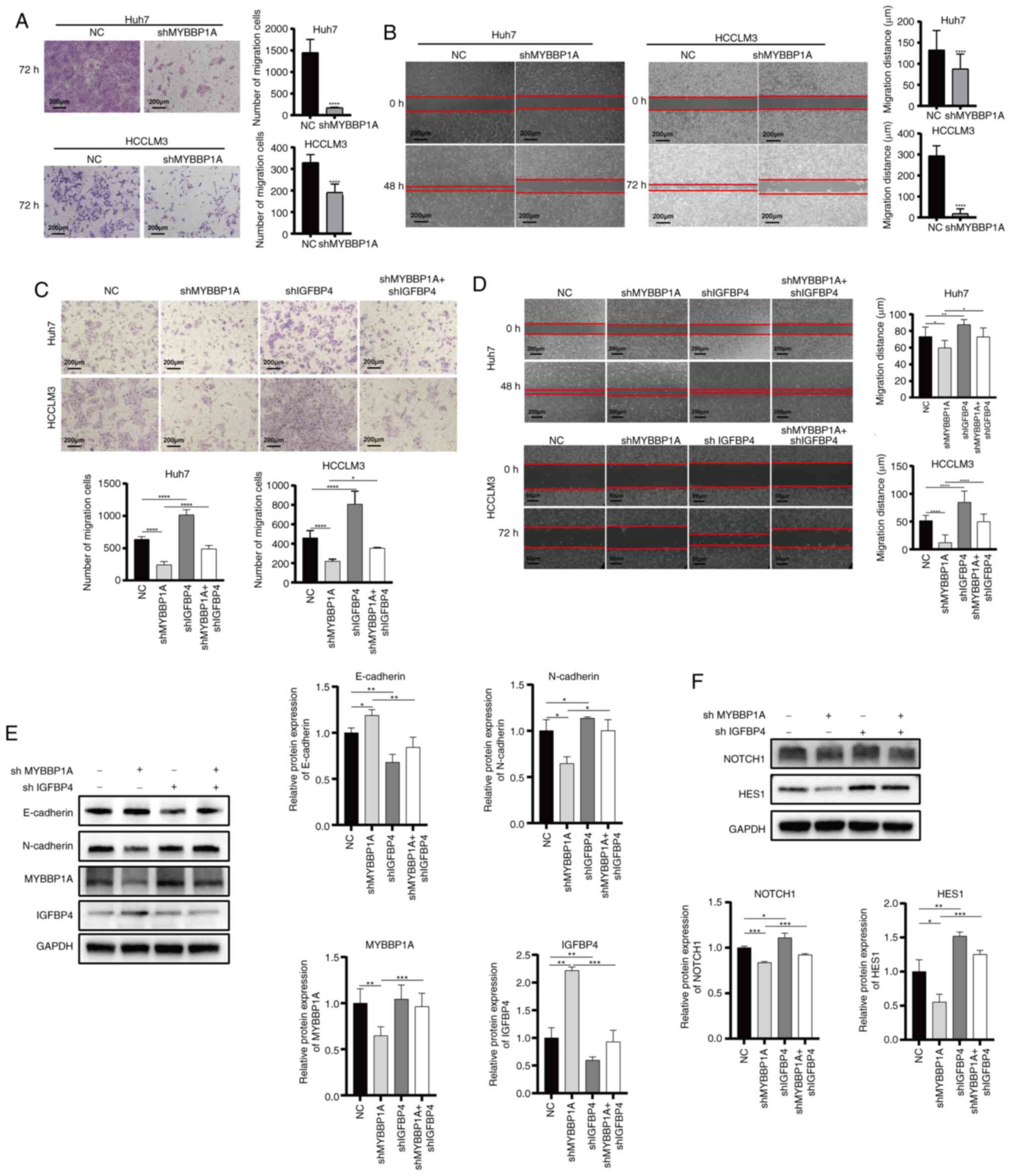 | Figure 6Knocking down IGFBP4 reduces the
metastasis ability of liver cancer cell with MYBBP1A low
expression. (A and B) The motile activities of shMYBBP1A Huh7 and
HCCLM3 cells based on Transwell migration assays and gap closure
assays, respectively. (original magnification, ×400;
****P<0.0001, Student's t-test). (C and D) The
migration capacity of Huh7 and HCCLM3 cell lines in the negative
control group, shMYBBP1A group, shIGFBP4 group, and shMYBBP1A +
shIGFBP4 group were detected by Transwell migration assay and gap
closure experiment (magnification, ×100 and ×400;
*P<0.05, **P<0.01 and
****P<0.0001, Student's t-test). (E) WB analysis of
the protein expression levels of MYBBP1A, IGFBP4, and EMT related
genes in the negative control group, shMYBBP1A group, shIGFBP4
group and shMYBBP1A + shIGFBP4 group (*P<0.05,
**P<0.01, ***P<0.001, Student's
t-test). (F) WB analysis was performed to detect the protein
expression levels of NOTCH pathway-related genes NOTCH1 and Hes1 in
the negative control group, shMYBBP1A group, shIGFBP4 group, and
shMYBBP1A + shIGFBP4 group (*P<0.05,
**P<0.01, ***P<0.001, Student's
t-test). IGFBP 4, insulin-like growth factor binding protein 4;
sh-, short hairpin; WB, western blotting; NC, negative control;
HES, hes family bHLH transcription factor 1. |
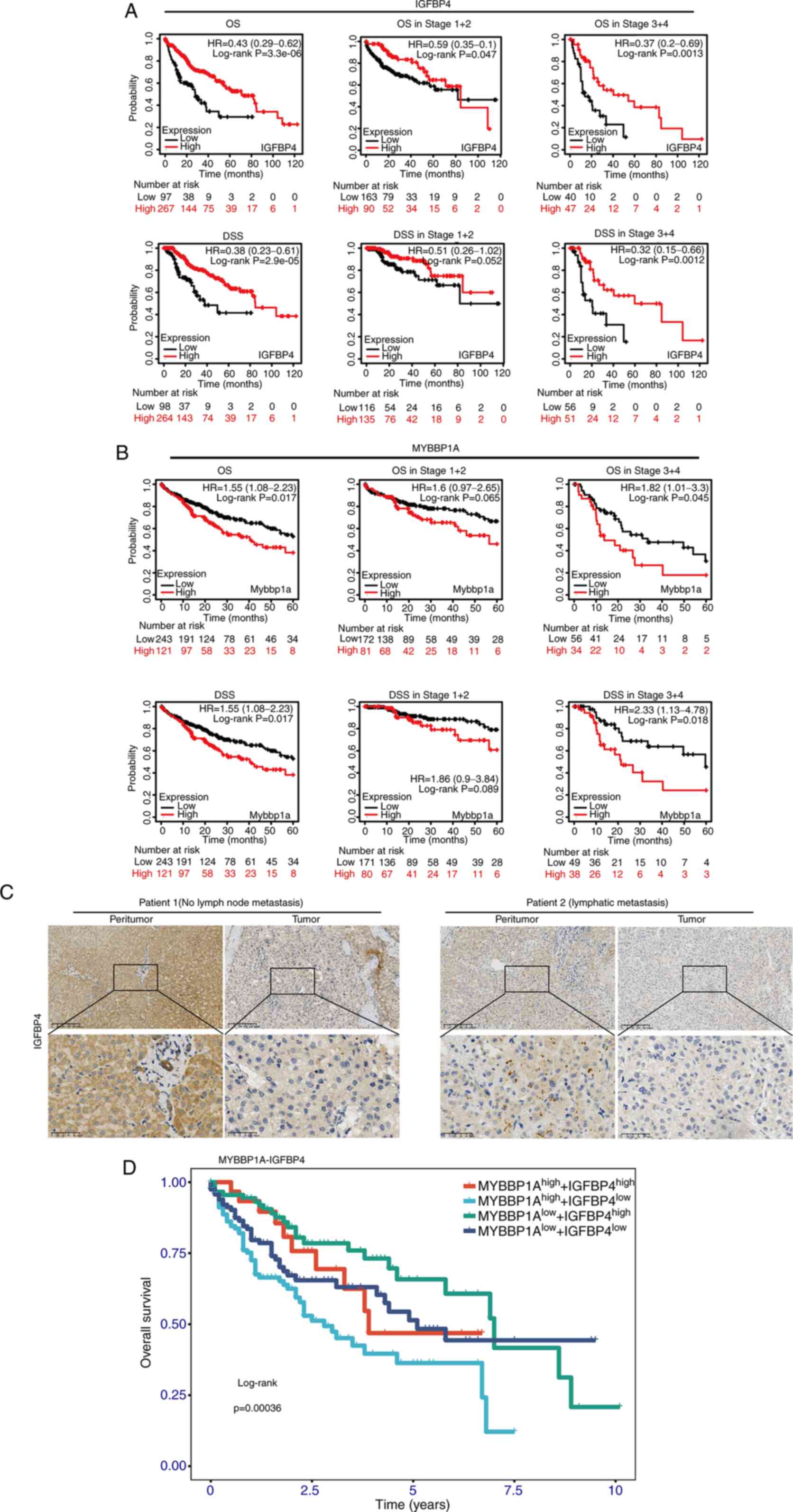
Clinical significance of IGFBP4 and
MYBBP1A in the prognosis of metastatic liver cancer
Overall survival (OS) time and disease-specific
survival time (DSS) of patients with liver cancer with different
IGFBP4 expression groups in different tumor grades and stages were
analyzed. The results demonstrated that high expression of IGFBP4
was correlated with longer OS [log-rank P=3.3×10−6,
hazard ratio (HR)=0.43] and DSS (log-rank P=2.9×10−5,
HR=0.38). This association was statistically significant in late
stages (stages III and IV; log-rank P=0.0013 in OS and log-rank
P=0.0012 in DSS) but not in early stages (stages I and II; log-rank
P=0.047 in OS and log-rank P=0.052 in DSS) in liver cancer
(Fig. 7A). Interestingly, the
negative correlation between MYBBP1A expression and OS (log-rank
P=0.0045) or DSS (log-rank P=0.018) was observed in late stages
(stages III and IV) rather than in early stages (stage I and II) in
liver cancer (Fig. 7B). IHC
staining was performed on tumor and peritumor tissue specimens of
patients with liver cancer with different metastatic status in The
First Affiliated Hospital of Zhejiang University. The results
suggested that IGFBP4 expression levels in the liver tissues of
patients with liver cancer were lower than those in normal
individuals, and even lower in patients with metastatic liver
cancer (Fig. 7C). Then, the
survival curves of patients with HCC with
MYBBP1AHighIGFBP4Low,
MYBBP1AHighIGFBP4High,
MYBBP1ALowIGFBP4High and
MYBBP1ALowIGFBP4Low were analyzed; the
results revealed that the OS of patients with
MYBBP1AHighIGFBP4Low was significantly poorer
than those with MYBBP1AlowIGFBP4High. The
MYBBP1ALowIGFBP4High group had the most
favorable OS in these four groups (Figs. 7D and S8). Therefore, it is concluded that
IGFBP4 and MYBBP1A expression could serve as potential biomarkers
in predicting the prognosis of liver cancer.
Discussion
The progression of HCC is associated with the
abnormal activation of multiple signaling pathways, including
angiogenesis, cell proliferation, apoptosis, invasion and
metastasis. Current therapies inhibit the proliferation and
survival of tumor cells by targeting specific molecules in these
abnormal signaling pathways, which rely on specific therapy under
molecular targeted diagnosis. While the therapy under the molecular
targeted diagnosis has made some progress in the treatment of
metastatic liver cancer, there are still challenges (27,28). Novel diagnosis and treatment are
getting increasing attention in clinical practice, and the
development of treatment strategies based on individualized
diagnosis will help improve the treatment effect and provide
patients with an improved chance of survival.
IGFBPs are a family of proteins that bind to IGFs
and are usually composed of seven high-affinity IGFBP isoforms,
namely IGFBP1 to IGFBP7 (7).
IGFBP4, the smallest protein in the IGFBP family (29), is a secreted protein mainly
produced by the liver (30). It
acts as a transport protein for IGF-I and IGF-II and regulates
their biological effects. The levels and expression of IGFBP4 in
various tissues are influenced by IGFBP proteinases, multiple
growth factors and hormones. The results of the present study
suggested that IGFBP4 may play a positive regulatory role in the
process of tumor cell metastasis and invasion. Overexpression of
IGFBP4 in glioblastoma cells leads to the upregulation of molecules
involved in tumor growth (31).
Inhibition of IGFBP4 could significantly reduce the invasion and
expression of mesenchymal markers in oral squamous cell carcinoma
(32). Several studies have found
that overexpression of IGFBP4 inhibits the invasiveness of cancer
cells, including lung cancer (33), colorectal cancer (34) and breast cancer (9). Previous research has shown that
transcriptional activation of IGFBP4 could inhibit the metastasis
of liver cancer (35,36). In the present study, the analysis
of the TCGA database and clinical samples from patients with HCC
showed that IGFBP4 was downregulated in liver cancer. In addition,
despite the lack of 5-10 years of survival data which is the
limitation in the present study, the five-year survival curve
analysis still showed that the high expression of IGFBP4 was
associated with favorable prognosis in patients with HCC. EMT was
considered one of the key steps in tumor metastasis and invasion.
During EMT, tumor cells could lose the adhesive properties of
epithelial cells, acquire the motility of mesenchymal cells, and
enter other organs through the blood or lymphatic system. In the
present study, comprehensive experimental methods such as gap
closure experiments and Transwell assays were used to verify that
overexpressing IGFBP4 could inhibit the migration ability of HCC
in vitro and reduce the tumor metastasis in vivo. It
was confirmed that overexpression of IGFBP4 in HCC cells leads to
upregulation of the epithelial marker protein E-cadherin,
downregulation of the mesenchymal marker protein N-cadherin and the
cytoskeletal protein Vimentin.
DNA methylation is a common epigenetic modification
(37). Aberrant regulation of DNA
methylation is closely associated with tumorigenesis and
progression in liver cancer (38). High methylation status is
associated with gene silencing and functional loss. Studies have
shown that some tumor suppressor genes undergo hypermethylation,
leading to the methylation of their promoter regions and subsequent
gene silencing, thereby causing tumor cells to lose normal
proliferation and growth control. Our previous research suggested
that IGFBP5 was regulated by MYBBP1A, inducing abnormal
hypermethylation of the CpG island in the IGFBP5 CDS region,
inhibiting IGFBP5 transcription and secretion which promoted HCC
progression (27). It was
hypothesized that IGFBP4, another important member of the IGFBP
family, may also be regulated by MYBBP1A. To validate this
hypothesis, it was firstly confirmed that MYBBP1A indeed promotes
the migratory and invasive abilities of liver cancer cells.
Secondly, it was demonstrated that IGFBP4 expression is negatively
regulated by MYBBP1A, and rescue experiments further confirmed this
relationship. Additionally, using Methprimer analysis, it was found
that there were CpG islands in the IGFBP4 promoter region, and the
methylation level of IGFBP4 was negatively correlated with poor
prognosis. To further verify that the high methylation status of
the IGFBP4 promoter is regulated by MYBBP1A, BSP was used to
confirm that knocking down MYBBP1A leads to a decrease in CpG
island methylation in the IGFBP4 promoter regions and an
enhancement of target gene transcription. Furthermore, treatment
with the methylation inhibitor 5-Aza resulted in a subsequent
decrease in IGFBP4 expression.
EMT is a dynamic process that can be achieved
through multiple signaling pathways such as IGR-1R receptor ligand
activation, NOTCH, Wnt/β-catenin and Hedgehog pathway activation,
which may also contribute to the EMT process. Studies have shown
that knocking down Wnt3a can affect the activation of the
Wnt/β-catenin signaling pathway, thereby affecting the EMT process
and inhibiting the metastatic ability of liver cancer cells
(39,40). Initially, it was considered that
IGFBP4 function was closely associated with the Wnt/β-catenin
pathway. WB assays were performed to assess the expression of
β-catenin/Wnt3a/Smad protein. However, no significant changes in
the expression of these proteins were observed following IGFBP4
overexpression (Fig. S3). This
result may be attributed to the complex crosstalk between signaling
pathways, suggesting that the β-catenin/Smad pathway may not serve
as the primary driver in this regulatory axis. Further
investigation supports that this pathway is likely not the
predominant mechanism by which IGFBP4 influences liver cancer
metastasis. Through GSEA analysis, differential gene enrichment was
found in the NOTCH pathway, which is another crucial pathway that
affects the EMT. The NOTCH signaling pathway was aberrantly
activated in various tumors and has been found to be associated
with tumor cell proliferation, invasion and metastasis (41). In pancreatic cancer, RHBDL2
stabilizes N1ICD through OTUD7B and activates the NOTCH signaling
pathway, promoting cell proliferation and migration (42). In the liver, activation of the
NOTCH-YAP1/TEAD-DNMT1 axis drives hepatocytes to develop into
intrahepatic cholangiocarcinoma (43). When the NOTCH1 receptor binds to
the DELTA ligand, the extracellular structure of the receptor
undergoes conformational changes, leading to the cleavage of the
receptor's intracellular domain (NICD) by γ-secretase. NICD is
further translocated into the nucleus and forms a transcriptional
regulatory complex with the key transcription factor CSL, promoting
the transcription of downstream target genes (44). Hes1 is one of the downstream
target genes of the NOTCH signaling pathway, and its expression is
regulated by the NICD-CSL complex. In the present study,
overexpression of IGFBP4 was found to significantly increase the
transcription and protein levels of NOTCH1 and its downstream
target gene Hes1. Some studies showed that EMT and IGR-1R
activation are involved in a positive feedback loop (45). IGFBP4 can bind to IGF1, inhibit
the binding of IGF ligands to the receptor, and prolong its
half-life in circulation (46).
The crosstalk between IGF-1R and NOTCH could jointly act on the EMT
phenotype of tumors. Some researchers considered that IGF-1R was a
target of NOTCH1, and NOTCH directly upregulated the expression of
IGF1R in human T-cell acute lymphoblastic leukemia cells,
significantly enhancing their sensitivity to environmental ligands
(47). It has also been suggested
that NOTCH1 may be transcriptionally activated by YAP (48,49) and the activity of this
transcription factor is triggered by the IGF-1R/PI3K/mTOR signaling
pathway. Additionally, the YAP-IGF1R signaling loop is also
involved in EMT-related proteins (50,51). The results of protein interaction
analysis showed that IGFBP4 had a strong interaction with
IGF1/IGF1R, and IGF1R also showed a strong interaction with NOTCH1
and YAP1 (Fig. S7). The present
study confirmed that the overexpression of IGFBP4 significantly
reduces the level of the transcription factor YAP, which is
positively correlated with the activation of the NOTCH pathway
(Fig. S4). Regrettably, the
specific mechanism by which IGFBP4 regulates the NOTCH pathway
through YAP was not investigated. Further studies, along with
additional clinical sample data, are required to confirm the
upstream and downstream regulatory effects of IGFBP4 on the NOTCH
pathway. This topic could serve as a significant focus for future
research.
The previous phase of our research (13) elucidated the mechanism of MYBBP1A
regulates IGFBP5 transcription and inhibits the AKT pathway, and it
was hypothesized that another member of the family, IGFBP4, may
have a similar function. Since IGFBP4 has been found to play an
important role in other cancers, experiments were designed to
verify the function of IGFBP4 and its upstream and downstream
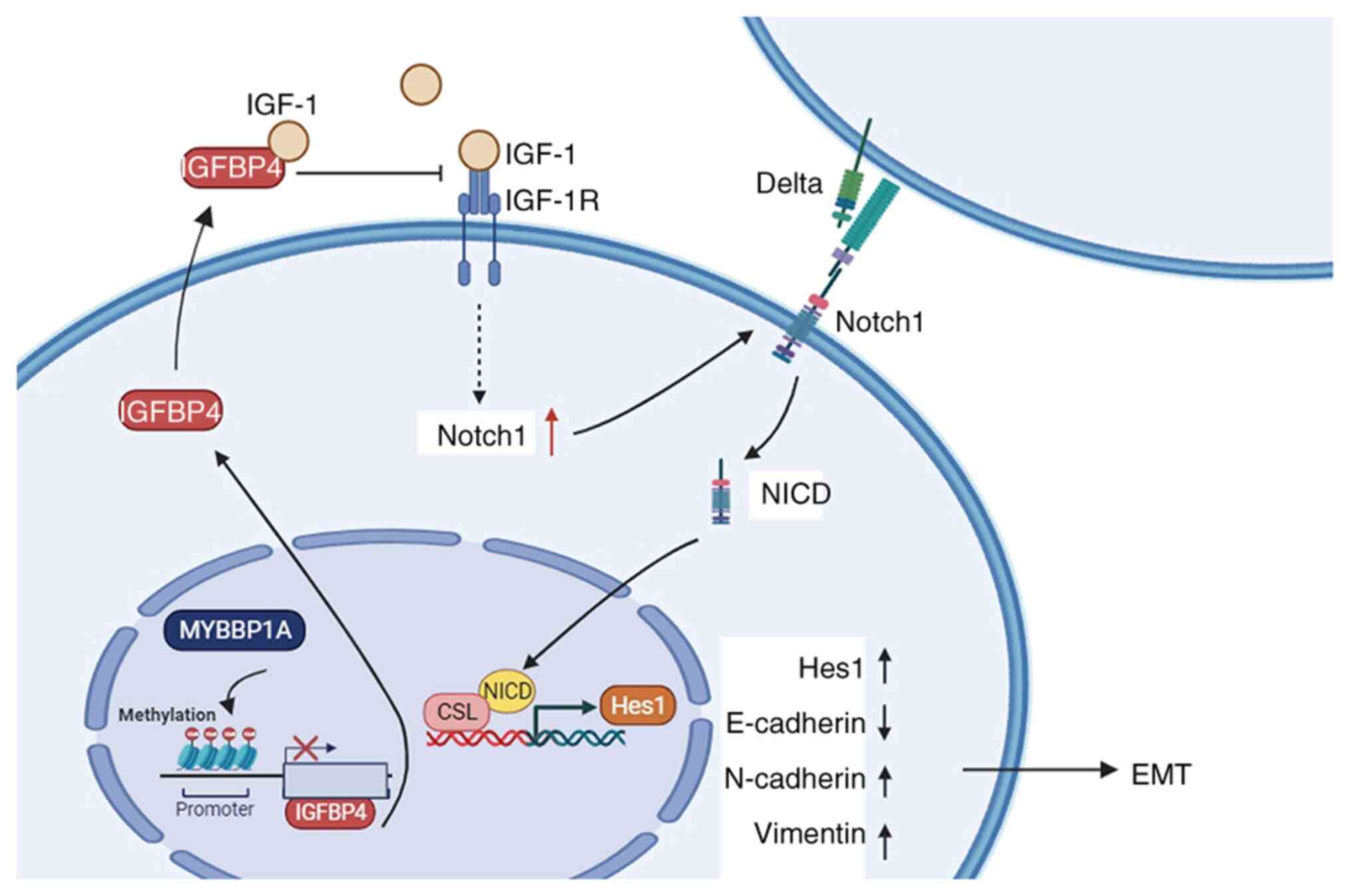
pathways in HCC. In conclusion, IGFBP4 is a potent biomarker that
plays a protective role in liver cancer, especially in metastatic
liver cancer. By exploring the upstream and downstream of IGFBP4,
it was revealed that MYBBP1A inhibits the transcription of IGFBP4
through high methylation levels of the CpG islands of the IGFBP4
promoter region mediated by DNMT1, which promotes the activation of
the NOTCH pathway, increasing the EMT of HCC and providing
migration ability for cancer cells (Fig. 8). A recent study by the authors
showed that liver cancer populations with high MYBBP1A expression
had a poor prognosis (27), and
subsequently, it was found that low expression of IGFBP4 in HCC was
associated with poor prognosis in patients with HCC. Therefore, a
combined prognostic analysis of MYBBP1A and IGFBP4 was performed.
It was found that MYBBP1AHighIGFBP4High
patients had an improved prognosis than
MYBBP1AHighIGFBP4Low, and in the population
with high MYBBP1A expression and poor prognosis, high IGFBP4
expression at the same time could prolong the OS time. It was also
found that MYBBP1AlowIGFBP4Low patients had
an improved prognosis than those with
MYBBP1AHighIGFBP4Low, and in populations with
high MYBBP1A expression and poor prognosis, simultaneous high
expression of IGFBP4 could prolong OS. Similarly, among the four
groups, MYBBP1ALowIGFBP4High patients had the
best prognosis, and the OS of patients with
MYBBP1ALowIGFBP4High was significantly longer
than those with MYBBP1AHighIGFBP4Low
(Fig. S8). Dual gene biomarker
combination prediction of MYBBP1A and IGFBP4 has great potential in
the prognosis of metastatic liver cancer, providing a theoretical
basis for the precise treatment and survival prediction of liver
cancer.
The primary limitations of the present study are the
restricted sample size and the absence of longitudinal prognostic
data. Future studies with larger cohorts will be essential to
confirm these observations. Further research should also aim to
elucidate the specific mechanisms by which IGFBP4 regulates the
NOTCH pathway in liver cancer and explore the roles of other IGFBP
family members in hepatocarcinogenesis, as they may contribute
through distinct or complementary mechanisms.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YS and XW conceived and designed the study,
acquired, analysed and interpreted the data, confirm the
authenticity of all the raw data, and participated in drafting or
revision of the submitted article. WC, JG, BD, JR and XH acquired,
analysed and interpreted the data, and revised the submitted
article. DM, YL, SC and RD collected clinical samples, analysed and
interpreted data. JR and BY conceived and designed the study, and
participated in drafting or revision of the submitted article. All
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Human studies (approval no. 2020-IIT-834) and
animal experiments (approval no. 2020-IIT-rapid-1132) were approved
by the Clinical Research Ethics Committee of the First Affiliated
Hospital, School of Medicine, Zhejiang University (Hangzhou,
China). Written informed consent was obtained by all patients
participating in the present study. Animal experiments were
performed in strict accordance with the NIH Guide for the Care and
Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
IGFBP
|
insulin-like growth factor binding
protein
|
|
IGF
|
insulin-like growth factor
|
|
MYBBP1A
|
Myb-binding protein 1A
|
|
EMT
|
epithelial-mesenchymal transition
|
|
TCGA
|
The Cancer Genome Atlas
|
|
GTEx
|
The Genotype-Tissue Expression
|
|
GSEA
|
Gene Set Enrichment Analysis
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
WB
|
western blotting
|
|
IHC
|
immunohistochemistry
|
|
OE
|
overexpression
|
|
WT
|
wild-type
|
|
BSP
|
bisulfite sequencing PCR
|
|
OS
|
overall survival
|
|
DSS
|
disease-specific survival
|
Acknowledgements
The authors appreciate the kind help from Mr
Yonghao Xu, a laboratory technician, from the Experimental Animal
Center of the First Affiliated Hospital of Zhejiang University
School of Medicine for providing technical support for animal
experiments.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 82103487 and 82302893) and the
Zhejiang Provincial Natural Science Foundation of China (grant nos.
LQ21H160018 and LQ24H160011).
References
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Llovet JM, Kelley RK, Villanueva A, Singal
AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J and
Finn RS: Hepatocellular carcinoma. Nat Rev Dis Primers. 7:62021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kulik L and El-Serag HB: Epidemiology and
management of hepatocellular carcinoma. Gastroenterology.
156:477–491.e1. 2019. View Article : Google Scholar
|
|
6
|
Yang JD, Hainaut P, Gores GJ, Amadou A,
Plymoth A and Roberts LR: A global view of hepatocellular
carcinoma: Trends, risk, prevention and management. Nat Rev
Gastroenterol Hepatol. 16:589–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Durai R, Davies M, Yang W, Yang SY,
Seifalian A, Goldspink G and Winslet M: Biology of insulin-like
growth factor binding protein-4 and its role in cancer (review).
Int J Oncol. 28:1317–1325. 2006.PubMed/NCBI
|
|
8
|
Yang B, Zhang L, Cao Y, Chen S, Cao J, Wu
D, Chen J, Xiong H, Pan Z, Qiu F, et al: Overexpression of lncRNA
IGFBP4-1 reprograms energy metabolism to promote lung cancer
progression. Mol Cancer. 16:1542017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen W, Hu L, Lu X, Wang X, Zhao C, Guo C,
Li X, Ding Y, Zhao H, Tong D, et al: The RNA binding protein MEX3A
promotes tumor progression of breast cancer by post-transcriptional
regulation of IGFBP4. Breast Cancer Res Treat. 201:353–366. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li C, Cao Y, Zhang L, Li J, Wu H, Ling F,
Zheng J, Wang J, Li B, He J, et al: LncRNA IGFBP4-1 promotes tumor
development by activating Janus kinase-signal transducer and
activator of transcription pathway in bladder urothelial carcinoma:
Retraction. Int J Biol Sci. 19:48332023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Conover CA: Insulin-like growth
factor-binding proteins and bone metabolism. Am J Physiol
Endocrinol Metab. 294:E10–E14. 2008. View Article : Google Scholar
|
|
12
|
Maridas DE, DeMambro VE, Le PT, Mohan S
and Rosen CJ: IGFBP4 is required for adipogenesis and influences
the distribution of adipose depots. Endocrinology. 158:3488–3500.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weng X, Wu J, Lv Z, Peng C, Chen J, Zhang
C, He B, Tong R, Hu W, Ding C, et al: Targeting MYBBP1A suppresses
HCC progression via inhibiting IGF1/AKT pathway by CpG islands
hypo-methylation dependent promotion of IGFBP5. EBioMedicine.
44:225–236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Ru J, Lu J, Ge J, Ding B, Su R, Jiang Y,
Sun Y, Ma J, Li Y, Sun J, et al: IRGM is a novel regulator of PD-L1
via promoting S6K1-mediated phosphorylation of YBX1 in
hepatocellular carcinoma. Cancer Lett. 581:2164952024. View Article : Google Scholar
|
|
16
|
Hu X, Chen G, Huang Y, Cheng Q, Zhuo J, Su
R, He C, Wu Y, Liu Z, Yang B, et al: Integrated multiomics reveals
silencing of has_circ_0006646 Promotes TRIM21-Mediated NCL
ubiquitination to inhibit hepatocellular carcinoma metastasis. Adv
Sci (Weinh). 11:e23069152024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Workman P, Aboagye EO, Balkwill F, Balmain
A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA,
Glennie MJ, et al: Guidelines for the welfare and use of animals in
cancer research. Br J Cancer. 102:1555–1577. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sugase T, Lam BQ, Danielson M, Terai M,
Aplin AE, Gutkind JS and Sato T: Development and optimization of
orthotopic liver metastasis xenograft mouse models in uveal
melanoma. J Transl Med. 18:2082020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Purohit A, Saxena S, Varney M, Prajapati
DR, Kozel JA, Lazenby A and Singh RK: Host Cxcr2-Dependent
regulation of pancreatic cancer growth, angiogenesis, and
metastasis. Am J Pathol. 191:759–771. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Seki K, Yamaguchi A, Goi T, Nakagawara G,
Matsukawa S, Urano T and Furukawa K: Inhibition of liver metastasis
formation by anti-CD44 variant exon 9 monoclonal antibody. Int J
Oncol. 11:1257–1261. 1997.PubMed/NCBI
|
|
21
|
Ohta T, Futagami F, Tajima H, Kitagawa H,
Kayahara M, Nagakawa T, Miwa K, Yamamoto M, Iseki S, Nakanuma Y and
Terada T: Inhibitory effect of a serine protease inhibitor, FOY-305
on the invasion and metastasis of human pancreatic cancers. Int J
Oncol. 11:813–817. 1997.PubMed/NCBI
|
|
22
|
Takesue S, Ohuchida K, Shinkawa T, Otsubo
Y, Matsumoto S, Sagara A, Yonenaga A, Ando Y, Kibe S, Nakayama H,
et al: Neutrophil extracellular traps promote liver micrometastasis
in pancreatic ductal adenocarcinoma via the activation of
cancer-associated fibroblasts. Int J Oncol. 56:596–605.
2020.PubMed/NCBI
|
|
23
|
Tauriello DVF, Palomo-Ponce S, Stork D,
Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M,
Ibiza S, Cañellas A, Hernando-Momblona X, et al: TGFβ drives immune
evasion in genetically reconstituted colon cancer metastasis.
Nature. 554:538–543. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Li Y, Kong D and Sarkar FH: The
role of NOTCH signaling pathway in epithelial-mesenchymal
transition (EMT) during development and tumor aggressiveness. Curr
Drug Targets. 11:745–751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang X, Bai Q, Chen W, Liang J, Wang F, Gu
W, Liu L, Li Q, Chen Z, Zhou A, et al: m(6) A-Dependent Modulation
via IGF2BP3/MCM5/NOTCH Axis Promotes Partial EMT and LUAD
Metastasis. Adv Sci (Weinh). 10:e22067442023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yuan X, Wu H, Han N, Xu H, Chu Q, Yu S,
Chen Y and Wu K: NOTCH signaling and EMT in non-small cell lung
cancer: Biological significance and therapeutic application. J
Hematol Oncol. 7:872014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Finn RS, Qin S, Ikeda M, Galle PR, Ducreux
M, Kim TY, Kudo M, Breder V, Merle P, Kaseb AO, et al: Atezolizumab
plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J
Med. 382:1894–1905. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu L, Shao Z, Fang X, Xin Z, Zhao S, Zhang
H, Zhang Y, Zheng W, Yu X, Zhang Z and Sun L: Exploring precision
treatments in immune-mediated inflammatory diseases: Harnessing the
infinite potential of nucleic acid delivery. Exploration.
202:301652024.
|
|
29
|
Sato H, Sakaeda M, Ishii J, Kashiwagi K,
Shimoyamada H, Okudela K, Tajiri M, Ohmori T, Ogura T, Woo T, et
al: Insulin-like growth factor binding protein-4 gene silencing in
lung adenocarcinomas. Pathol Int. 61:19–27. 2011. View Article : Google Scholar
|
|
30
|
Mazerbourg S, Callebaut I, Zapf J, Mohan
S, Overgaard M and Monget P: Up date on IGFBP-4: Regulation of
IGFBP-4 levels and functions, in vitro and in vivo. Growth Horm IGF
Res. 14:71–84. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Praveen Kumar VR, Sehgal P, Thota B, Patil
S, Santosh V and Kondaiah P: Insulin like growth factor binding
protein 4 promotes GBM progression and regulates key factors
involved in EMT and invasion. J Neurooncol. 116:455–464. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma X, Zhao D, Liu S, Zuo J, Wang W, Wang
F, Li Y, Ding Z, Wang J and Wang X: FERMT2 upregulation in CAFs
enhances EMT of OSCC and M2 macrophage polarization. Oral Dis.
30:991–1003. 2024. View Article : Google Scholar
|
|
33
|
Diehl D, Hoeflich A, Wolf E and Lahm H:
Insulin-like growth factor (IGF)-binding protein-4 inhibits colony
formation of colorectal cancer cells by IGF-independent mechanisms.
Cancer Res. 64:1600–1603. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li W, Sun D, Lv Z, Wei Y, Zheng L, Zeng T
and Zhao J: Insulin-like growth factor binding protein-4 inhibits
cell growth, migration and invasion, and downregulates COX-2
expression in A549 lung cancer cells. Cell Biol Int. 41:384–391.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tao L, Wang Y, Shen Z, Cai J, Zheng J, Xia
S, Lin Z, Wan Z, Qi H, Jin R, et al: Activation of IGFBP4 via
unconventional mechanism of miRNA attenuates metastasis of
intrahepatic cholangiocarcinoma. Hepatol Int. 18:91–107. 2024.
View Article : Google Scholar
|
|
36
|
Lee YY, Mok MT, Kang W, Yang W, Tang W, Wu
F, Xu L, Yan M, Yu Z, Lee SD, et al: Loss of tumor suppressor
IGFBP4 drives epigenetic reprogramming in hepatic carcinogenesis.
Nucleic Acids Res. 46:8832–8847. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang N, Lin C, Huang X, Kolbanovskiy A,
Hingerty BE, Amin S, Broyde S, Geacintov NE and Patel DJ:
Methylation of cytosine at C5 in a CpG sequence context causes a
conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine
adduct in DNA from a minor groove alignment to intercalation with
base displacement. J Mol Biol. 346:951–965. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hernandez-Meza G, von Felden J,
Gonzalez-Kozlova EE, Garcia-Lezana T, Peix J, Portela A, Craig AJ,
Sayols S, Schwartz M, Losic B, et al: DNA methylation profiling of
human hepatocarcinogenesis. Hepatology. 74:183–199. 2021.
View Article : Google Scholar
|
|
39
|
Qi L, Sun B, Liu Z, Cheng R, Li Y and Zhao
X: Wnt3a expression is associated with epithelial-mesenchymal
transition and promotes colon cancer progression. J Exp Clin Cancer
Res. 33:1072014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang L, He S, Guan H, Zhao Y and Zhang D:
Circulating RNA ZFR promotes hepatocellular carcinoma cell
proliferation and epithelial-mesenchymal transition process through
miR-624-3p/WEE1 axis. Hepatobiliary Pancreat Dis Int. 23:52–63.
2024. View Article : Google Scholar
|
|
41
|
Jackstadt R, van Hooff SR, Leach JD,
Cortes-Lavaud X, Lohuis JO, Ridgway RA, Wouters VM, Roper J,
Kendall TJ, Roxburgh CS, et al: Epithelial NOTCH signaling rewires
the tumor microenvironment of colorectal cancer to drive
poor-prognosis subtypes and metastasis. Cancer Cell. 36:319–336.e7.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen S, Cai K, Zheng D, Liu Y, Li L, He Z,
Sun C and Yu C: RHBDL2 promotes the proliferation, migration, and
invasion of pancreatic cancer by stabilizing the N1ICD via the
OTUD7B and activating the NOTCH signaling pathway. Cell Death Dis.
13:9452022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hu S, Molina L, Tao J, Liu S, Hassan M,
Singh S, Poddar M, Bell A, Sia D, Oertel M, et al:
NOTCH-YAP1/TEAD-DNMT1 axis drives hepatocyte reprogramming into
intrahepatic cholangiocarcinoma. Gastroenterology. 163:449–465.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kawaguchi K and Kaneko S: NOTCH signaling
and liver cancer. Adv Exp Med Biol. 1287:69–80. 2021. View Article : Google Scholar
|
|
45
|
Sivakumar R, Koga H, Selvendiran K,
Maeyama M, Ueno T and Sata M: Autocrine loop for IGF-I receptor
signaling in SLUG-mediated epithelial-mesenchymal transition. Int J
Oncol. 34:329–338. 2009.PubMed/NCBI
|
|
46
|
Baxter RC: Signaling pathways of the
insulin-like growth factor binding proteins. Endocr Rev.
44:753–778. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Medyouf H, Gusscott S, Wang H, Tseng JC,
Wai C, Nemirovsky O, Trumpp A, Pflumio F, Carboni J, Gottardis M,
et al: High-level IGF1R expression is required for
leukemia-initiating cell activity in T-ALL and is supported by
NOTCH signaling. J Exp Med. 208:1809–1822. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Totaro A, Castellan M, Di Biagio D and
Piccolo S: Crosstalk between YAP/TAZ and NOTCH Signaling. Trends
Cell Biol. 28:560–573. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Engel-Pizcueta C and Pujades C: Interplay
between NOTCH and YAP/TAZ pathways in the regulation of cell fate
during embryo development. Front Cell Dev Biol. 9:7115312021.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhu H, Wang DD, Yuan T, Yan FJ, Zeng CM,
Dai XY, Chen ZB, Chen Y, Zhou T, Fan GH, et al: Multikinase
inhibitor CT-707 targets liver cancer by interrupting the
hypoxia-activated IGF-1R-YAP axis. Cancer Res. 78:3995–4006. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ngo MT, Peng SW, Kuo YC, Lin CY, Wu MH,
Chuang CH, Kao CX, Jeng HY, Lin GW, Ling TY, et al: A
yes-associated protein (YAP) and insulin-like growth factor 1
receptor (IGF-1R) signaling loop is involved in sorafenib
resistance in hepatocellular carcinoma. Cancers (Basel).
13:38122021. View Article : Google Scholar : PubMed/NCBI
|