Introduction
Neurofibromatosis type 1 (NF1) is a rare autosomal
dominant disorder caused by mutations in NF1 gene, with a
global incidence of ~1 in 3,000 live births (1). Patients are at risk of developing
benign and malignant tumors throughout their lives, and typical
symptoms include plexiform neurofibroma (PNF), which occurs in
20-50% of patients with NF1 mutation (2-4).
Patients often present with painful, oversized masses that
interfere with function in early childhood that rapidly progress
during childhood and adolescence (5). PNF tumors may spread extensively and
invade surrounding tissue, causing severe physical defects and
functional impairment, resulting in high rates of disability and
malformation. As the patient ages, risk of the tumors transforming
into malignant peripheral nerve sheath tumors (MPNSTs) increases,
posing a threat to the patient life and health (4). However, current clinical treatments
for PNF are limited because the tumors grow along nerves, and some
types are poorly circumscribed from surrounding tissue, frequently
causing difficulties in surgical resection (6). Furthermore, the indications (such as
patient age and tumor progression) for surgery are often unclear
and risky and the tumors are prone to recurrence (6). In 2020, selumetinib, a
mitogen-activated protein kinase inhibitor, became the first
effective targeted therapy approved for PNF and is currently the
preferred treatment option for pediatric patients with inoperable
PNF (7,8). However, issues remain, including the
30% rate of primary drug resistance, secondary drug resistance
following long-term drug use and lack of treatment options in
adulthood (7,8). Therefore, development of novel
targeted drugs is still urgently needed.
Mass spectrometry and non-targeted metabolomics
analysis found that SUCLG1 and citric acid (CA) in the catalytic
enzyme pathway of the tricarboxylic acid cycle (TCA) are highly
expressed in PNF (9). SUCLG1 is
responsible for converting succinyl-CoA into succinate (10). Mutations in SUCLG1 are implicated
in metabolic disorders, fatal infantile lactic acidosis and
mitochondrial DNA depletion (10,11). In a recent study, SUCLG1 was found
to be associated with leukemia progression (12). To the best of our knowledge,
however, no other tumors have been studied in relation to SUCLG1.
CA is an important signaling molecule in cell metabolism. CA is
synthesized by citrate synthase (CS) and transported from the
mitochondria to the cytoplasm via a CA carrier (SLC25A1). It is
then broken down by ATP citrate lyase (ACLY) into oxaloacetate and
acetyl CoA, which are used to synthesize pro-inflammatory factors
such as reactive oxygen species (ROS) and nitric oxide (NO), as
well as lipids (13,14). To the best of our knowledge,
neither the SUCLG1 gene nor changes in tumor metabolism have been
studied in relation to PNF.
The present study aimed to examine the role of
SUCLG1 in the function and metabolism of PNF cells and explore its
potential as an effective target for treatment of PNF.
Materials and methods
Tissue collection and cell culture
A total of three pairs of PNF and normal human skin
tissue and four pairs of serum samples were obtained from the
Department of Plastic Surgery, Shandong Provincial Hospital, Jinan,
Shandong, China with informed written consent and approval from the
Human Research Ethics Review Committee of Shandong Provincial
Hospital (approval no. SWYX2024-556). The samples were collected
from four patients (three male, one female; mean age, 18.25±3.40
years) from April 2021 to April 2022. The inclusion criteria were
as follows: i) Patients with PNF; ii) the lesion involved skin
tissue and iii) no other disease. According to the inclusion
criteria, the lesions invaded the skin tissue; therefore, normal
skin was used as the control group. Demographic characteristics are
shown in Table SI. PNF cells
(ipNF95.6 and ipNF05.5; American Type Culture Collection; cat. nos.
CRL 3389 and CRL 3387, respectively) were donated by the Department
of Plastic Surgery, Shanghai Ninth People's Hospital, Jinan, China.
The human Schwann cell (HSC) line was purchased from Zhong Qiao Xin
Zhou Biotechnology Co., Ltd. and cultured in HSC immortalization
medium (ZMY106; Zhong Qiao Xin Zhou Biotechnology Co., Ltd.) at
37°C in an atmosphere containing 5% CO2. PNF cells and
293T cells were cultured in high-glucose DMEM (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Procell Life Science &
Technology Co., Ltd.) and 1% penicillin/streptomycin at 37°C in an
atmosphere containing 5% CO2.
Proteomics and metabolomics analysis
Proteomics and non-targeted metabolomics analysis
were performed with skin tissue and serum samples from patients
with PNF and healthy individuals, respectively. Proteomics was
performed using conventional high performance liquid chromatography
(LC20AD, Shimadzu) for processing samples with a column temperature
of 40°C and a flow rate of 1 ml/min. Instruments used for
metabolomics are Mass Spectrometer (Q Exactive™ HF, Thermo Fisher,
Germany), chromatograph (Vanquish UHPLC, Thermo Fisher, Germany)
and chromatographic column (Hypesil Gold column(C18), Thermo
Fisher, USA). Positive and negative ionisation modes were used, the
scanning range was 100-1,500 m/z, the Sheath gas flow rate was 35
psi, and the Aux gas heater temp was 350°C. P<0.05 and
fold-change (FC)≥1.2 or ≤0.83 in the protein sample and FC ≥2 or
≤0.5 and variable importance in the projection ≥1 in the serum
metabolite samples were considered to indicate a statistically
significant difference. R1.6.20, VennDiagram package
(omicstudio.cn/tool.) was used to visualize data. Kyoto
Encyclopedia of Genes and Genomes(KEGG; genome.jp/kegg) pathway was
plotted using the OmicStudio (omicstudio.cn/tool.)
Reverse transcription-quantitative
(RT-q)PCR
Total RNA of cells was extracted using RNAiso Plus
(Takara Bio, Inc.), and cDNAs were reverse-transcribed from 1
μg total RNA using HiScript RT SuperMix for qPCR (Vazyme
Biotech Co., Ltd.) at 4°C for 20 min. DNA of cells was extracted
with a DNA Isolation Mini kit (Vazyme Biotech Co., Ltd.). qPCR was
performed using SYBR Green Master Mix (Vazyme Biotech Co., Ltd.) on
the LightCycler 480 II (Roche Diagnostics). Thermocycling
conditions were as follows: Pre-denaturation at 95°C for 30 sec,
followed by 40 cycles at 95°C for 5 sec and 60°C for 30 sec and
final extension at 95°C for 15 sec, 60°C for 60 sec and 95°C for 15
sec. β-actin was selected as the internal control.
2−∆∆Cq was used to calculate relative expression
(14). To measure mtDNA content,
we used 1 μg DNA and primers to the Journal Pre-prooD-loop
region of the mitochondrial genome. G6PC primers served as genomic
DNA control to normalize the mitochondrial to the genomic DNA
ratio. The primers are listed in Table SII.
Western blotting (WB)
Total protein of cells was extracted using the
Minute Total Protein Extraction kit (Invent Biotechnologies, Inc.)
or RIPA Lysis Buffer (Beyotime Institute of Biotechnology). Protein
concentration was determined using a BCA Protein Assay Kit
(Beyotime Institute of Biotechnology). A total of 20 μg/lane
protein was separated by 10% SDS-PAGE, proteins were placed in
rapid QuickBlock Blocking Buffer for WB (Beyotime Institute of
Biotechnology; cat. no. P0252) and blocked for 20 min at 25°C, then
transferred to polyvinylidene fluoride membranes and incubated with
primary antibodies at 4°C overnight. The primary antibodies
included β-actin (Proteintech Group, Inc.; 66009-1-I; 1:1,000),
α-tubulin (Wuhan Servicebio Technology Co., Ltd.; cat. no. GB15201;
1:2,000), SUCLG1 rabbit mAb (Cell Signaling Technology, Inc.; cat.
no. 8071; 1:1,000), CS rabbit pAb (cat. no. A5713; 1:1,000),
SLC25A1 Rabbit pAb (cat. no. A24754; 1:5,000), mitofusin-1 (MFN1)
rabbit pAb (cat. no. A9880; 1:1,000), MFN2 rabbit mAb (cat. no.
A19678; 1:1,000), OPA1 (optic Atrophy Protein 1) rabbit pAb (all
ABclonal Biotech Co., Ltd.; cat. no. A9833; 1:4,000) and ACLY
rabbit pAb (Proteintech Group, Inc.; cat. no. 15421-1-AP; 1:2,000).
Then, the membranes were incubated with HRP-conjugated goat
anti-rabbit secondary antibodies (cat. no. SA00001-2; 1:10,000) or
anti-mouse secondary antibodies (both Proteintech Group, Inc.; cat.
no. SA00001-1; 1:10,000) at 25°C for 1 h. Enhanced chemiluminescent
solution (Sparkjade ECL plus, ED0016-B; Sparkjade) and ChemiDoc
Imaging System (Bio-Rad Laboratories, Inc.) was used to quantify
the expression of proteins. ImageJ 1.53e (National Institutes of
Health) analyses the greyscale values and performs
calculations.
Immunofluorescence staining
Tissues were fixed with 4% polydoxaldehyde at room
temperature for 24 h, dip-waxed at ~60°C for 4.5 h and then
embedded in paraffin. The pre-cooled wax blocks were sectioned
(thickness, ~3 μm). The slices were placed in the oven at
60°C for 1 h. Slices were placed in Eco-friendly dewaxing solution
I for 10 min-Eco-friendly dewaxing solution II for 10
min-Eco-friendly dewaxing solution III (Wuhan Servicebio Technology
Co., Ltd.; cat. no. G1128) for 10 min-anhydrous ethanol I for 5
min-anhydrous ethanol II for 5 min-70% alcohol for 5 min-and washed
with distilled water. Sections were placed in citric acid antigen
repair solution (pH, 6; Wuhan Servicebio Technology Co., Ltd.; cat.
no. G1202), microwaved for 10 min on medium heat, ceased for 5 min,
transferred to medium-low heat for 5 min, ceased for 2 min and
finally medium-low heat for 5 min, and recovered at room
temperature for 30 min. Dewaxed sections were placed in 3% hydrogen
peroxide and incubated for 20 min at room temperature, then closed
with 3% BSA (Wuhan Servicebio Technology Co., Ltd.; cat. no.
GC305010) for 30 min at room temperature. Dewaxed sections were
stained with primary antibodies against SUCLG1 (Cell Signaling
Technology, Inc.; cat. no. 8071; 1:100) and recombinant anti-160
kDa neurofilament medium antibody (Wuhan Servicebio Technology Co.,
Ltd.; cat. no. GB15763-100; 1:500) at 4°C overnight, and then
incubated with CY3-labelled goat anti-rabbit and Alexa Fluor 488
labelled goat anti-mouse IgG. (cat. nos. GB21303 and GB25301; both
1:300; both Wuhan Servicebio Technology Co., Ltd) at room
temperature in the dark for 50 min, and DAPI stain solution was
added at room temperature in the dark for 10 min before
fluorescence microscopy (NIKON ECLIPSE C1; Nikon Corporation).
Magnification is 50x. Data were analyzed using ImageJ 1.53e
(National Institutes of Health, USA).
Knockdown and overexpression (OE) of
SUCLG1
Recombinant packaging plasmids (PG-P1-VSVG,
PG-P2-REV and PG-P3-RRE) and vector plasmids (lentivirus vector)
were prepared by GenePharma Co., Ltd. A total of 1 μg third
generation lentiviral packaging system package mix was prepared in
the ratio of PG-P1-VSVG:PG-P2-REV:PG-P3-RRE 1:2:3 for the
lentiviral plasmid packaging experiments in 60-mm cell culture
dishes. RNAi-Mate (GO4001, GenePharma Co., Ltd, Shanghai, China) to
co-transfect 293T cells (GenePharma Co., Ltd, Shanghai, China). OE
SUCLG1 plasmid backbone was LV5(EF-1a/GFP&Puro) and its
sequence was 5′-TTCTCCGAACGTGTCACGT-3′. ShSUCLG1-containing plasmid
backbone was LV3(H1/GFP&Puro) and its sequence was
5′-AGATCTGGCACCCTGACTTAT-3′. Sequence of shSUCLG1 negative control
was 5′-TTCTCCGAACGTGTCACGT-3′. The multiplicity of infection of
both the knockdown and overexpression lentivirus was 100. ipNF05.5
and ipNF95.6 2×105 cells were seeded onto six-well
plates (Corning, Inc.). After adding short hairpin negative control
(shNC) and shSUCLG1 to ipNF05.5 and OE-SUCLG1 and NC to ipNF95.6,
the cells were infected for 24 h at 37°C and then the medium was
replaced with a fresh high-glucose DMEM (Gibco; Thermo Fisher
Scientific, Inc.) containing 10% FBS (Procell Life Science &
Technology Co., Ltd.). Transfection efficiency was assessed based
on the expression of green fluorescent protein 72 h after
transfection by fluorescence microscopy. Successfully infected
cells were immediately subjected to subsequent experiments.
Inhibitor-treated cells
50 μM of the inhibitor of SLC25A1 (CTPI-2;
Cat. No. HY-123986; MedChemExpress) was added to the cells of
OE-SUCLG1 group, which were then incubated in a 37°C incubator for
24 h before subsequent functional experiments.
Cell Counting Kit 8 (CCK-8) assay
ipNF95.6 and ipNF05.5 2×103 cells were
seeded onto 96-well plates (Corning, Inc.). At 2 h (37°C after
seeding, 10 μl CCK-8 (Wuhan Elabscience Biotechnology Co.,
Ltd.) was added to each well. The optical density was measured
after 2, 12, 24 and 36 h at 450 nm by Multiskan FC Microplate
Photometer (Catalog #1410101, Thermo Fisher, USA).
Wound healing assay
A total of 2×105 virus-infected cells
were seeded onto six-well plates to 90% confluence and scratched.
After rinsing the free cells in PBS, the medium was replaced with
fresh DMEM without FBS. Cell migration was observed at 0 and 24 h
by light microscope. Magnification is 100x. Migration rate was
calculated using ImageJ 1.53e (National Institutes of Health) as
follows: (Width at 0 h-width at 24 h)/width at 0 h.
Flow cytometry
Virus-infected cells were washed twice with cold PBS
and resuspended in 1X Binding Buffer (PE Annexin V Apoptosis
Detection kit I; BD Biosciences; cat. no. 559763) at a
concentration of 1×106 cells/ml, according to the
manufacturer's instructions. PE Annexin V and 5 μl 7-AAD
were added to stain the cells. After incubation at 25°C for 15 min
in the dark, cell apoptosis was detected via flow cytometry. The
sum of early and late apoptotic cells gives the total percentage of
apoptotic cells in the sample. When determining cell cycle phase
using the Cell Cycle Detection kit (Nanjing KeyGen Biotech Co.,
Ltd.; cat. no. KGA512), a cell suspension containing
1×106 cells/ml was washed with PBS, centrifuged (380 x
g, 5 min) at 4°C and fixed with 70% ethanol at 4°C overnight. The
cells were stained with pre-prepared PI/RNase A Staining Solution
(1:9 ratio) at 25°C for 30 min in the dark before loading into
CytoFLEX S (Beckman Coulter, Inc.) for detection. Data were
analyzed using FlowJo 10.8.1 (BD Biosciences).
Transmission electron microscopy
After washing with PBS, the virus-infected cells
were pre-fixed with 3% glutaraldehyde at 4°C overnight and re-fixed
with 1% osmium tetroxide at 25°C for 2 h. This was followed by
dehydration, infiltration and embedding with Epon-812 (45345,
MerckMillipore) at 70°C overnight. Sections (60-90 nm) were cut
using an ultrathin sectioning machine (cat. no. UC7rt; Leica GmbH).
The sections were stained with uranyl acetate for 10-15 min at room
temperature, then with lead citrate for 1-2 min at room
temperature. Cells were observed and photographed using a
transmission electron microscope (JEOL Ltd.; cat. no.
JEM-1400FLASH).
ROS measurement
Complete medium of 1×106 virus-infected
cells was centrifuged (860 x g) at 4°C for 20 min, and the
supernatant was collected. ROS ELISA Research kit (Jiangsu ELISA
Industry Co., Ltd.; cat. no. MM-1893H2) was used to assess ROS
levels by measuring the absorbance of each well at 450 nm,
according to the manufacturer's instructions. The concentration of
the sample was calculated based on the standard curve.
Determination of metabolite content and
activity
A total of 1×106 virus-infected cells was
collected, and 1 ml extract solution (Keyybio; cat. no. ADS-W-S002)
was added. The cells were disrupted by ultrasonication at low power
on an ice bath (3 sec followed by 7 sec; total time, 3 min). After
centrifugation (13,680 x g) at 4°C for 10 min, the supernatant was
discarded. CA Content kit (Keyybio; cat. no. ADS-W-S002) was used
according to the manufacturer's instructions. After incubation at
room temperature for 20 min, absorbance A was measured at
470 nm to calculate ΔA=A blank-A measured.
Values from the standard curve were used to calculate the CA
content. Activity assays were performed using CS (cat. no.
MM-63621H2) and ACLY ELISA Research kits (both Keyybio; cat. no.
KYY-62138H1), according to the manufacturer's instructions.
Absorbance of each well was measured at 450 nm. The standard curve
was plotted, and the concentration of the samples was
calculated.
Metabolic energy assay
A total of 8 ml of 5×104/ml
virus-infected cell suspension was plated on XF96 cell culture
plates (Agilent Technologies, Inc.) and cultured at 37°C overnight.
The probe plate was hydrated with XF Calibrant and placed in a
non-CO2 cell culture incubator at 37°C overnight. On the
second day, the probe plate was re-hydrated with sterile water and
the detection solution was prepared to wash the cells. The
detection solution consisted of 97 ml 103575-100 Seahorse XF DMEM
(PH 7.4, Agilent Technologies, Inc.), 1 ml glucose (103577-100,
Agilent Technologies, Inc.), 1 ml pyruvate (103578-100, Agilent
Technologies, Inc.) and 1 ml glutamine (103579-100, Agilent
Technologies, Inc.) Seahorse XF Cell Mito Stress Test kit (cat. no.
103015-100) and Seahorse XF Glycolysis Seahorse XF (both Agilent
Technologies, Inc.; cat. no. 103020-100) were used according to the
manufacturer's instructions. The Agilent Seahorse XFe96 Analyzer
was used to measure oxygen consumption rate and extracellular
acidification rate.
Statistical analysis
The data were analyzed using GraphPad Prism 9.4.1
(Dotmatics, Inc.). Data are presented as the mean and standard
deviation, after three independent experimental replications.
One-way ANOVA and Tukey's post hoc test was performed to evaluate
the differences. Paired Student's t test was performed to evaluate
the differences between two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
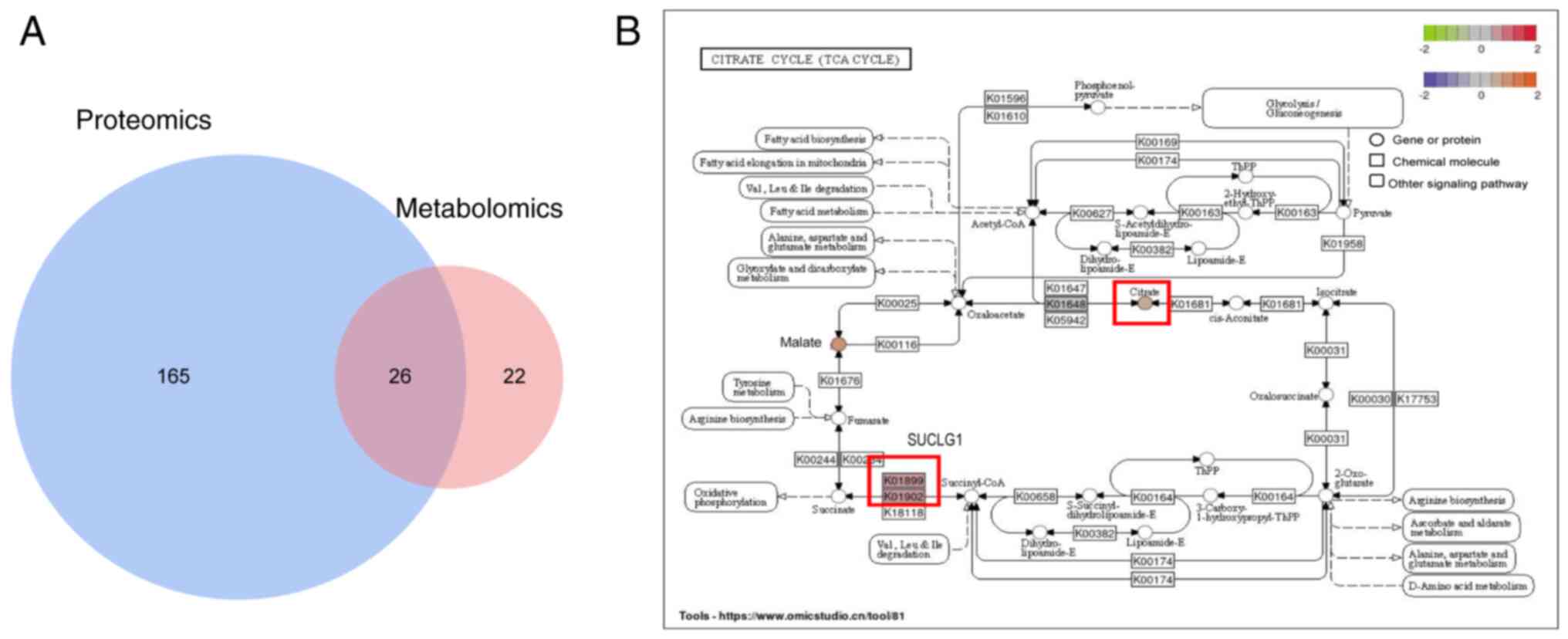
Proteomics and metabolomics results
Proteomic analysis was performed with the
tissues of patients with PNF and healthy individuals, whereas
non-targeted metabolomic analysis was performed with serum samples.
Table SIII shows differentially
expressed proteins measured by mass spectrometry sequencing and
metabolites measured by non-targeted sequencing. A total of 26
pathways were enriched (Fig. 1A).
Among these 26 pathways, the protein with the highest expression
was SUCLG1 (Table SIII). Based
on the results of the co-analysis, SUCLG1 affected PNF in the TCA
cycling pathway, which targeted CA (Fig. 1B).
Verification of SUCLG1 and CA levels
Immunofluorescence indicated that the levels of
SUCLG1 were higher in PNF tumor tissues than in normal skin tissues
(Fig. 2A and B). SUCLG1 was
distributed along the nerve. WB showed that at a cellular level,
expression of SUCLG1 in PNF cells was higher than that in HSC.
ipNF05.5 cells showed higher levels of SUCLG1 expression than
ipNF95.6 cells (Fig. 2C and D).
In addition, the content of CA in PNF cells was higher than that in
the cells of normal skin samples (Fig. 2E).
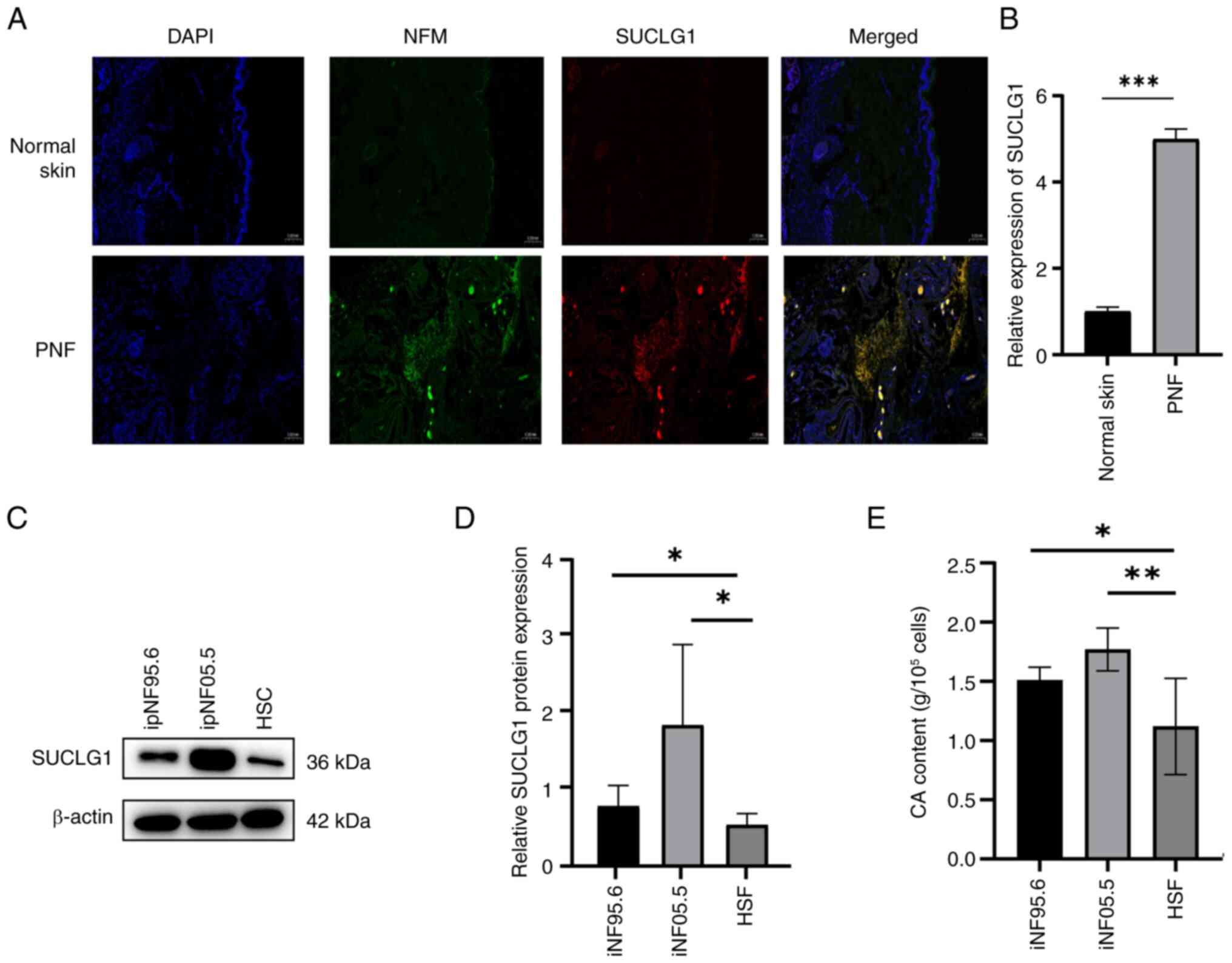 | Figure 2SUCLG1 and CA are highly expressed in
PNF. (A) Representative immunofluorescence staining of (B) SUCLG1
expression in normal skin and PNF. Magnification, x50x. (C) Western
blotting for (D) SUCLG1 expression in ipNF95.6, ipNF05.5 and HSCs.
(E) CA content in ipNF95.6, ipNF05.5 and HSCs.
*P<0.05, **P<0.01,
***P<0.001. SUCLG1, succinate-CoA ligase
GDP/ADP-forming subunit α; CA, citric acid; PNF, plexiform
neurofibroma; HSC, human Schwann cell; NFM, neurofilament
medium. |
Regulation of PNF cell function by
SUCLG1
SUCLG1 knockdown in ipNF05.5 cells and
overexpression in ipnF95.6 cells were induced to explore the in
vitro effects of SUCLG1. Transfection efficiency was verified
by fluorescence photography. Knockdown and overexpression
efficiency were confirmed by RT-qPCR and WB (Fig. 3A-C). CCK-8 assay indicated that
the proliferative capacity of PNF cells decreased following SUCLG1
knockdown and increased upon overexpression (Fig. 3D and E). Similarly, wound healing
assay revealed that the migratory ability of PNF cells decreased
following SUCLG1 knockdown and increased after SUCLG1
overexpression (Fig. 3F and G).
SUCLG1 knockdown resulted in a decrease in the proportion of cells
in S phase and a concomitant increase in the proportion of total
apoptotic cells (Q2 + Q3; Fig.
3H-K). By contrast, overexpression resulted in an increased
proportion of cells in S phase and suppressed the proportion of
total apoptotic cells.
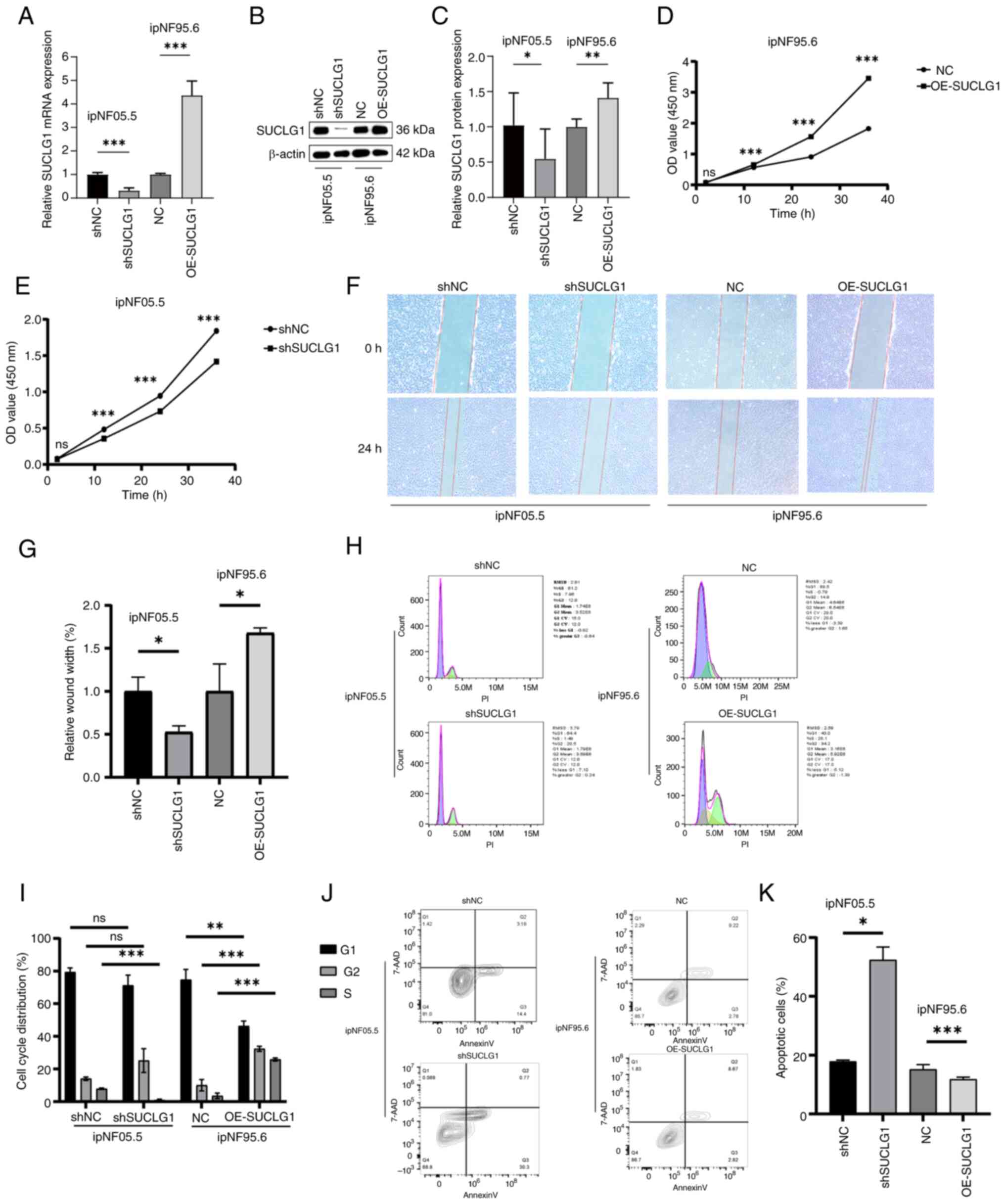 | Figure 3Regulation of PNF cell function by
SUCLG1 expression. (A) Reverse transcription-quantitative PCR
analysis of SUCLG1 in ipNF05.5 and ipNF95.6 cells after
transfection. (B) Western blotting of (C) SUCLG1 expression in
ipNF05.5 and ipNF95.6 after transfection. Proliferation of (D)
ipNF95.6 and (E) ipNF05.5 cells. (F) Wound healing assay showing,
Magnification, x100x. (G) Wound width of PNF cells after
transfection. (H) Representative flow cytometry of (I) cell cycle
distribution in PNF cells after transfection. (J) Representative
flow cytometry of (K) apoptosis in PNF cells after transfection.
*P<0.05, **P<0.01,
***P<0.001 vs. 0 h. ns, not significant; PNF,
plexiform neurofibroma; SUCLG1, succinate-CoA ligase
GDP/ADP-forming subunit α; sh, short hairpin; NC, negative control;
OE, overexpression; OD, optical density. |
Effect of SUCLG1 on intracellular
metabolism
To examine the effect of SUCLG1 on intracellular
metabolism, the present study used a mitochondrial stress test
(Fig. 4A). Basal respiration, ATP
production and maximum respiratory rate decreased after SUCLG1
knockdown. However, these parameters also increased following OE,
suggesting an improvement in the levels of mitochondrial aerobic
respiration (Fig. 4B and C).
There no significant differences in glycolysis levels between
groups (Fig. 4D and E). This
implies that SUCLG1 mainly regulated the level of mitochondrial
aerobic respiration in PNF cells and had no effect on the levels of
glycolysis.
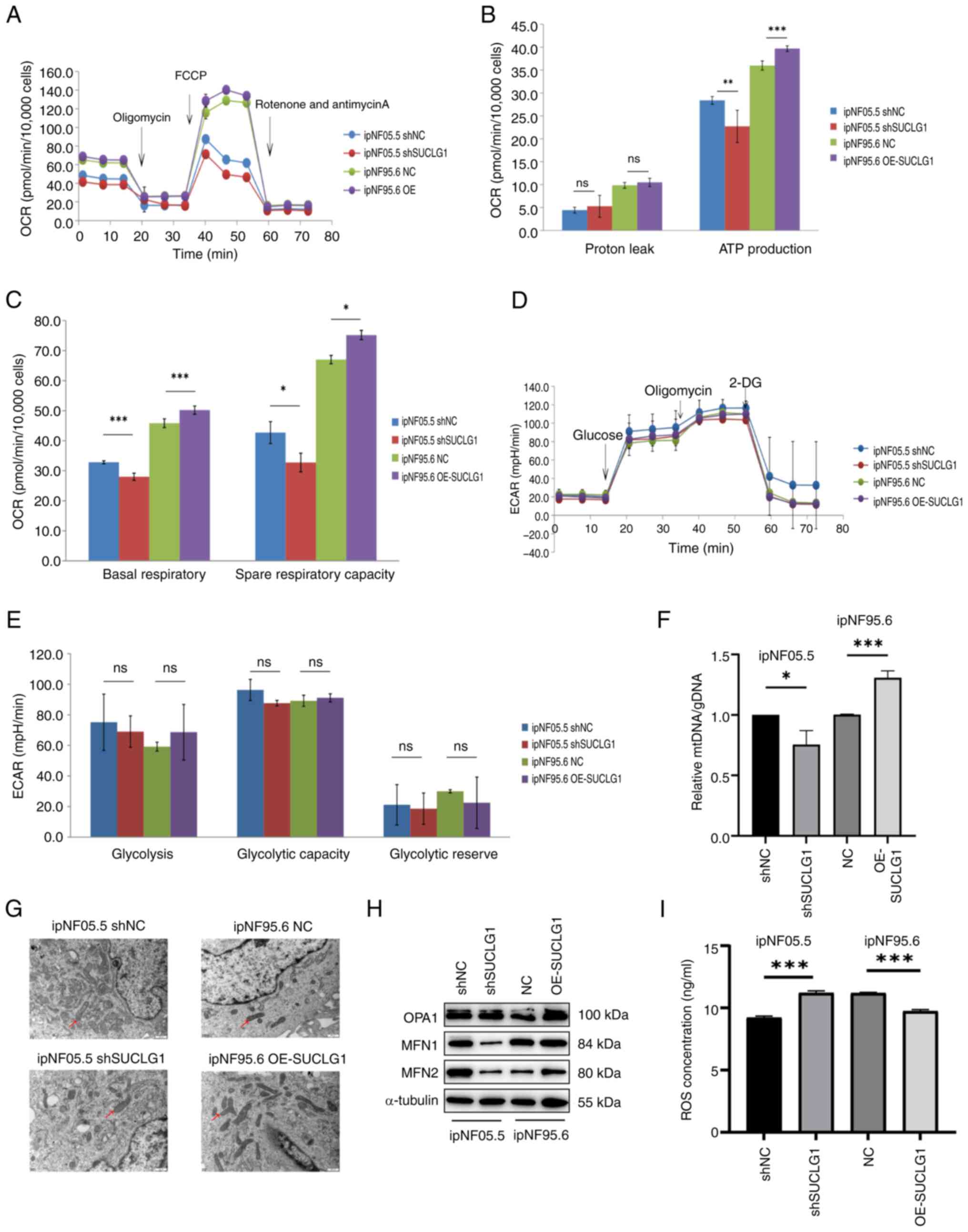 | Figure 4Effect of SUCLG1 on intracellular
metabolism and mitochondrial fusion. (A) OCR. (B) Changes in proton
leak and ATP production in mitochondrial respiration. (C) Changes
in basal respiration and spare respiratory capacity in
mitochondrial respiration. (D) Glycolysis stress test. (E)
Parameters of glycolytic function. (F) Mitochondrial mass was
estimated by ratio of mtDNA to gDNA. (G) Representative
transmission electron microscopy images of mitochondria.
Magnification: 500x. Arrows indicate elongated mitochondria. (H)
Representative western blotting of mitochondrial fusion proteins
OPA1, MFN1 and MFN2. (I) ROS concentration. *P<0.05,
**P<0.01, ***P<0.001. ns, not
significant; OCR, oxygen consumption rate; ECAR, extracelluar
acidification rate; SUCLG1, succinate-CoA ligase GDP/ADP-forming
subunit α; ROS, reactive oxygen species; mtDNA, mitochondrial DNA;
OPA, Optic Atrophy Protein ; MFN, mitochondrial fusion protein; sh,
short hairpin; NC, negative control; FCCP, trifluoromethoxy
carbonylcyanide phenylhydrazone; DG, deoxy-glucose; OE,
overexpression. |
Effect of SUCLG1 on mitochondrial quality
control
To assess mitochondrial changes supporting increased
aerobic respiration., RT-qPCR was performed to determine the
relative copy number of mtDNA) with respect to the genomic DNA
(gDNA) to determine mitochondrial DNA content. Ratio of mtDNA to
gDNA increased when SUCLG1 was overexpressed and decreased when
SUCLG1 expression was knocked down (Fig. 4F). As aerobic respiration may have
been regulated by changes in mitochondrial dynamics (16), transmission electron microscopy
was performed to evaluate mitochondrial morphology. When SUCLG1 was
highly expressed, mitochondria appeared elongated and deformed,
which was hypothesized to be a sign of fusion (Fig. 4G). Compared with NC, cells in the
OE-SUCLG1 had higher levels of mitochondrial fusion proteins (MFN1
and 2 and OPA1; Fig. 4H-K).
Therefore, ROS measurement was performed to evaluate mitochondrial
damage. The intracellular ROS levels were lower in the OE-SUCLG1,
indicating reduced damage, but higher in shSUCLG1, indicating
increased damage (Fig. 4L).
Effect of SUCLG1 expression on
SLC25A1
Proteomics and metabolomics showed that in addition
to SUCLG1, expression of CA was also affected in the TCA pathway
(Table SIII). Thus, proteins
that were associated with CA (CS, ACLY and SLC25A1) were
investigated. Based on the WB results, SLC25A1 expression was
increased when SUCLG1 expression was increased, and when SUCLG1
expression was decreased, then SLC25A1 was similarly decreased,
whereas ACLY and CS were not affected by changes in SUCLG1
(Fig. 5A-D). Enzymatic activity
assays also indicated no significant changes in the activities of
the latter two proteins (Fig. 5E and
F). For SLC25A1, partial functional recovery was observed.
Compared with the OE-SUCLG1, cell function was inhibited in the
OE-SUCLG1 inhibitor group (Fig.
5G-M). Cells in the OE group exhibited higher proliferation and
wound healing rates (Fig. 5G-I).
However, after adding the inhibitor, these abilities decreased but
did not return to baseline, indicating that the function was only
partially restored. The proportion of cells in S phase was
significantly lower in cells with the addition of the inhibitor
than in OE group but did not return to baseline (Fig. 5J and K). Apoptosis was also
partially restored by the inhibitor (Fig. 5L and M).
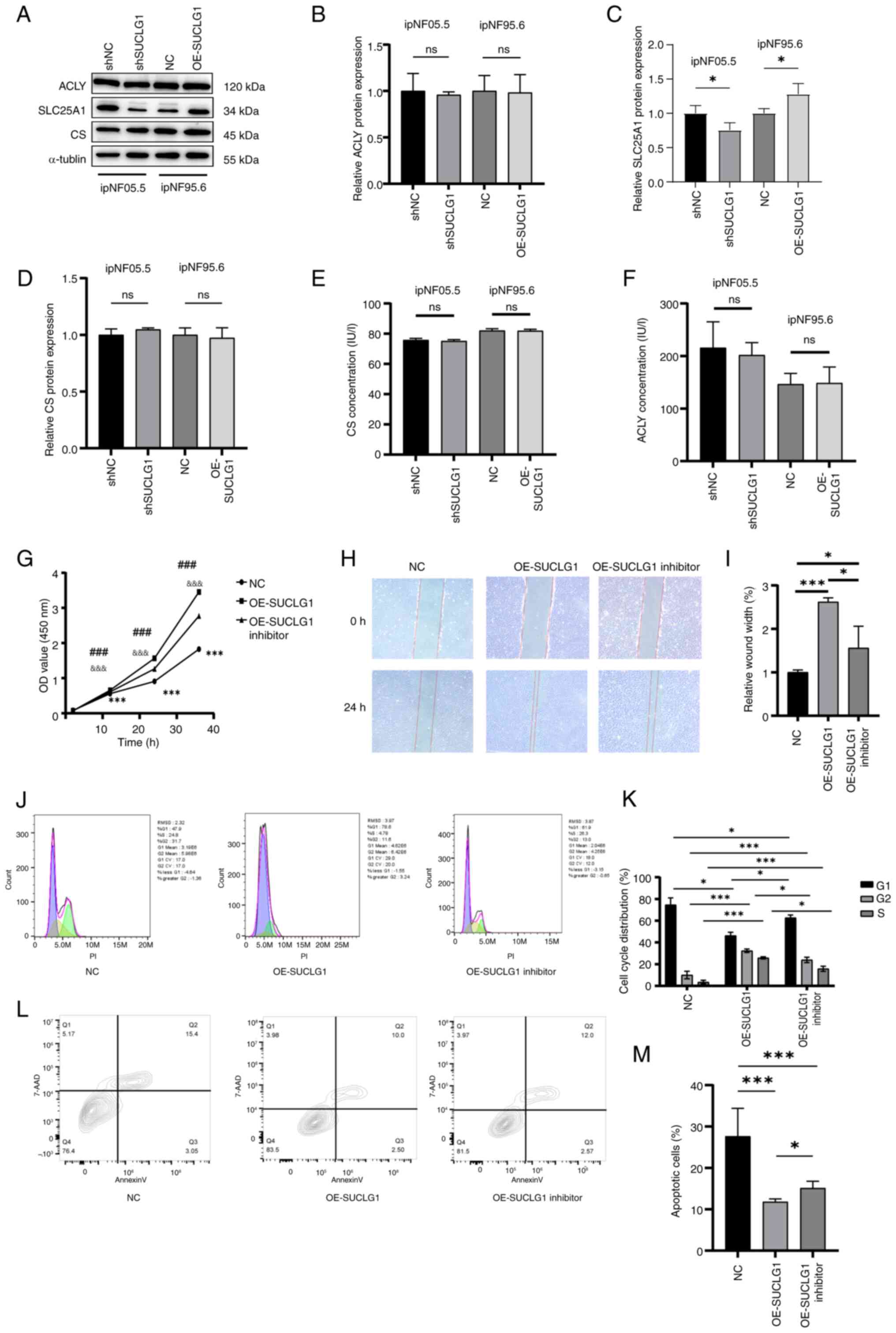 | Figure 5Effect of SUCLG1 expression on
SLC25A1. (A) Representative Western blotting for (B) ACLY, (C)
SLC25A1 and (D) CS. ELISA determination of (E) CS and (F) ACLY
protein activity. (G) Cell proliferation was evaluated by the Cell
Counting Kit-8 assay. (H) Wound healing assay. Magnification, x100.
(I) Wound width of ipNF95.6 after transfection with NC and
OE-SUCLG1 viruses and addition of SLC25A1 inhibitor. (J) Flow
cytometry of (K) cell cycle distribution following transfection
with NC and OE-SUCLG1 viruses and adding SLC25A1 inhibitor. (L)
Flow cytometry of (M) apoptosis following transfection and adding
SLC25A1 inhibitor. &&&P<0.001 vs.
OE-SUCLG1 inhibitor. ***P<0.001 vs. NC,
###P<0.001 vs. OE-SUCLG1, *P<0.05 vs.
NC. SUCLG1, succinate-CoA ligase GDP/ADP-forming subunit α; ns, no
significance; OD, optical density; SLC25A1, ;Solute Carrier Family
25 Member 1; ACLY, ATP Citrate Lyase; CS, Citrate Synthase; NC,
negative control; OE, overexpression; sh, short hairpin. |
Discussion
PNF, a common clinical manifestation of the rare
genetic disorder NFI (17,18),
develops in ~1/3 of patients (3).
Although it is a benign tumor, there is a risk of transformation
into MPNST, which occurs at the highest risk in childhood and
adolescence and has a poor prognosis (19,20). A total of 8-13% of PNFs are at
risk of developing Malignant Peripheral Nerve Sheath Tumor
(21). Conventional radiotherapy
is ineffective for PNF and surgery remains the only potentially
effective treatment (6). In April
2020, the U.S. Food and Drug Administration approved the use of
simetinib for pediatric patients aged ≥3 years for the treatment of
inoperable symptomatic and/or progressive PNF; this provides
symptomatic relief and tumor shrinkage in certain patients, but the
drug remains ineffective in some patients, and the disease
continues to progress (22,23). Although other MEK inhibitors have
entered clinical trials (24-29), the role of metabolic changes in
the disease is not yet known.
SUCLG1 is the α-subunit encoding the
heterodimerization enzyme succinate coenzyme A ligase (30); previous studies (9,10)
did not find the gene to be associated with tumors, but its
mutation was associated with mitochondrial DNA depletion syndrome
(11). However, a recent study by
Yan et al (12), found
that SUCLG1 restricts Polymerase (RNA) Mitochondrial (DNA Directed)
succinylation to enhance mitochondrial biogenesis and leukemia
progression. This suggests a potential association between SUCLG1
and tumors but to the best of our knowledge, this has been little
studied (10-12). Here, SUCLG1 expression was
upregulated in PNF, indicated by tissue mass spectrometry, and its
expression was confirmed in tissue and cells. In vitro
experiments showed that SUCLG1 promoted tumor cell proliferation
and migration, inhibited apoptosis and affected the cell cycle,
with an increased proportion of cells in S phase when expression is
elevated.
The present mass spectrometry and metabolic analyses
revealed that high expression of SUCLG1 and CA occurs in the TCA
cycle pathway. The TCA cycle occurs in mitochondria; dysfunctional
mitochondrial quality control is associated with development of
numerous types of diseases, including tumors (31). Mitochondrial quality control
involves numerous mechanisms that are largely dependent on the
extent of mitochondrial damage, activating appropriate repair
pathways (32). In this process,
cells implement quality control through mitochondrial fusion and
fission (33-35). SUCLG1 is a regulatory factor
acting on mitochondria and previous studies (12,30) have revealed that it has a positive
regulatory effect on mitochondrial quality, which was further
validated in the present study (12). When the expression of SUCLG1
increased, there was an increase in the ratio of mitochondrial DNA
to total genomic DNA and mitochondrial mass and fusion. ROS
production was effectively suppressed, potentially because
mitochondrial fusion decreased the degree of mitochondrial damage
(36,37). These results suggested that in PNF
cells, SUCLG1 exerts a key influence on mitochondrial quality
control and promotes mitochondrial fusion.
The Warburg effect-where cells produce energy
through aerobic glycolysis in the presence of sufficient oxygen and
with intact mitochondrial function-is a key factor in driving
cancer progression, leading to resistance to conventional therapy
and poor patient prognosis (38,39). In certain tumor cases,
mitochondrial defects, due to certain mutations in TCA cycle
enzymes and overproduction of mitochondrial ROS, serve a key role
in promoting the Warburg effect and tumor progression (38). By contrast, testing mitochondrial
and glycolytic stress here demonstrated that SUCLG1, when expressed
at elevated levels, primarily promoted mitochondrial respiratory
capacity, with little effect on glycolytic processes. This result
is different from previous results (38-40), which may be because the Warburg
effect has been studied primarily in cancer and the tumors in the
present study were benign and did not develop as fast as malignant
tumors, therefore not requiring the Warburg effect to promote tumor
development. It is possible that when PNFs are transformed into
MPNATs, the Warburg pathway is initiated, providing a favorable
microenvironment for malignant tumor progression. Here, more
undamaged mitochondria provided sufficient energy for PNF cells.
However, to demonstrate the promotion of tumor progression through
SUCLG1 via aerobic respiration, further experiments is still
needed.
When SUCLG1 levels increased, levels of metabolite
CA, which is involved in the TCA cycle, were elevated. Key enzymes
affecting its content, CS, ACLY and SLC25A1, may be directly
responsible for this phenomenon. The first two enzymes are key
enzymes in the TCA cycle and their expression was here unaffected.
While SLC25A1 is a transporter protein on the mitochondrial
membrane, SUCLG1 increased the mitochondrial mass, so it is likely
that expression of SLC25A1 protein increased as well. Metabolite
assay also revealed that the expression of CA increased with
elevated expression of SLC25A1. Thus, it was demonstrated that
SUCLG1 affected CA expression by regulating SLC25A1 expression.
Following addition of an inhibitor of SLC25A1 to cells
overexpressing SUCLG1, function of PNF cells was partially
restored, demonstrating that SUCLG1 affected the development of PNF
cells via SLC25A1.
In vitro experiments proved that SUCLG1
served a key role in the progression of PNF and promoted aerobic
respiration metabolism, but this needs to be verified in
vivo. The present results not only provide a new potential
target for the treatment of PNF, but also lay a preliminary
foundation for further study of metabolic mechanisms.
Supplementary Data
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to ongoing study but
may be requested from the corresponding author.
Authors' contributions
QL and RH confirm the authenticity of all the raw
data. ZZ conducted the experiments and analyzed the data. RH, QL
and ZZ designed the study. RH and QL supervised the study. ZZ wrote
the manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by Committee for
Ethical Review of Research involving Human Subjects of Shandong
Provincial Hospital, Jinan, China (approval no. SWYX2024-556). All
human specimens were collected with informed written consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science
Foundation of China (grant no. 82172227).
References
|
1
|
Miller DT, Freedenberg D, Schorry E,
Ullrich NJ, Viskochil D and Korf BR; Council on Genetics and
American College of Medical Genetics and Genomics: Health
supervision for children with neurofibromatosis type 1. Pediatrics.
143:e201906602019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Acar S, Armstrong AE and Hirbe AC:
Plexiform neurofibroma: Shedding light on the investigational
agents in clinical trials. Expert Opin Investig Drugs. 31:31–40.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fisher MJ, Blakeley JO, Weiss BD, Dombi E,
Ahlawat S, Akshintala S, Belzberg AJ, Bornhorst M, Bredella MA, Cai
W, et al: Management of neurofibromatosis type 1-associated
plexiform neurofibromas. Neuro Oncol. 24:1827–1844. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gross AM, Glassberg B, Wolters PL, Dombi
E, Baldwin A, Fisher MJ, Kim A, Bornhorst M, Weiss BD, Blakeley JO,
et al: Selumetinib in children with neurofibromatosis type 1 and
asymptomatic inoperable plexiform neurofibroma at risk for
developing tumor-related morbidity. Neuro Oncol. 24:1978–1988.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu B, Zheng T, Wang W, Gu Y, Wei C, Li Q
and Wang Z: Genotype-phenotype correlations of neurofibromatosis
type 1: A cross-sectional study from a large Chinese cohort. J
Neurol. 271:1893–1900. 2024. View Article : Google Scholar
|
|
6
|
Zhu B, Wei C, Wang W, Gu B, Li Q and Wang
Z: Treatment and progress of cutaneous neurofibroma. Zhongguo Xiu
Fu Chong Jian Wai Ke Za Zhi. 36:1064–1071. 2022.In Chinese.
PubMed/NCBI
|
|
7
|
Dombi E, Baldwin A, Marcus LJ, Fisher MJ,
Weiss B, Kim A, Whitcomb P, Martin S, Aschbacher-Smith LE, Rizvi
TA, et al: Activity of selumetinib in neurofibromatosis type
1-related plexiform neurofibromas. N Engl J Med. 375:2550–2560.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gross AM, Wolters PL, Dombi E, Baldwin A,
Whitcomb P, Fisher MJ, Weiss B, Kim A, Bornhorst M, Shah AC, et al:
Selumetinib in children with inoperable plexiform neurofibromas. N
Engl J Med. 382:1430–1442. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eniafe J and Jiang S: The functional roles
of TCA cycle metabolites in cancer. Oncogene. 40:3351–3363. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen YM, Chen W, Xu Y, Lu CS, Zhu MM, Sun
RY, Wang Y, Chen Y, Shi J and Wang D: Novel compound heterozygous
SUCLG1 variants may contribute to mitochondria DNA depletion
syndrome-9. Mol Genet Genomic Med. 10:e20102022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ramsheh S, Omidvar ME, Tabasinezhad M,
Alipoor B, Salmani TA and Ghaedi H: SUCLG1 mutations and
mitochondrial encephalomyopathy: A case study and review of the
literature. Mol Biol Rep. 47:9699–9714. 2020. View Article : Google Scholar
|
|
12
|
Yan W, Xie C, Sun S, Zheng Q, Wang J, Wang
Z, Man CH, Wang H, Yang Y, Wang T, et al: SUCLG1 restricts POLRMT
succinylation to enhance mitochondrial biogenesis and leukemia
progression. EMBO J. 43:2337–2367. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Icard P, Simula L, Zahn G, Alifano M and
Mycielska ME: The dual role of citrate in cancer. Biochim Biophys
Acta Rev Cancer. 1878:1889872023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arnold PK, Jackson BT, Paras KI, Brunner
JS, Hart ML, Newsom OJ, Alibeckoff SP, Endress J, Drill E, Sullivan
LB and Finley LWS: A non-canonical tricarboxylic acid cycle
underlies cellular identity. Nature. 603:477–481. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Thoudam T, Chanda D, Sinam IS, Kim BG, Kim
MJ, Oh CJ, Lee JY, Kim MJ, Park SY, Lee SY, et al: Noncanonical
PDK4 action alters mitochondrial dynamics to affect the cellular
respiratory status. Proc Natl Acad Sci USA. 119:e21201571192022.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Well L, Döbel K, Kluwe L, Bannas P,
Farschtschi S, Adam G, Mautner VF and Salamon J: Genotype-phenotype
correlation in neurofibromatosis type-1: NF1 whole gene deletions
lead to high tumor-burden and increased tumor-growth. PLOS Genet.
17:e10095172021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bernier A, Larbrisseau A and Perreault S:
Cafe-au-lait macules and neurofibromatosis type 1: A review of the
literature. Pediatr Neurol. 60:24–29.e1. 2016. View Article : Google Scholar
|
|
19
|
Nicoli TK, Saat R, Tarkkanen J, Kinnunen
I, Mäkitie AA and Jero J: Challenging management of plexiform
schwannoma and plexiform neurofibroma. J Craniofac Surg.
33:803–808. 2022. View Article : Google Scholar
|
|
20
|
Avery RA, Katowitz JA, Fisher MJ, Heidary
G, Dombi E, Packer RJ and Widemann BC; OPPN Working Group:
Orbital/periorbital plexiform neurofibromn children with
neurofibromatosis type 1: Multidisciplinary recommendations for
care. Ophthalmology. 124:123–132. 2017. View Article : Google Scholar
|
|
21
|
Wang W, Gu Y, Zhu B, et al: Retrospective
study of surgical treatment in 121 patients with head and neck
plexiform neurofibromas. Chin J Plast Surg. 35:169–178. 2024.
|
|
22
|
Jackson S, Baker EH, Gross AM, Whitcomb P,
Baldwin A, Derdak J, Tibery C, Desanto J, Carbonell A, Yohay K, et
al: The MEK inhibitor selumetinib reduces spinal neurofibroma
burden in patients with NF1 and plexiform neurofibromas. Neurooncol
Adv. 2:vdaa0952020.PubMed/NCBI
|
|
23
|
Veres K, Bene J, Hadzsiev K, Garami M,
Pálla S, Happle R, Medvecz M and Szalai ZZ: Superimposed mosaicism
in the form of extremely extended segmental plexiform neurofibroma
caused by a novel pathogenic variant in the NF1 gene. Int J Mol
Sci. 24:121542023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang D, Ge L, Guo Z, Li Y, Zhu B, Wang W,
Wei C, Li Q and Wang Z: Efficacy and safety of trametinib in
neurofibromatosis type 1-associated plexiform neurofibroma and
low-grade glioma: A systematic review and meta-analysis.
Pharmaceuticals (Basel). 15:9562022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Armstrong AE, Belzberg AJ, Crawford JR,
Hirbe AC and Wang ZJ: Treatment decisions and the use of MEK
inhibitors for children with neurofibromatosis type 1-related
plexiform neurofibromas. BMC Cancer. 23:5532023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weiss BD, Wolters PL, Plotkin SR, Widemann
BC, Tonsgard JH, Blakeley J, Allen JC, Schorry E, Korf B, Robison
NJ, et al: NF106: A neurofibromatosis clinical trials consortium
Phase II trial of the MEK inhibitor Mirdametinib (PD-0325901) in
adolescents and adults with NF1-related plexiform neurofibromas. J
Clin Oncol. 39:797–806. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rizzo D, Ruggiero A, Amato M, Maurizi P
and Riccardi R: BRAF and MEK inhibitors in pediatric glioma: New
therapeutic strategies, new toxicities. Expert Opin Drug Metab
Toxicol. 12:1397–1405. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fisher MJ, Shih CS, Rhodes SD, Armstrong
AE, Wolters PL, Dombi E, Zhang C, Angus SP, Johnson GL, Packer RJ,
et al: Cabozantinib for neurofibromatosis type 1-related plexiform
neurofibromas: A phase 2 trial. Nat Med. 27:165–173. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gross AM, Glassberg B, Wolters PL, Dombi
E, Baldwin A, Fisher MJ, Kim A, Bornhorst M, Weiss BD, Blakeley JO,
et al: Selumetinib in children with neurofibromatosis type 1 and
asymptomatic inoperable plexiform neurofibroma at risk for
developing tumor-related morbidity. Neuro Oncol. 24:1978–1988.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chinopoulos C, Batzios S, van den Heuvel
LP, Rodenburg R, Smeets R, Waterham HR, Turkenburg M, Ruiter JP,
Wanders RJA, Doczi J, et al: Mutated SUCLG1 causes mislocalization
of SUCLG2 protein, morphological alterations of mitochondria and an
early-onset severe neurometabolic disorder. Mol Genet Metab.
126:43–52. 2019. View Article : Google Scholar
|
|
31
|
Ho GT and Theiss AL: Mitochondria and
inflammatory bowel diseases: Toward a stratified therapeutic
intervention. Annu Rev Physiol. 84:435–459. 2022. View Article : Google Scholar :
|
|
32
|
Sugiura A, Mclelland GL, Fon EA and
Mcbride HM: A new pathway for mitochondrial quality control:
Mitochondrial-derived vesicles. EMBO J. 33:2142–2156. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Z, Li TE, Chen M, Xu D, Zhu Y, Hu
BY, Lin ZF, Pan JJ, Wang X, Wu C, et al: MFN1-dependent alteration
of mitochondrial dynamics drives hepatocellular carcinoma
metastasis by glucose metabolic reprogramming. Br J Cancer.
122:209–220. 2020. View Article : Google Scholar :
|
|
34
|
You MH, Jeon MJ, Kim SR, Lee WK, Cheng SY,
Jang G, Kim TY, Kim WB, Shong YK and Kim WG: Mitofusin-2 modulates
the epithelial to mesenchymal transition in thyroid cancer
progression. Sci Rep. 11:20542021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Wang Y, Liu W, Ding L, Zhang X,
Wang B, Tong Z, Yue X, Li C, Xu L, et al: TIM-4 orchestrates
mitochondrial homeostasis to promote lung cancer progression via
ANXA2/PI3K/AKT/OPA1 axis. Cell Death Dis. 14:1412023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Roca-Portoles A and Tait SWG:
Mitochondrial quality control: From molecule to organelle. Cell Mol
Life Sci. 78:3853–3866. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yao CH, Wang R, Wang Y, Kung CP, Weber JD
and Patti GJ: Mitochondrial fusion supports increased oxidative
phosphorylation during cell proliferation. ELife. 8:e413512019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vaupel P and Multhoff G: Revisiting the
Warburg effect: Historical dogma versus current understanding. J
Physiol. 599:1745–1757. 2021. View Article : Google Scholar
|
|
39
|
Wang Y and Patti GJ: The Warburg effect: A
signature of mitochondrial overload. Trends Cell Biol.
33:1014–1020. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhong X, He X, Wang Y, Hu Z, Huang H, Zhao
S, Wei P and Li D: Warburg effect in colorectal cancer: The
emerging roles in tumor microenvironment and therapeutic
implications. J Hematol Oncol. 15:1602022. View Article : Google Scholar : PubMed/NCBI
|