Introduction
Multiple myeloma (MM) is a hematologic malignancy
characterized by clonal proliferation and focal proliferation of
terminally differentiated plasma cells in the bone marrow (BM),
which produce monoclonal immunoglobulin in the blood or urine
(1). The primary clinical
manifestations of MM are bone dise(ase, including osteolytic bone
lesions and pathological fractures (2). Bone disease occurs in up to 90% of
patients. MM cell proliferation is highly dependent on the BM
microenvironment (BMME) and its adhesive interactions with
extracellular matrix components, including fibronectin and collagen
(3). Enhanced adhesion of MM
cells promotes their homing to the BM (4), followed by malignant proliferation
that exacerbates bone destruction (5). Therefore, understanding the
mechanisms of MM cell proliferation, migration and homing into the
BM is essential for developing new strategies for MM treatment.
As the fourth member of the G protein-coupled
receptors, LGR4 is involved in multiple physiological and
pathological processes, including embryonic development (6), stem cell maintenance (7), bone remodeling (8) and tumorigenesis. A previous research
by the authors has demonstrated that LGR4 plays a role in
regulating the number of fetal liver hematopoietic stem cells
(9). It has been reported that
LGR4-deficient mice exhibit multiple organ defects, such as eye
(10), bone (8) and reproductive organs (11), and exhibit abnormal energy
metabolism (12). Additionally,
LGR4 is known as a key regulator of osteoblast and osteoclast
differentiation (8). The high
expression of LGR4 has been associated with poor prognosis of
multiple cancers. LGR4 is upregulated in cancer and is involved in
regulating tumorigenic processes. LGR4 promotes cell migration,
invasion and proliferation in prostate, colorectal and cervical
cancers (13). In colorectal
cancer, LGR4 directly induces cell ferroptosis and drug resistance
through Wnt-βcatenin signaling (14). Moreover, LGR4 enhances
osteoclastic premetastatic niche formation and promotes bone
metastasis in breast cancer cells through the Gαq and β-catenin
signaling pathways (15). These
findings suggest potential crosstalk between tumor cell receptors
and BMME during cancer progression. Additionally, it has been
indicated that the LGR4/R-spondin axis plays a crucial role in
activating Wnt signaling in MM (16). LGR4 is highly expressed in
patients with MM, promoting MM cell proliferation (17). A previous study has reported that
LGR4 can activate the NF-κB signaling pathway. It has been reported
that activated NF-κB signaling enhances the ability of
hematopoietic stem cell homing (18). However, the unique function and
mechanism of LGR4 in MM remain unclear. It remains unclear whether
LGR4, a membrane protein, increases the interaction between MM
cells and BMME, thereby promoting cell homing to BM and
accelerating MM progression.
In the present study, it was demonstrated that LGR4
was positively associated with cell adhesion molecules, and its
high expression was associated with poor prognosis in MM. It was
aimed to investigate the effects of LGR4 on MM progression through
its role in cell adhesion, migration and BM homing both in
vitro and in vivo. The present findings suggest that
targeting LGR4 can offer a potential therapeutic strategy for MM
treatment.
Materials and methods
Clinical samples
BM samples derived from healthy donors (HD; n=5) and
newly diagnosed patients with MM (n=9) were obtained from Xiangya
Hospital, the Second Xiangya Hospital of Central South University
(Changsha, China), from January 2020 to June 2023.
CD138+ plasma cells were isolated by using anti-human
CD138 magnetic beads (Miltenyi Biotec GmbH) and incubated in 4°C
for 15 min with monocytes isolated from BM samples using lymphocyte
separation medium (cat. no. LTS1077; TBD; https://www.tbdscience.com/). The patients with MM
enrolled in the present study were newly diagnosed. International
Myeloma Working Group criteria (19) were processed by hematologists for
the diagnosis of MM. Patients with monoclonal gammopathy of
undetermined significance (MGUS) and smoldering MM were excluded,
together with patients with MM combined with other diseases. The
clinical information of the enrolled patients with MM in the
present study is included in Table
SI. The present study was approved by Cancer Research Institute
Review Board of Central South University (Changsha, China).
Cell culture
The human MM cell lines ARP1 (20), KMS28-BM (21), KMS28-pleural effusion (PE)
(20) and OCI-My5 (20,22), were utilized to explore the
function of LGR4 in MM. Cell lines were cultured in Roswell Park
Memorial Institute (RPMI)-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
cat. no. FSP500; Shanghai ExCell Biology, Inc.) and 1% penicillin
and streptomycin (P/S; Gibco; Thermo Fisher Scientific, Inc.). The
cell lines have been used in previous studies (20,22). The human MM cell line
OCI-T3rd-luc (derived from OCI-My5-luc) was established
by our group using tail vein injection into NCG mice with three
rounds of homing transplantation and enriched from BM. KMS28-BM
(21) and KMS28-PE cells are the
paired cell lines originated from BM and PE of MM patients with
immortalization. The human BM stromal cell line HS5 (a gift from
Dr. Jiaxi Zhou, Institute of Hematology, Chinese Academy of Medical
Sciences) was maintained in DMEM low glucose supplemented with 10%
FBS and 1% P/S.
Reagents and antibodies
Reagents included QNZ (cat. no. EVP4593; Selleck
Chemicals) and doxycycline (DOX; cat. no. 24390-14-5;
MilliporeSigma). The antibodies were as follows: Anti-LGR4
(1:1,000; cat. no. A12657; for western blotting), anti-β-actin
(1:5,000; cat. no. AC004), anti-nuclear factor kappa B (NF-κB) 2
(1:1,000; cat. no. A19605), anti-P-NF-κB2-S866 (1:1,000; cat. no.
Ap0418), anti-IκBα (1:1,000; cat. no. A19714), anti-P-IκBα-S36
(1:1,000; cat. no. A191714), anti-Snail (1:1,000; cat. no. A5243)
and anti-TNFRSF1B (1:1,000; cat. no. A13556) were all obtained from
ABclonal Biotech Co., Ltd. Anti-GAPDH (1:5,000; cat. no.
10494-1-AP), anti-MUC2 (1:1,000; cat. no. 27675-1-AP), anti-Caspase
(1:1,000; cat. no. 319677-1-AP) and anti-Zinc Finger E-Box Binding
Homeobox 1 (ZEB1; 1:1,000; cat. no. 21544-1-AP) were all obtained
from Proteintech Group, Inc. Anti-poly-(ADP-ribose) polymerase
(PARP) antibody (1:1,000; cat. no. 9532), anti-cleaved caspase 3
antibody (1:1,000; cat. no. 9664) and anti-Vimentin (1:1,000; cat.
no. D21H3) were obtained from Cell Signaling Technology, Inc.
Anti-N-cadherin (1:800; cat. no. WL011047) was obtained from
Wanleibio Co., Ltd. PE anti-human CD138 (cat. no. 352306) was
obtained from BioLegend, Inc. and CXCR12 (cat. no. 350-NS) from
R&D Systems, Inc.
Vectors and transfections
LGR4-overexpression (LGR4-OE) constructs were
constructed by cloning LGR4 cDNA into a pSIN-EF2-Puro (23) lentiviral vector using EcoRI (cat.
no. R3101S; New England Biolabs) and BamHI (cat. no. R3136S; New
England Biolabs). LGR4-knockdown constructs using two pairs of
short hairpin RNA sequences (shRNA1 and shRNA2) were ligated into a
pLKO-tet-on lentiviral vector. Aim Lentiviruses (5 μg) were
packaged in 293T cells (a gift from Dr. Rong Chang, Kunming
Institute of Zoology, the Chinese Academy of Sciences) using pMD2G
(1.25 μg) and psPAX2 (3.75 μg) helper vectors and
polybrene (8 μg/ml)-mediated transduction (cat. no.
H9268-5G; MilliporeSigma). After 60 h the 10 ml virus was collected
and 1ml virus was used to transfect 1×106 ARP1 or
OCI-My5 cell lines in 1 ml medium. A total of 48-72 h after the
transfection, puromycin (1 μg/ml) was added to screen the
positive cells. The final concentration of siRNA transfection was
50 nM. Transient transfection was performed using a Nefect DNA
Transfection reagent (cat. no. TF20121201; Neofect; http://www.neofect.cn/) according to the
specification. All primer and siRNA sequences are listed in
Tables SII and SIII.
Western blotting
Western blot analysis was performed as previously
described (20). Proteins were
separated by 10% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to a 0.45-μm
polyvinylidene fluoride membrane. The blots were then probed with
specific primary antibodies overnight at 4°C, followed by
HRP-conjugated secondary antibodies incubation for 1 h at room
temperature (RT). Protein signals were developed with SuperSignalTM
West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific,
Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
To detect mRNA expression in MM cells, RT-qPCR was
performed as previously described (22). Total RNA was extracted using
TRIzol® (cat. no. 15596026; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Total RNA was
reverse transcribed using the SuperScript™ II Reverse Transcriptase
kit (cat. no. 18064071; Thermo Fisher Scientific, Inc.). qPCRs were
performed by using ABsolute qPCR SYBR Green Mixes (cat. no.
AB1163A; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. All primer sequences are listed in
Table SIV.
Immunofluorescence analysis
A total of 4×104 CD138+ cells
were spun down on glass slides and then fixed with methanol for 15
min at 20°C. Diluted LGR4 antibodies (1:200) and ZEB1 antibodies
(1:200) were placed on glass slides and incubated overnight at 4°C.
Then, the cells were incubated with secondary antibodies (1:1,000)
conjugated with Goat anti-Rabbit Alexa Fluor™ 488 (cat. no.
A-11008; Thermo Fisher Scientific, Inc.) or Goat anti-Mouse Alexa
Fluor™ 594 (cat. no. A-11012; Thermo Fisher Scientific, Inc.) for 2
h at RT (protected from light). Nuclei were labeled with
4′,6-diamidino-2-phenylindole (DAPI, 1 mg/ml; 1:5,000) (cat. no.
S2110; Beijing Solarbio Science & Technology Co., Ltd.).
Fluorescence was observed under a Leica fluorescence microscope.
The fluorescence intensity was quantified using ImageJ 1.54
software (National Institutes of Health).
Cell proliferation and viability
assay
To determine cell proliferation, MM cells were
plated in 24-well plates with 5,000 cells per well by counting
alive cells after trypan blue exclusion. Cell numbers were counted
using a cell counting chamber for six days. To determine cell
viability, MM cells were plated in 96-well plates at a density of
5,000 cells per well. The cells were treated with QNZ for 48 h and
counted using Cell Counting Kit-8 (cat. no. B34302; Bimake). For
each well, 10 μl of reagent was added, followed by
incubation at 37°C for 2-3 h. The optical density (OD) was measured
at 450 nm. Each test was repeated three times. IC50 was
calculated using GraphPad Prism 9 software.
Soft-agar colony formation assay
Soft agar colony formation assay was performed as
previously described (22). The
colonies were treated with RPMI-1640 complete medium in the
presence or absence of DOX twice every week. One colony was defined
if >50 cells were observed. Plates were imaged, and colonies
were enumerated using ImageJ software.
Flow cytometry
For BrdU assay, cells were starved with 2%
FBS-RPMI1-640 for 12 h and recovered in 10% FB-RPMI1640 for 24 h
for cell synchronization. Cells were labeled with BrdU in culture
medium for 1 h. All procedures followed the standard protocol with
the allophycocyanin (APC) BrdU Flow Kit (cat. no. 552598; BD
Biosciences). Cell cycle progression was determined by propidium
iodide (PI) staining. Cells were fixed in 75% ethanol at −20°C
overnight and incubated with PI/RNase Staining Buffer (cat. no.
550825; BD Biosciences) for 15-20 min at RT (protecting the cells
from light). For apoptosis assay, cells were labeled by
APC-conjugated Annexin V and PI/7-aminoactinomycin D (cat. no.
A7313020; Shanghai Yeasen Biotechnology Co., Ltd.) according to the
manufacturer's protocol. Stained cells were analyzed by flow
cytometry (DxP Athena B4-R2; Cytek Biosciences) and analyzed with
FlowJo 10.0 software (FlowJo LLC).
Cell migration, invasion and homing
assay
The Transwell assay was performed using the
Transwell chambers (Corning, Inc.) with a filter membrane (aperture
8 μm). For the migration assay, the chemoattractant was 10%
FBS. The invasion experiment required the addition of Matrigel to
the upper chamber and incubation at 37°C for 2 h, followed by the
same steps as the migration assay. The chemoattractant of the cell
homing assay was CXCR12 (40 ng/ml). Cells were starved with 2%
FBS-RPMI1640 for 12 h. Then 5×105 cells/400 μl
serum-free medium/well were inoculated into the upper chamber, and
600 μl/well medium with chemoattractant was added into the
lower chamber. After incubation in 37°C for 24 h, the migratory
cells in the lower chamber were harvested and counted through flow
cytometry. After removing the cells in the upper chambers, chambers
were stained with 0.2% crystal violet (CV) for 20 min. The filter
membrane was cut and placed on the glass slide for counting using a
Leica light microscope.
Cell adhesion assay
A co-culture system was used to detect the direct
adhesion ability of MM cells. 96-well plates were incubated at 37°C
with fibronectin (FN; 50 μg/ml; 100 μl/well; cat. no.
354008; Corning, Inc.) or seeded HS5 cells (2×104
cells/well) overnight. LGR4-OE or LGR4-knockdown ARP1 and OCI-My5
were harvested and resuspended with serum-free RPMI-1640 medium,
seeded with 1×105 MM cell/100 μl into 96-well
plate. To detect the adhesion ability of MM cells after being
treated with QNZ for 48 h, MM cells were harvested and seeded into
the 96-well pre-coated with FN or HS5 cells. After co-culture for 4
h, non-adherent MM cells were removed. Adherent cells were stained
with 0.2% CV for 2 h at RT. Superfluous CV was washed off with
distilled water, and the plates were dried overnight at RT. The dye
was dissolved with 2% SDS for 2 h on the shaking platform, and the
plates were measured at 570 nm using a microplate reader. The
optical densities (ODs) from HS5s cultured alone were tested as
background absorption.
Homing assay and Xenograft mouse models
of MM
All animal experiments were performed in accordance
with the guidelines of the Institutional Animal Care and local
Veterinary Office and Ethics Committee of the Animal Center of
Hunan Normal University School of Medicine (approval no. D2021013;
Changsha, China). The SPF housing conditions were maintained at
20-26°C, with relative humidity at 40-70% and a 12/12-h light/dark
circadian rhythm. OCI-My5 cells (1×106 cells in 200
μl PBS) were injected by tail vain intravenously into 8
weeks-old female NCG mice (n=22; weight, 19-24 g)
(NOD/ShiLtJGptPrkdcem26Cd52Il2rgem26Cd22/Gpt,
GemPharmatech). MM progression in the mice was monitored by
measuring the tumor burden through live imaging. Homing assay of
injected MM cells to the BM was measured by flow cytometry. Mice
were euthanized using sodium pentobarbital at a dose of 100 mg/kg
via intraperitoneal injection, and living imaging of leg bones was
performed at week 6. Homing MM cells were flushed out of the BM
with 1X PBS. After lysis of the erythrocyte lysate (ACK lysis
Buffer; cat. no. SL1070; Coolaber science & technology Co.,
Ltd.), the cells were labeled with PE-anti-human CD138 (1:100; cat.
no. 352306; BioLegend, Inc.). Flow cytometry was used to analyze
the proportion of the homing cells to BM.
Radiography and micro-computed tomography
(micro-CT)
Micro-CT scanning was performed as previously
described (22). The mouse tibia
was fixed in 4% paraformaldehyde (PFA) in 4°C for 48 h. Tibia scans
were performed by High-resolution Micro-CT (Skyscan 1176; Bruker,
https://www.bruker.com/zh.html?ao=1)
at a resolution of 6.5 μm per pixel. Measuring the bone
parameters, including trabecular bone volume fraction (Tb. BV/TV),
trabecular number (Tb.N), trabecular thickness (Tb.Th) and
trabecular separation (Tb.Sp). These bone parameters were analyzed
using DataViewer 1.4.1.9 (CTAn version 1.11) and (μCTVol
version 2.2; (both from Bruker Corporation).
Tartrate-resistant acid phosphatase
(TRAP) staining
The mouse tibia was fixed in 5 ml of 4% PFA solution
at 4°C overnight and then decalcified the tibia in 10 ml of 0.5 M
EDTA at 4°C for 24 h. Paraffin sections (6 μm) were stained
with (TRAP; cat. no. 387a-1KT; MilliporeSigma). Images of TRAP were
obtained through a light microscope (Keyence Corporation).
Immunohistochemistry (IHC)
The experimental mice tibia was fixed with PFA in
4°C, embedded in paraffin following gradient ethanol dehydration,
and sliced into 3 μm for IHC. The slides were dewaxed with
xylene, rehydrated, and subjected to antigen retrieval treatment
using an IHC kit (cat. no. KIT-9720; MXB Biotechnologies;
http://www.maxim.com.cn/sitecn/myzhjcxthsjh/981.html).
Subsequently, the slides were incubated with anti-CD138 antibody at
a 1:400 dilution overnight at 4°C. Next, the slides were incubated
in 25°C with HRP-conjugated antibody (Reagent 3 in IHC kit) and
stained with 3,30-diaminobenzidine tetrahydrochloride hydrate (DAB)
for 3 min. Finally, cell nuclei were counterstained with
hematoxylin.
RNA sequencing (RNA-seq) and
analysis
Total RNA was extracted from MM cells using TRIzol
(cat. no. 15596018CN; Invitrogen; Thermo Fisher Scientific, Inc.),
and its quality and quantity were assessed using a Fragment
Analyzer (Agilent Technologies, Inc.). Library preparation was
performed using Optimal Dual-mode mRNA Library Prep Kit (BGI;
https://www.bgi.com/). The loading concentration
of the final library is 300g/lane quantified by Qubit (Thermo
Fisher Scientific, Inc.). Sequencing was performed on T7 platform
(BGI) using paired-end 150-base reads are generated. MM cell lines,
including OCI-EV, OCI-LGR4-OE, OCI-Ctrl and OCI-LGR4-shRNA1, were
used for RNA-seq. Gene set enrichment analysis (GSEA) enrichment
from differentially expressed genes between OCI-EV and OCI-LGR4-OE
was performed using clusterProfiler_4.2.2 (24) function of R language, and the
threshold was set as P<0.05.
Statistical analysis
Data were analyzed and represented using GraphPad
Prism software (version 9; Dotmatics). All data are presented as
the mean ± standard deviation (SD). The statistical significance of
the data was determined using the two-tailed unpaired Student's
t-test, one-way or two-way ANOVA with Tukey or Dunnett post-hoc
test, or Kruskal-Wallis test. Overall survival was measured using
the Kaplan-Meier method, and the log-rank test was used for group
comparison based on GraphPad Prism 9 software. The statistical
significance of the data in the Table SVI was determined through the
Fisher-Freeman-Hanlton test. Each experiment was performed three
times. The analysis of the Gene Expression Programming database
utilized dataset GSE2658 and GSE24080, accessed from the GEO
database (https://www.ncbi.nlm.nih.gov/geo/). *P<0.05 was
considered to indicate a statistically significant difference.
Results
High expression of LGR4 is associated
with cell adhesion and links to poor prognosis in MM
To identify the role of LGR4 in MM, the dataset
GSE2658 (25) was analyzed, which
contains data from 22 HD, 44 patients with MGUS and 351 newly
diagnosed patients with MM. The analysis revealed a gradual
increase in LGR4 expression (Fig.
S1A). Higher expression of adhesion-associated molecules was
observed in patients with MM compared with both HD and MGUS on the
dataset GSE2658, same as LGR4 (Fig.
1A). These results were further validated using
immunofluorescence in CD138+ cells derived from HD (n=5)
and patients with MM (n=9) (Fig.
S1B). The results confirmed a higher expression of LGR4 in MM,
consistent with previous studies (16,17).
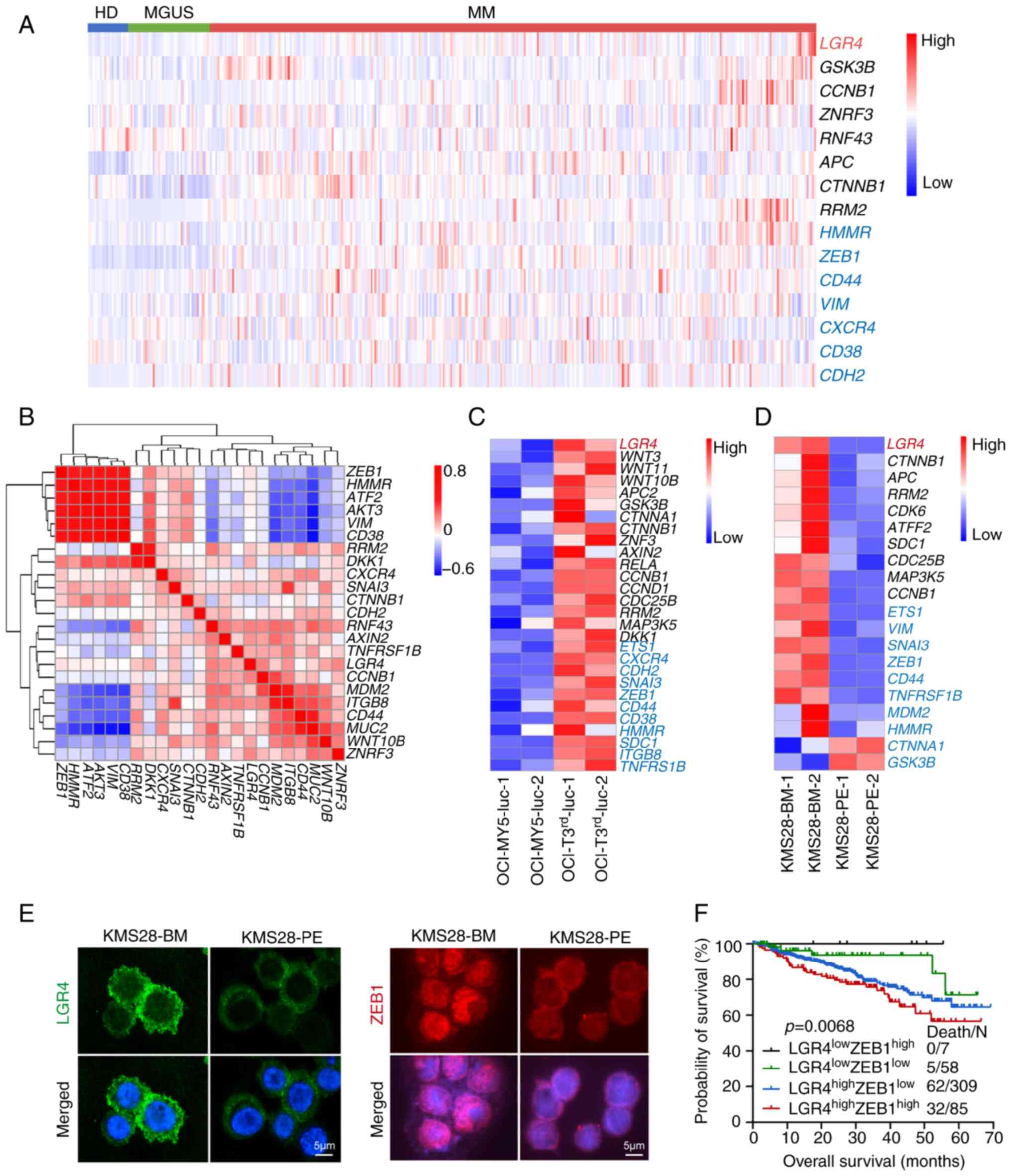 | Figure 1High expression of LGR4 is associated
with cell adhesion and poor prognosis in multiple myeloma. (A) Gene
expression heatmap of LGR4 (Red) and Wnt/β-catenin signal related
genes (Black), Cell adhesion associated genes (Blue) in
CD138+ cells from HD (n=22), MGUS (n=44) and MM (n=351).
(B) Pearson's correlation analysis of the relationship between LGR4
and cell adhesion genes. Red for positive, blue for negative. (C)
Gene expression heatmap of OCI-My5 and OCI-T3rd-luc with
LGR4 (Red), Wnt/β-catenin signal genes (Black), Cell adhesion
associated genes (Blue). (D) Gene expression heatmap of KMS28-BM
and KMS28-PE with LGR4 (Red), Wnt/β-catenin signal genes (Black),
Cell adhesion associated genes (Blue). (E) Representative images of
immunofluorescence images of LGR4 and ZEB1 protein expression in
KMS28-BM and KMS28-PE. Scale bars, 50 μm. (F) Kaplan-Meier
analyses of overall survival in MM patients with
LGR4lowZEB1low (n=58),
LGR4lowZEB1high (n=7),
LGR4highZEB1low (n=309) and
LGR4highZEB1high (n=85) from GSE2658. HD,
healthy donors; MGUS, monoclonal gammopathy of undetermined
significance; MM, multiple myeloma; BM, bone marrow; PE, pleural
effusion; ZEB1, Zinc Finger E-Box Binding Homeobox 1. |
The focal proliferation of MM cells in the BM is a
hallmark of MM, where increased adhesion enhances cell homing
(9). To investigate whether LGR4
influences adhesion and homing, the association of LGR4 and
adhesion-associated molecules was examined. Pearson's correlation
heatmap analysis exhibited a positive correlation between LGR4 and
adhesion-associated molecules (Fig.
1B). Subsequently, RNA-seq data of OCI-T3rd-luc
cells revealed significant upregulation of LGR4 and adhesion genes
compared with OCI-My5-luc cells (Fig.
1C). Additionally, the high expression of LGR4 along with
adhesion genes was verified using RNA-seq data from paired KMS28-BM
and KMS28-PE cell lines (Fig.
1D). Among the positive correlated gene, it was found that
ZEB1, a transcription factor associated cell adhesion (26), is associated with worse overall
survival; as MM patients with
LGR4highZEB1high (n=85) exhibited
significantly worse overall survival (P=0.068) (Figs. 1F and S1F). Furthermore, ZEB1 expression was
significantly increased in KMS28-BM compared with KMS28-PE cells,
at both mRNA (Fig. S1D) and
protein levels (Figs. 1E and
S1C-E). These results suggested
that LGR4 is associated with cell adhesion and promoted MM cell
homing to the BM. In summary, high expression of LGR4 is associated
with increased cell adhesion and is associated with poor prognosis
in MM.
LGR4 overexpression promotes cell
adhesion, migration and homing in MM cells in vitro
To explore whether high expression of LGR4 promotes
MM progression, LGR4 was the BrdU incorporation assay (Figs. 2B and S2C) and a marked increase in colony
formation in the soft-agar colony formation assay (Fig. 2C) compared with the EV group.
Additionally, cell cycle assays indicated that LGR4 overexpression
increased the percentages of both the S phase and G2/M phase in
ARP1 and OCI-My5 cells (Fig.
S2D). The aforementioned results confirmed that LGR4
overexpression promotes the proliferation of MM cells.
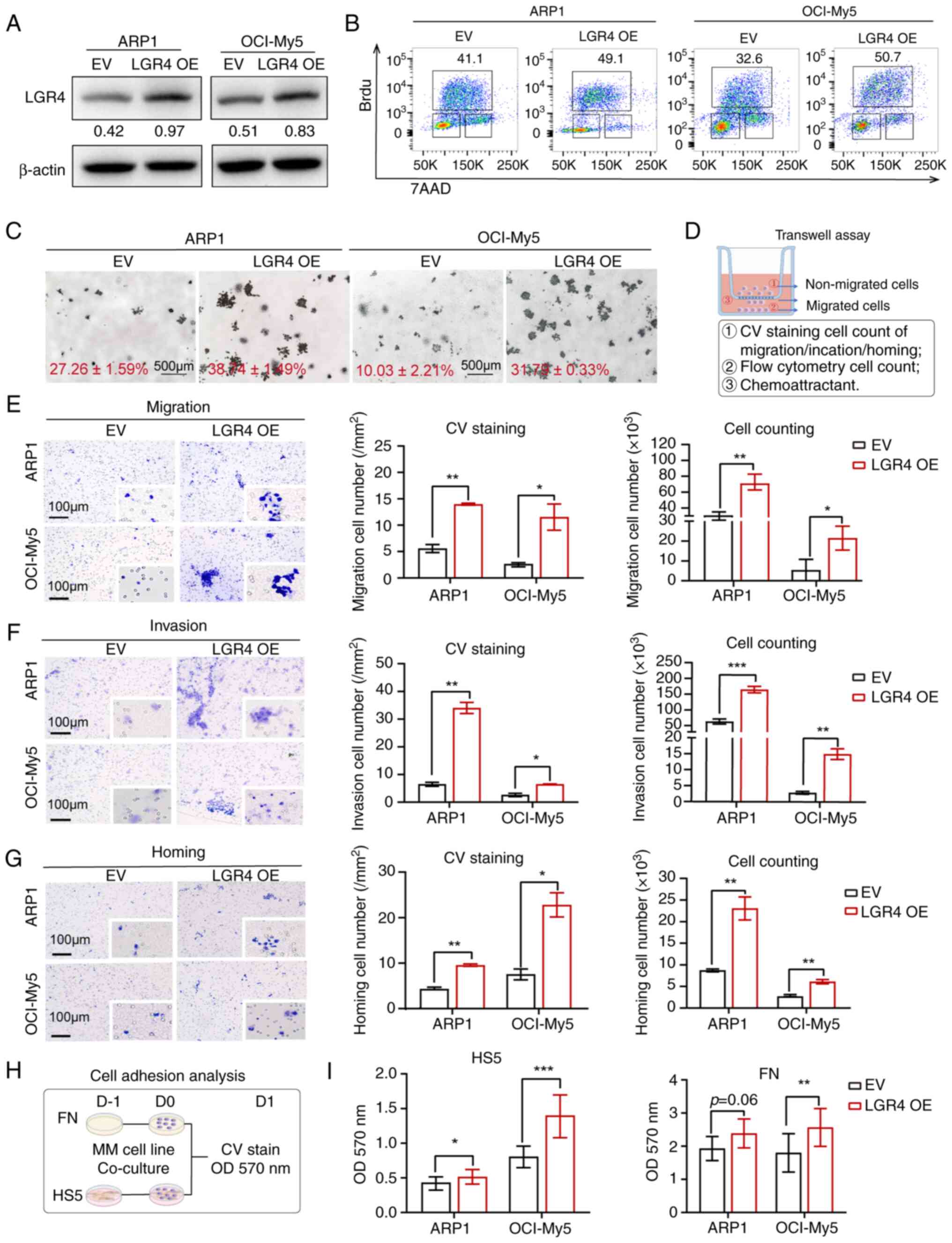 | Figure 2LGR4 overexpression promotes cell
adhesion, migration and homing in MM cells in vitro. (A)
Western blots of LGR4-OE in ARP1 and OCI-My5 MM cell lines,
compared with EV. (B) Representative flow cytometry dot plots of
the number of BrdU-positive cells. (C) Representative images of
clonogenic analysis in ARP1-EV, ARP1-LGR4-OE, OCI-EV and
OCI-LGR4-OE cells. Scale bars, 500 μm. (D) Schematics of
Transwell experiments. (E) Transwell migration assays were
conducted with LGR4-OE ARP1 and OCI-My5 cells. The quantification
of the number of migratory cells is presented in the column graph.
Scale bars, 100 μm. (F) Matrigel invasion assays were
conducted with LGR4-OE ARP1 and OCI-My5 cells. The quantification
of the number of invasive cells is presented in the column graph.
Scale bars, 100 μm. (G) Transwell cell homing assays
conducted with LGR4-OE ARP1 and OCI-My5 cells. The quantification
of the number of homing cells is presented in the column graph.
Scale bars, 100 μm. (H) Schematic of cell adhesion assay.
(I) Adhesion assay of LGR4-OE ARP1 and OCI-My5 co-cultured with HS5
cells or FN. Statistical analyses were performed using Student's
t-test. *P<0.05, **P<0.01 and
***P<0.001. LGR4-OE, LGR4 overexpression; MM,
multiple myeloma; EV, empty vector; FN, fibronectin; CV, crystal
violet. |
Next, the effect of LGR4 was examined on the
migration, invasion and homing abilities of MM cells. A Transwell
assay using different chemo-attractants was performed to assess
cell migration (Fig. 2D). The
results indicated that LGR4 overexpression significantly promoted
cell migration and invasion, which was quantified by counting
migratory cells (Fig. 2E and F).
Additionally, a cell homing assay using CXCR12, a chemokine known
to induce immune cell homing to BM (27), demonstrated that more cells homed
to the BM in the LGR4-OE group (Fig.
2G). As previously reported, FN acts as a connection between
cells and matrix (26), and the
HS5 cell line mimics the bone marrow stromal cells (BMSCs),
promoting MM cell proliferation and adhesion (28). To determine whether LGR4 improves
the interaction between MM cells and the BMME, a cell adhesion
co-culture assay was performed using FN and HS5 cells (29)(Fig.
2H). The OD value at 570 nm indicated a direct increase in
adhesion to both FN and HS5 in the LGR4-OE group (Fig. 2I). Even though in ARP1-LGR4-OE
cells was observed increase tend of adhesion (P=0.06), which may
cause by the complex genetic characteristics such as
TP53del (30),
the aforementioned results demonstrated that LGR4-OE enhances the
cell homing and adhesion ability in MM cells.
LGR4 knockdown impairs cell
proliferation, adhesion, migration and homing in MM cells in
vitro
To further investigate the function of LGR4 on
adhesion, migration and homing of MM cells, two shRNA sequences
(shRNA1 and shRNA2) targeting LGR4 were designed. A
DOX-inducible lentiviral expression system expressing LGR4 shRNA
was used to knock down LGR4 in MM cell lines. LGR4 knockdown was
confirmed at both mRNA and protein levels (Figs. 3A, S3A and C). Among these, LGR4-shRNA1
silencing was confirmed to be more effective. Growth curves
indicated that LGR4 knockdown significantly inhibited proliferation
in MM cells following DOX induction (Fig. S3B). Additionally, the proportion
of BrdU-positive cells in the LGR4-shRNA1 groups was significantly
lower than in the control group (Fig.
3B and C). The colony formation of LGR4-shRNA1 cells exhibited
a significant inhibition (Fig.
3D). Cell cycle assays revealed that LGR4 silencing decreased
the proportion of cells in the S and G2/M phases in both ARP1 and
OCI-My5 (Fig. S3D). Furthermore,
the proportion of Annexin-V-positive cells was significantly
increased (Fig. S3E), and
cleaved caspase 3 and PARP were significantly upregulated in the
LGR4-knockdown group (Fig. S3F and
G), indicating that LGR4-knockdown induced apoptosis in MM
cells. These results suggested that LGR4-knockdown inhibited the
proliferation and induced apoptosis in MM cells.
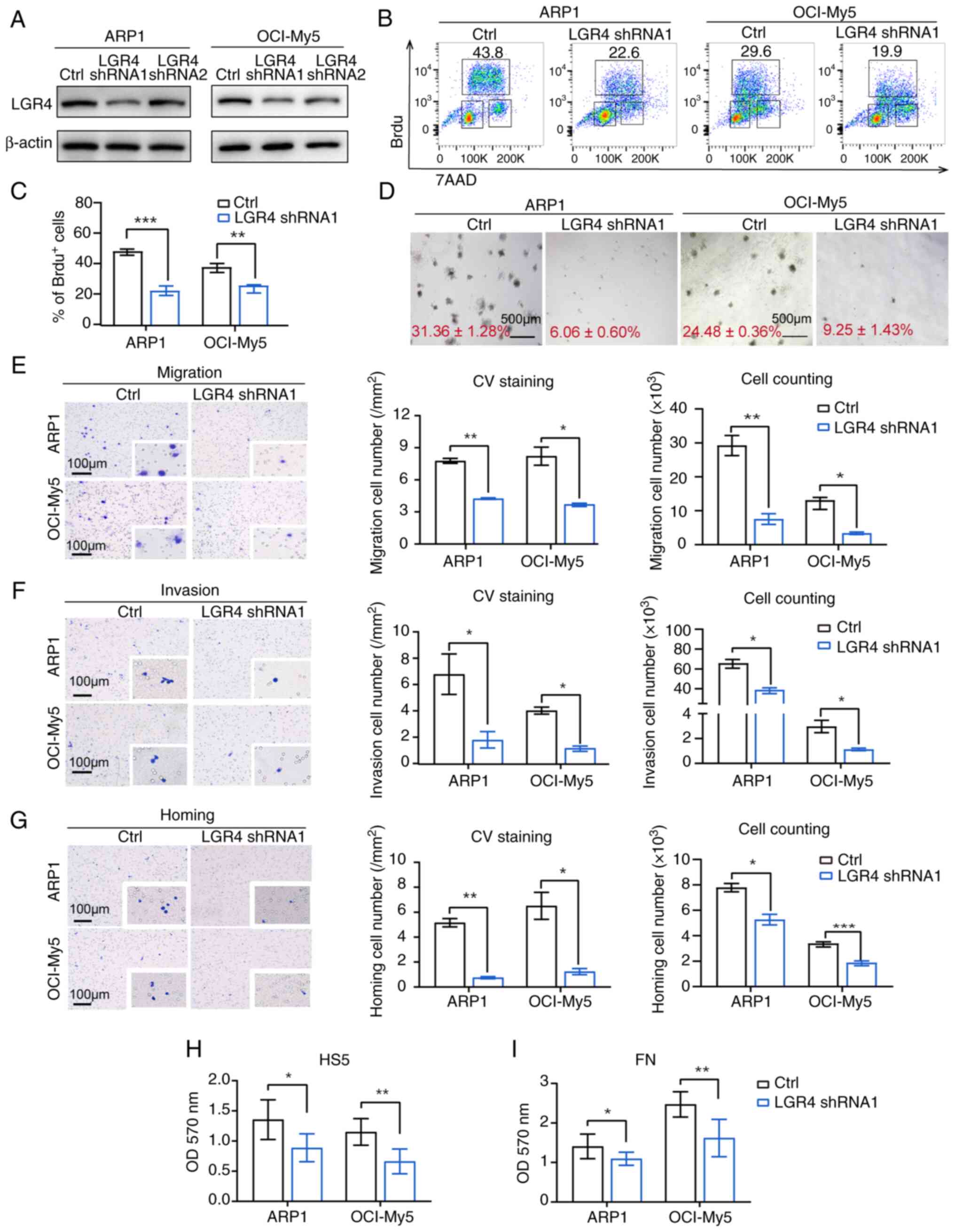 | Figure 3LGR4 knockdown impairs cell
proliferation, adhesion, migration and homing in MM cells in
vitro. (A) Western blots of LGR4-silencing in ARP1 and OCI-My5,
compared with the controls. (B) Representative flow cytometry dot
plots for detection of BrdU-positive cells. (C) Statistical
analysis of the number of BrdU-positive cells among LGR4-silencing
MM cells. (D) Representative images of clonogenic analysis in
ARP1-Ctrl, ARP1-LGR4-shRNA1, OCI-Ctrl and OCI-LGR4-shRNA1 cells
cultured in RPM1-1640 media. Scale bars, 500 μm. (E)
Transwell migration assays were conducted with LGR4-knockdown ARP1
and OCI-My5 cells. The quantification of the number of migratory
cells is presented in the column graph. Scale bars, 100 μm.
(F) Matrigel invasion assays were conducted with LGR4-knockdown
ARP1 and OCI-My5 cells. The quantification of the number of
invasive cells is illustrated in the column graph. Scale bars, 100
μm. (G) Cell homing assays were conducted with
LGR4-knockdown ARP1 and OCI-My5 cells. Scale bars, 100 μm.
(H) The quantification of the number of homing cells is presented
in the column graph. (I) Adhesion assay of LGR4-knockdown ARP1 and
OCI-My5 co-cultured with HS5 cells or FN. Statistical analyses were
performed using Student's t-test. *P<0.05,
**P<0.01 and ***P<0.001. MM, multiple
myeloma; shRNA, short hairpin RNA. |
Next, Transwell migration and invasion assays
revealed that LGR4-knockdown reduced both migration and invasion,
with a corresponding decrease in the number of migratory cells
(Fig. 3E and F). Additionally,
LGR4-knockdown decreased the homing of MM cells induced by the BM
chemokine CXCR12, as confirmed by statistical analysis (Fig. 3G). Using cell-adhesion co-cultured
assay, the absorbance at 570 nm exhibited that LGR4-knockdown
suppressed the adhesion ability of MM cells to FN and BMSCs
(Fig. 3H and I). The
aforementioned results confirmed that LGR4-mediated interaction
between malignant plasma cells and BMME is crucial for cell
adhesion and homing to BM niches.
LGR4 overexpression promotes cells'
homing to BM and MM progression in vivo
To further explore the role of LGR4 in MM cell
homing in vivo, OCI-My5 cells with LGR4-OE were generated
and injected through the tail vein into NCG mice. MM cells are
typically home to BM, where they proliferate and cause symptoms,
including hindlimb paralysis (31). Tumor burden was monitored through
whole-animal live imaging, evaluating the proportion of human MM
cells in BM and their homing efficiency (Fig. 4A). Compared with the control mice,
the LGR4-OE mice significantly demonstrated an increased
tumor-associated luminescence intensity at weeks 4 and 6 (Fig. 4B and C). LGR4-OE mice exhibited
60% of paralysis, while control mice had no expression at week 6
(Fig. S4). Due to reaching the
humane endpoint, the mice were euthanized at week 6. Flow
cytometric analysis revealed that LGR4 overexpression significantly
increased the proportion of homing MM cells in the BM. As a result,
LGR4-OE significantly increased the proportion of human MM cells
(AVG 55%) in the BM compared with the control mice (AVG 23.8%)
(Fig. 4D).
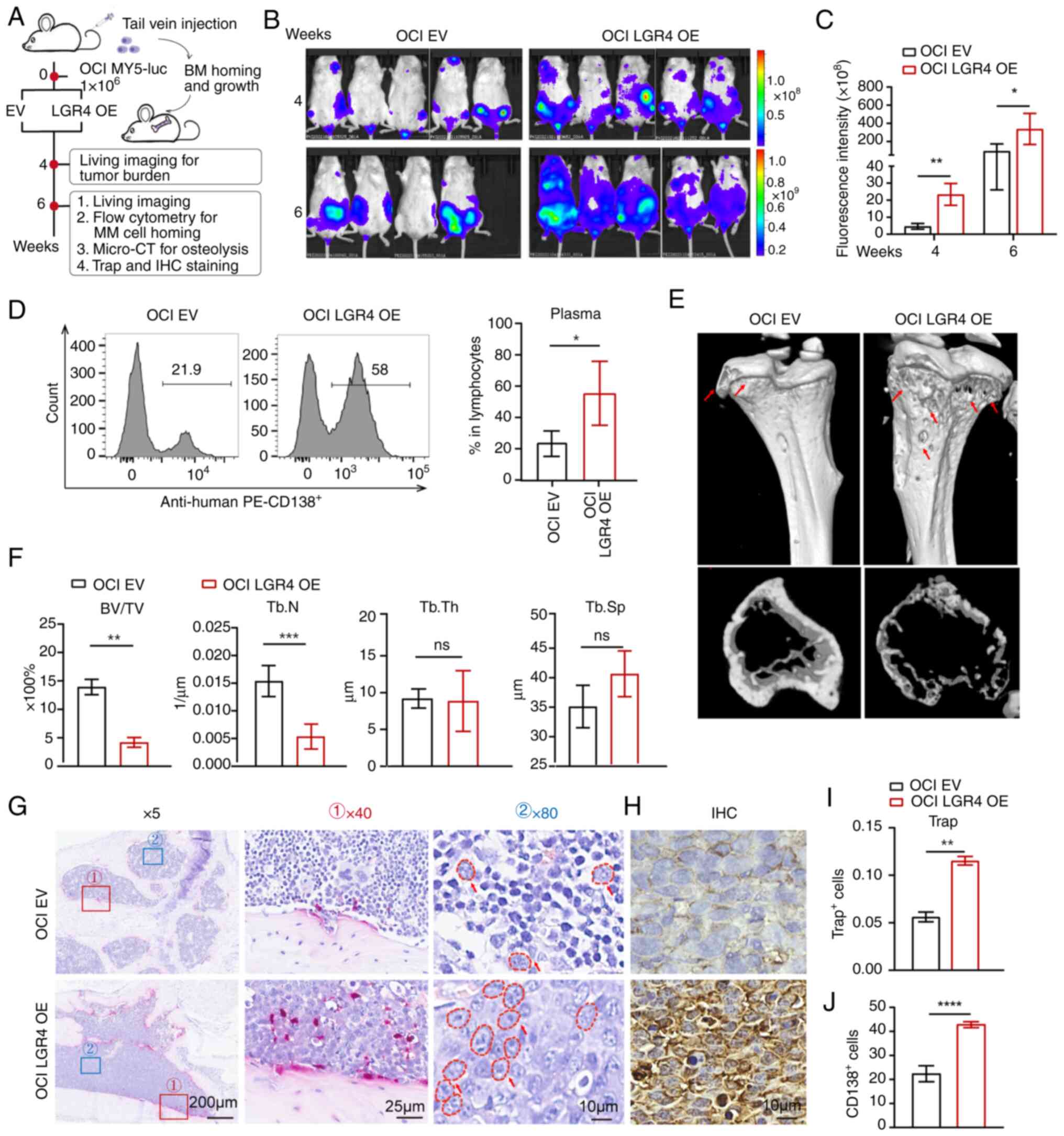 | Figure 4Overexpression of LGR4 promotes
cells' homing to the BM and MM progression in vivo. (A)
Schematic of in vivo experiments. (B) Tumor-associated live
imaging of NCG mice injected with OCI-Ctrl or OCI-LGR4-OE cells at
4 and 6 weeks (n=5 for each group). (C) Quantification of
luminescence intensity in live NCG mice. (D) Flow cytometric
analysis images and statistics of the human MM cell proportion in
the bone marrow after sacrificing NCG mice. (E) Micro-CT images of
tibia derived from NCG mice. (F) Quantification of bone
microstructural parameters, namely BV/TV, Tb.N, Tb.Th and Tb.Sp
(n=3). (G) TRAP staining for NCG xenografted mice bone marrow
section. Scale bars, 200, 25 and 10 μm. (I) The
quantification of the number of Trap-positive osteoclast cells is
presented in the column graph. (H) Neoplastic CD138-positive plasma
cells. Scale bars, 10 μm. (J) The quantification of the
number of neoplastic CD138 positive plasma cells is presented in
the column graph. Statistical analyses were performed using
Student's t-test. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. BM, bone marrow; MM,
multiple myeloma; LGR4-OE, LGR4 overexpression; BV/TV, trabecular
bone volume fraction; Tb.N, trabecular number; Tb.Th, trabecular
thickness; Tb.Sp, trabecular separation; TRAP, tartrate-resistant
acid phosphatase; EV, empty vector; ns, not significant
(P>0.05). |
Since MM cell homing and proliferation within the BM
are key drivers of bone disease (21), bone health was further assessed by
evaluating bone fractures and CD138 immunostaining. Micro-CT
scanning was used to detect bone damage, finding that LGR4-OE had
markedly more severe trabecular bone loss compared with the control
mice (Fig. 4E). Quantitative bone
microstructure parameters exhibited that LGR4-OE in mice had a
lower BV/TV and Tb.N, while Tb.Sp was markedly higher compared with
the controls (Fig. 4F).
Consistent with these results, TRAP staining indicated the
increased positive osteoclast number in the LGR4-OE mice femora
compared with control mice (Fig. 4G
and I). Moreover, plasma cell morphology was clearly visible at
×80 magnification and exhibited enrichment in LGR4-OE mouse bones
(Fig. 4G). CD138 immunostaining
revealed the increased plasma cell number in the LGR4-OE mice
compared with the control (Fig. 4H
and J). Additionally, the potential association between LGR4
expression and clinical characteristics was assessed, using the
GSE24080 dataset. It was found that LGR4 expression correlated
strongly with the percentage of plasma cells in the BM (P=0.037),
and the number of magnetic resonance imaging (MRI)-defined focal
lesions (P=0.019) (Table SVI).
These findings suggest that high expression of LGR4 is linked to
myeloma cell homing, promotes bone destruction and contributes to
malignant progression in patients with MM. In summary, these
results demonstrated that overexpression of LGR4 enhances MM cell
homing to the BM and accelerates disease progression.
Cell-adhesion association genes and NF-κB
signaling pathway are upregulated in LGR4-OE MM cells
To further understand the signaling pathways
regulated by LGR4 in MM, RNA-seq was performed on OCI-LGR4-OE and
OCI-LGR4-shRNA1, along with the control cells. Moreover, GSEA
exhibited major types of gene signatures in LGR4-OE cells that were
enriched in the regulation of cell migration and cell adhesion
(Figs. 5A-C, S5A and B). Then, the changes in the
expression of key cell-adhesion genes were verified at both Mrna
and protein levels, including N-Cadherin, Snail, Vimentin, MUC2,
ZEB1, TNFRSF1B in LGR4-OE, LGR4-shRNA1 in ARP1 and OCI-My5 cells
(Figs. 5D and E and S5C). The results indicated that LGR4-OE
significantly increased the cell-adhesion molecules while
LGR4-knockdown resulted in a significant decrease (Figs. 5F and G and S5D). A recent study has suggested that
LGR4 regulates intestinal epithelial cell proliferation and
development through C-terminal activation of NF-κB signaling
(32). R-spondin signals drive
NF-kB activity through LGR4 and stimulate the proliferation of stem
cells (33). It has been reported
that activated NF-κB signaling enhances the ability of
hematopoietic stem cell homing (9). Besides, NF-κB signaling can activate
endothelial cell adhesion molecules (34). Therefore, it was hypothesized that
LGR4 promotes MM cell homing by activating NF-κB signaling. To
investigate whether LGR4 influences MM cell adhesion through NF-κB
activation, the protein level of key NF-κB genes was determined,
including p65, phosphorylated (p-)p65, IκBα and p-IκBα. The results
indicated that LGR4-OE significantly activated NF-κB signaling,
while LGR4-knockdown decreased as western blotting illustrated
(Figs. 5H and I, S5E and F). The aforementioned data
indicate that LGR4 promotes cell homing to BM through activating
NF-κB signaling.
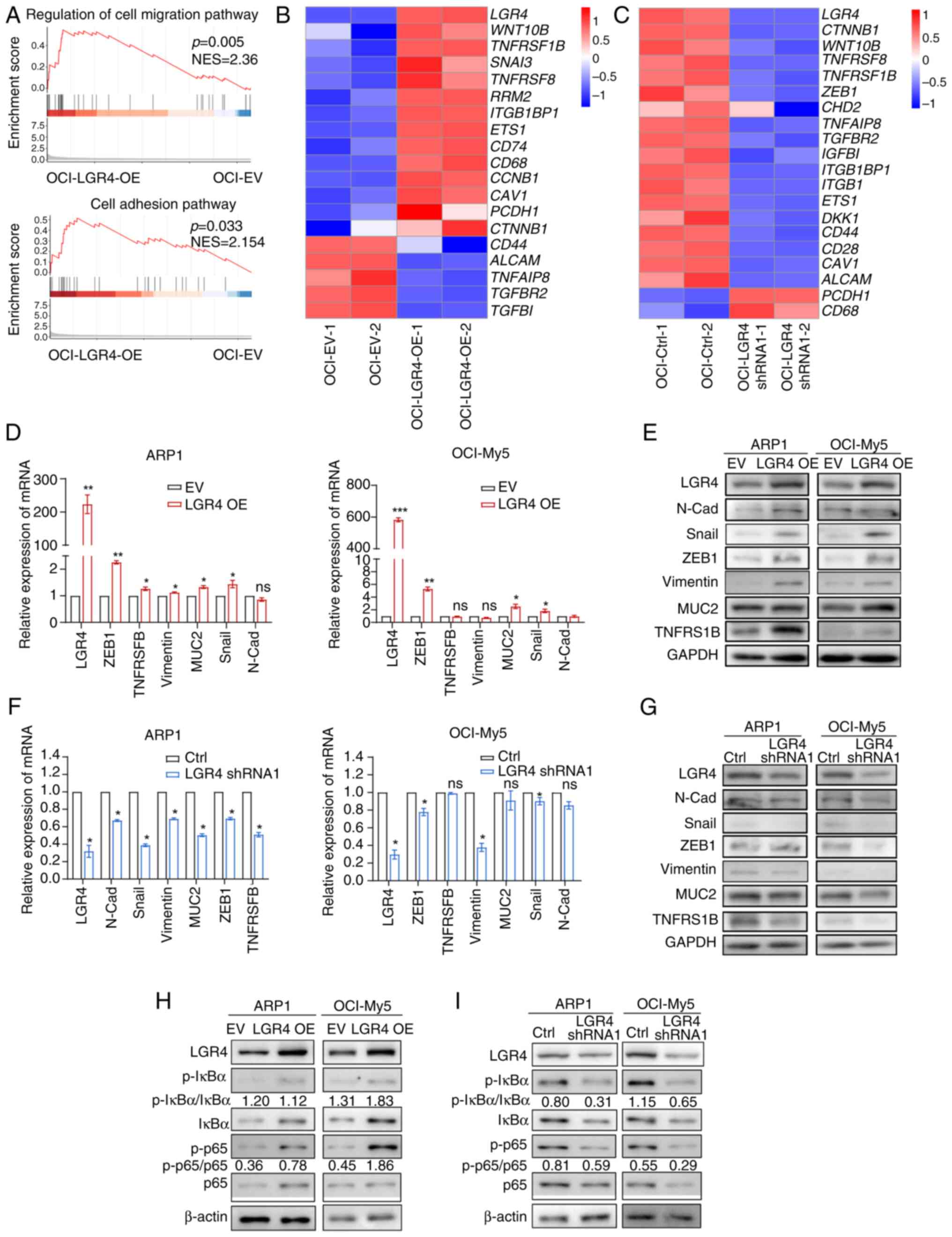 | Figure 5Cell-adhesion association genes and
NF-κB signaling pathway are upregulated in LGR4-OE multiple myeloma
cells. (A) Gene Set Enrichment Analysis of cell-adhesion
pathway-related genes from differentially expressed genes between
OCI-Ctrl and OCI-LGR4-OE. (B and C) Heatmap of RNA sequencing
analysis of adhesion-related gene expression in OCI-EV,
OCI-LGR4-OE, OCI-Ctrl, and OCI-LGR4-shRNA1. (D and E) Relative mRNA
and protein levels of cell-adhesion genes in OCI-EV and OCI-LGR4-OE
cells, respectively. (F and G) Relative mRNA and protein levels of
cell-adhesion genes in OCI-Ctrl and OCI-LGR4-shRNA1 cells,
respectively. (H and I) The protein level of NF-κB signal genes was
detected in (H) OCI-EV and OCI-LGR4-OE cells; and (I) in OCI-Ctrl
and OCI-LGR4-shRNA1 cells (I). Statistical analyses were performed
using Student's t-test. *P<0.05,
**P<0.01 and ***P<0.001. LGR4-OE, LGR4
overexpression; EV, empty vector; shRNA, short hairpin RNA; p-,
phosphorylated; ZEB1, Zinc Finger E-Box Binding Homeobox 1; ns, not
significant (P>0.05). |
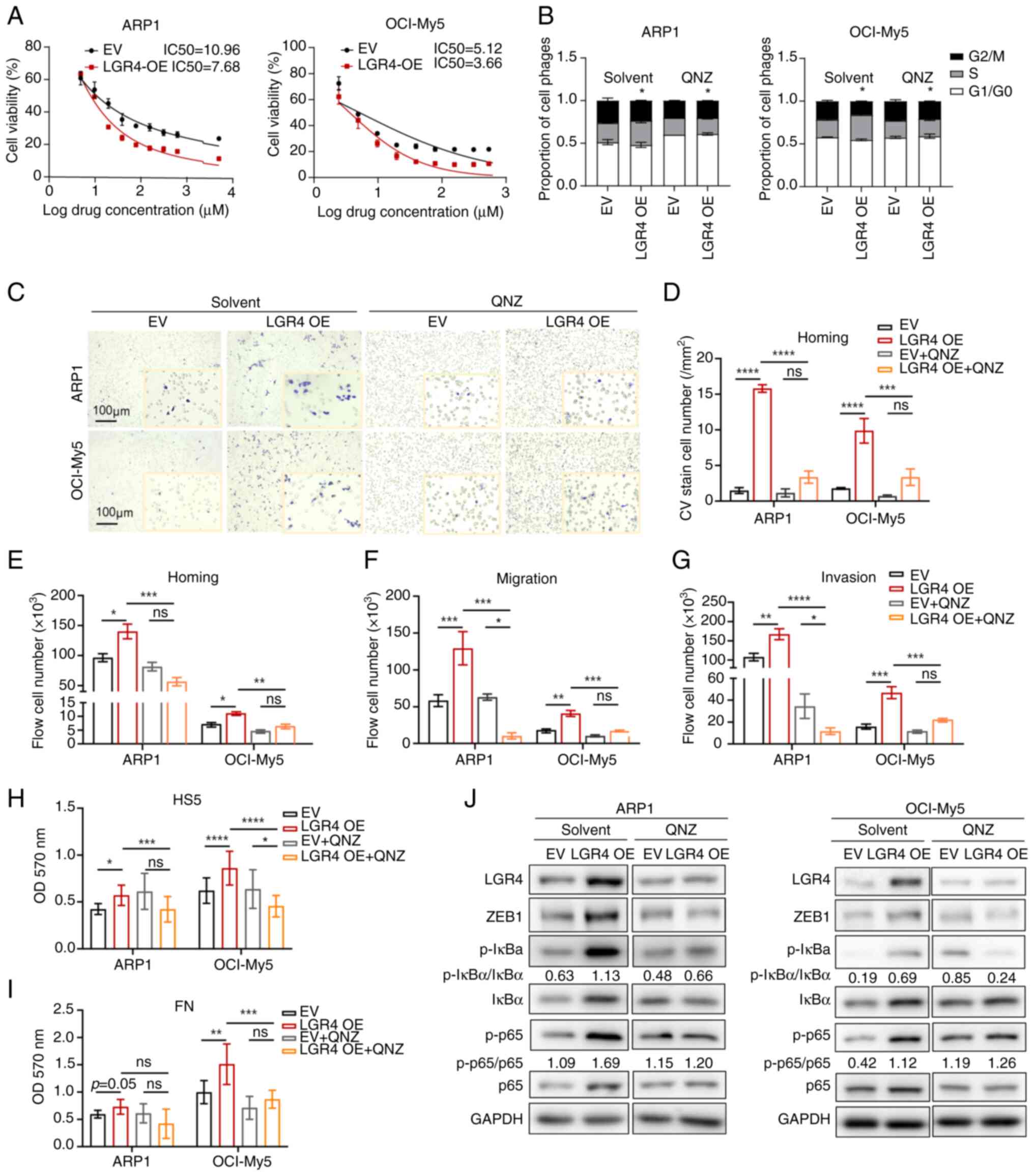
Inhibition of NF-κB pathway suppresses
cell homing and MM progression in vitro
It was further investigated whether QNZ, an NF-κB
signaling pathway inhibitor, could suppress cell proliferation and
homing in MM. The IC50 of QNZ was significantly lower in
LGR4-OE ARP1 and OCI-My5 cells compared with the control group
(Fig. 6A). Growth curves
indicated that the proliferation was inhibited in the LGR4-OE cells
treated with QNZ (Fig. S6C and
E). Cell cycle assays exhibited that QNZ alleviated the
increased proportion of S-phase cells caused by the overexpression
of LGR4 in ARP1 and OCI-My5 cells (Fig. 6B). Subsequently, to investigate
whether QNZ inhibits LGR4-induced cell adhesion and homing,
Transwell assay of cell homing was performed. The results revealed
that cell homing ability induced by LGR4-OE was inhibited after 48
h of QNZ treatment (Figs. 6C and
D). Additionally, the Transwell migration and Matrigel invasion
assays confirmed that QNZ reduced cell migration and invasion of
cells, with the quantification of migratory cells supporting these
results (Figs. 6F and G, and
S6A and B). Consistently, the
results of the cell co-culture adhesion assay exhibited that MM
cell adhesion ability was reduced after 48 h of QNZ treatment in
LGR4-OE ARP1 and OCI-My5 cells (Fig.
6H and I). Furthermore, western blot analysis revealed that QNZ
treatment significantly inhibited NF-κB signaling in LGR4-OE cells,
with a corresponding decrease in the expression of
adhesion-associated molecules ZEB1 (Figs. 6J, S6D and F). As a critical function
subunit of NF-κB signal, RELA (p65) contains transcriptional
activation domains of gene transcription and facilitates the
binding of p50 to DNA (35,36). Subsequently, to verify interaction
molecules of NF-κB signaling activation by LGR4, siRNA was used to
knock down RELA in LGR4-OE MM cells. Protein levels
(Fig. S7A and B) confirmed a
significant knockdown of RELA in LGR4-OE ARP1 and OCI-My5
cells, compared with the same cell lines transduced with SiNC
serving as controls. The results indicated that siRNA1 and siRNA3
were more pronounced. Knockdown of RELA reversed the LGR4-induced
proliferation and homing effects, as demonstrated by growth curve
analysis (Fig. S7C), cell cycle
assays (Fig. S7D), Transwell
analysis about cell homing (Fig. S7E
and F) and migration (Fig. S7G
and H). By activating the NF-κB pathway, LGR4 facilitates the
entry of the p50/RELA dimer into the nucleus to initiate gene
transcription. The aforementioned results confirmed that inhibition
of the NF-κB pathway alleviated the promoting effect of LGR4-OE on
adhesion ability and MM cell homing to BM.
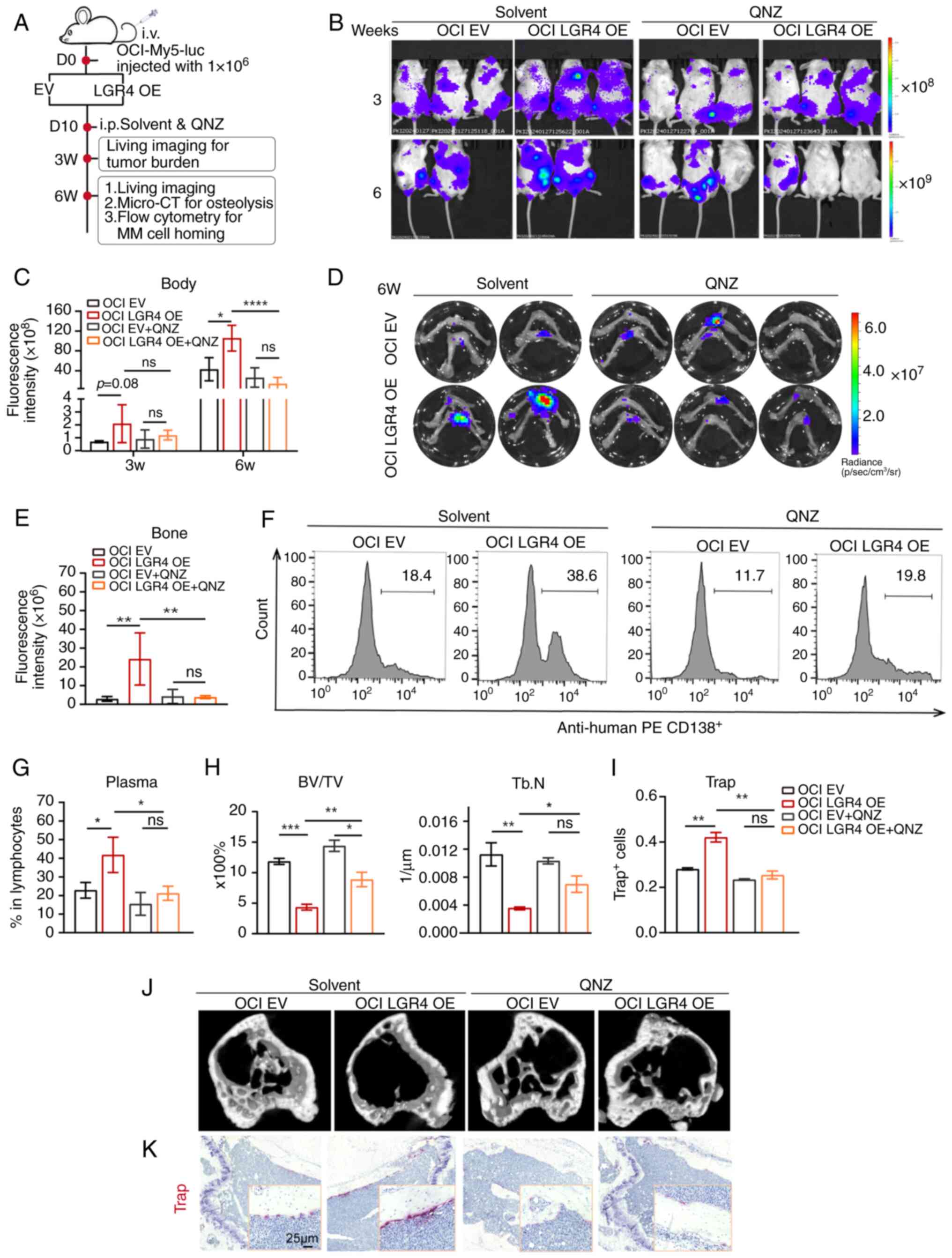
Inhibition of the NF-κB pathway relieves
the effect of LGR4-OE on MM cell proliferation and cell homing in
vivo
To explore whether QNZ has similar effects in
vivo as observed in vitro, 1×106 OCI-ctrl and
OCI-LGR4-OE cells were injected through the tail vein into NCG mice
to establish a xenografts mouse model. A total of 10 days
post-transplantation, QNZ was injected intraperitoneally every two
days (Fig. 7A). As compared with
the solvent-treated mice (Fig.
7B), the mice treated with QNZ exhibited a reduction in
tumor-associated luminescence intensity in the LGR4-OE group at
weeks 3 and 6 (Fig. 7C). Due to
reaching the humane endpoint at week 6, one mouse from each in the
solvent group was observed to exhibit paralysis, then succumbed
unexpectedly the following day, therefore the mice were euthanized.
To verify whether QNZ affected cell homing, femur and tibia were
dissected for image of bone tissue. The fluorescence intensity in
the bone of the LGR4-OE group was higher, whereas QNZ treatment
significantly reduced this intensity, particularly in the LGR4-OE
group (Fig. 7D and E). Flow
cytometric analysis confirmed that QNZ treatment significantly
decreased the proportion of human MM cells in the BM of LGR4-OE
mice compared with the solvent group (Fig. 7F and G).
Next, micro-CT scanning was used to detect bone
damage in the tibia, revealing that QNZ treatment rescued severe
trabecular bone loss caused by LGR4-OE, compared with the
solvent-treated mice (Figs. 7J).
Quantitative analysis of bone microstructure parameters exhibited
that both Tb.BV/TV and Tb.N were improved (Fig. 7H). TRAP staining revealed that the
number of osteoclasts was reduced in QNZ-treated LGR4-OE mice in
contrast to solvent mice (Fig.
7K). Quantitative analysis of TRAP-positive osteoclast cells
confirmed this reduction (Fig.
7I). In summary, these results demonstrated that inhibition of
the NF-κB pathway relieves the effects of LGR4 overexpression on MM
cell proliferation and cell homing in vivo.
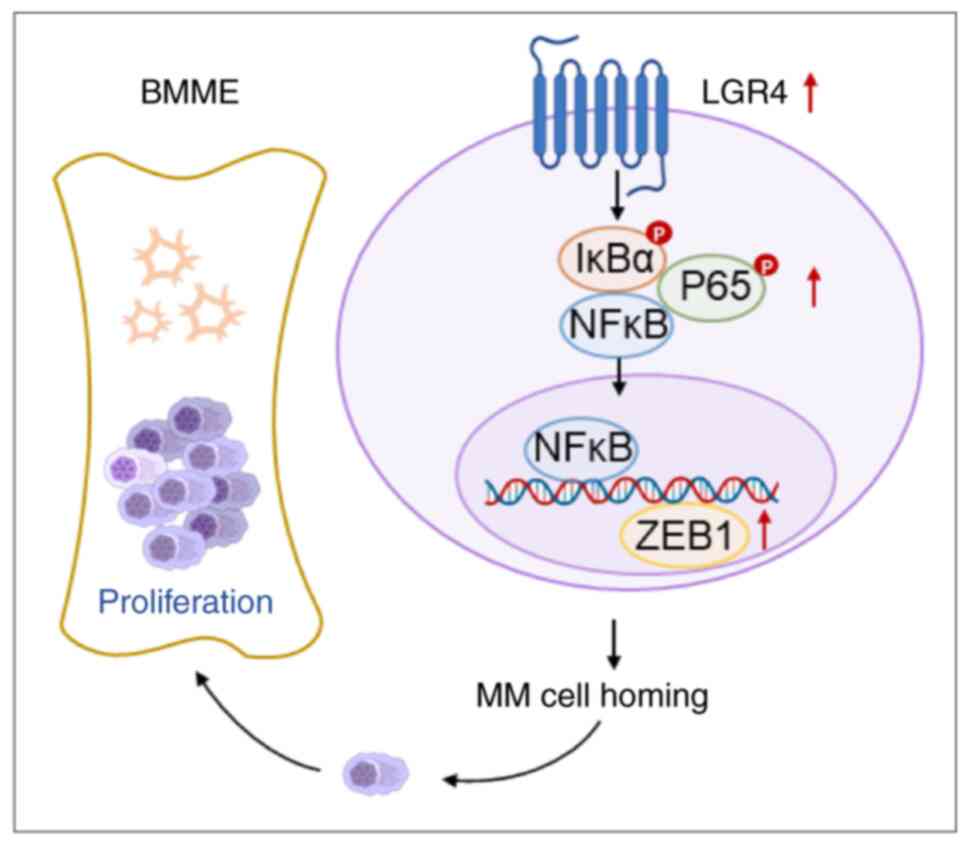
Discussion
The interaction between MM cells and BMME is
essential to MM malignant proliferation and bone destruction
(4,5). The present study provides direct
evidence, using genetic approaches, that LGR4 plays a critical role
in regulating MM cell proliferation, migration and homing.
Mechanistically, it was demonstrated that elevated LGR4 expression
in MM cells activates the NF-κB signaling pathway and upregulates
the migration-related adhesion molecule ZEB1, thus facilitating MM
cell homing into BM (Fig. 8).
Exploring the role of LGR4 in cell homing and tumorigenesis offers
valuable insights into the molecular evolution of MM, which is
vital for optimizing both current and future treatment
strategies.
The physiological role of LGR4 is associated with
the development of multiple organs. LGR4-deficient mice exhibit
developmental defects in various organs, including the eyes, bones
and reproductive system (10-13). Our previous study demonstrated
that LGR4 plays a role in early hematopoietic cell differentiation
(9). Under pathological
conditions, aberrant RSPO3-LGR4 signaling enhances tumor
aggressiveness through increased epithelial-mesenchymal transition
(EMT) in lung adenocarcinomas (37,38). Moreover, LGR4 facilitates breast
cancer cell metastasis (15),
which is an essential self-renewal gene in leukemia stem cells
(39). Consistently, studies
indicated that aberrant R-spondin/LGR4 signaling contributes to MM
progression (16,17). In the present study, it was first
demonstrated that the high expression of LGR4 associated
significantly with myeloma cell homing, promoted bone destruction,
and contributed to malignant progression in patients with MM using
clinical information analysis. Furthermore, the current study
confirms that LGR4 significantly enhances MM cell homing in
vitro and exacerbates osteolytic bone destruction in
vivo.
Normally, LGR4 has been reported to promote tumor
progression through the activation of the Wnt signaling pathway.
LGR4 activates Wnt-β catenin, which promotes EMT in lung cancer
(37), and activates GSK3β to
support tumor stem cell survival in acute myeloid leukemia
(38). Additionally, LGR4
promoted aberrant MM proliferation through Wnt signaling (data not
shown). Previous studies have demonstrated that NF-κB signaling
enhances the homing ability of hematopoietic stem cells (18) and activates endothelial cell
adhesion molecules (34). In the
present study, the GSEA analysis revealed that regulating the cell
migration pathway was enriched in RNA-seq data from LGR4-OE cells.
It was observed that LGR4 overexpression activates the NF-κB
signaling pathway and upregulates the migration-related adhesion
molecule ZEB1, thereby promoting MM cell homing and tumor
progression. Treating MM cells with an NF-κB inhibitor suppressed
tumor progression, proliferation, cell migration and homing.
Furthermore, the inhibitor's effective concentration at the
nanomolar level presents significant potential for clinical
translation. siRNA was used to suppress the expression of RELA
(which encodes p65) to validate the results obtained from the NF-κB
inhibitor in LGR4-OE cells. In summary, these findings suggest that
the NF-κB inhibitor QNZ impairs MM cell homing.
Moreover, it was found that LGR4 expression was
significantly correlated with the proportion of plasma cells in the
BM and the number of MRI-defined focal lesions (40) that established a clinical
correlation. Elevated LGR4 expression can serve as an early
indicator of aggressive MM associated with severe bone fractures.
Furthermore, LGR4, as a G-protein-coupled membrane receptor,
suggests it could be a potential therapeutic target in MM.
Recently, a humanized monoclonal antibody was developed, LGR4-mAb,
which effectively inhibits LGR4/Wnt signaling by blocking LGR4
(14). The aforementioned
antibody has been investigated for the treatment of colorectal
cancer. Consequently, conjugating a monoclonal antibody targeting
LGR4 with a proteasome inhibitor could be a promising approach for
further investigation in MM. However, due to the structural
diversity and the cross-reactivity of similarity receptors such as
LGR5/6, the limitation of the use of anti-LGR4 antibodies may
overcome by focus on improving binding affinity (41,42). The present findings suggest that
targeting LGR4 holds significant potential as a therapeutic
strategy for inhibiting MM progression.
Since LGR4 is recognized as a key regulator of
osteoblast and osteoclast differentiation (8), its high expression in MM can be
correlated with osteoclast differentiation, promoting MM
progression by inducing bone disease. The role of highly expressed
LGR4 on MM cells and the tumor microenvironment, such as the
possible promotion of MM bone disease by promoting osteoclast
differentiation, remains to be further explored. Additionally,
NF-κB inhibitors can suppress MM cell proliferation and homing and
can be tested in combination with frequently used clinical
therapies, including proteasome inhibitors, to evaluate their
therapeutic efficacy. The current findings reveal that LGR4
influences MM by activating NF-κB, as shown in the use of NF-κB
inhibitors and RELA knockdown. The specific molecules and
mechanisms involved in LGR4-mediated NF-κB signaling remain to be
fully elucidated. Therefore, the potential role and mechanism of
LGR4 in MM require further investigation.
In conclusion, it was demonstrated that the
elevated LGR4 contributes to MM progression by modulating cell
adhesion, thereby promoting cell homing to BM. LGR4 activates NF-κB
signaling, which enhances cell homing. The findings of the present
study suggest that targeting LGR4 holds significant potential as a
therapeutic strategy for inhibiting MM progression.
Supplementary Data
Availability of data and materials
The data generated in the present study are
included in the figures and/or tables of this article. The data
generated in the present study may be found in the National
Genomics Data Center under accession number HRA007584 or at the
following URL: https://ngdc.cncb.ac.cn/search/specific?db=hra&q=HRA007584.
Authors' contributions
WZ and GZ designed the study. NH, ZL and XL
performed experiments and analyzed the data. ZL and FS collected
clinical samples. QY, JG, CK, YZ, XC, GA and XF provided technical
assistance. WZ and NH wrote and revised the manuscript. WZ, NH and
QY confirm the authenticity of all the raw data. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
For using human samples, the Cancer Research
Institute Review Board of Central South University (Changsha,
China) approved the present study (date, 2019/03/12; approval no.
2022-KT188). All patients provided written informed consent. All
animal experiments were performed in accordance with the guidelines
of the Institutional Animal Care and local Veterinary Office and
Ethics Committee of the Animal Center of Hunan Normal University
School of Medicine (approval no. D2021013; Changsha, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
The authors express their gratitude to professor
Kaiqun Ren (Hunan Normal University School of Medicine) for
technical assistance. They also thank the Animal Center of Hunan
Normal University School of Medicine for providing the experimental
platform to perform the animal experiments. The authors are
grateful to Professor Jiaxi Zhou (Institute of Hematology, Chinese
Academy of Medical Sciences) for providing the HS5 cells, and to
Professor Rong Chang (Kunming Institute of Zoology, the Chinese
Academy of Sciences) for providing 293T cells.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 82130006) and the Scientific
Research Program of FuRong Laboratory (grant no. 2023SK2085-2).
References
|
1
|
Palumbo A and Anderson K: Multiple
myeloma. N Engl J Med. 364:1046–1060. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Slovak ML: Multiple myeloma: Current
perspectives. Clin Lab Med. 31:699–724. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chauhan D, Uchiyama H, Akbarali Y,
Urashima M, Yamamoto K, Libermann TA and Anderson KC: Multiple
myeloma cell adhesion-induced interleukin-6 expression in bone
marrow stromal cells involves activation of NF-kappa B. Blood.
87:1104–1112. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neri P, Ren L, Azab AK, Brentnall M,
Gratton K, Klimowicz AC, Lin C, Duggan P, Tassone P, Mansoor A, et
al: Integrin β7-mediated regulation of multiple myeloma cell
adhesion, migration, and invasion. Blood. 117:6202–6213. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu L, Mohammad KS, Wu H, Crean C, Poteat
B, Cheng Y, Cardoso AA, Machal C, Hanenberg H, Abonour R, et al:
Cell adhesion molecule CD166 drives malignant progression and
osteolytic disease in multiple myeloma. Cancer Res. 76:6901–6910.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kinzel B, Pikiolek M, Orsini V, Sprunger
J, Isken A, Zietzling S, Desplanches M, Dubost V, Breustedt D,
Valdez R, et al: Functional roles of Lgr4 and Lgr5 in embryonic
gut, kidney and skin development in mice. Dev. Biol. 390:181–190.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mao B, Huang S, Lu X, Sun W, Zhou Y, Pan
X, Yu J, Lai M, Chen B, Zhou Q, et al: Early development of
definitive erythroblasts from human pluripotent stem cells defined
by expression of glycophorin A/CD235a, CD34, and CD36. Stem Cell
Reports. 7:869–883. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G,
Huang J, Dai W, Li C, Zheng C, et al: LGR4 is a receptor for RANKL
and negatively regulates osteoclast differentiation and bone
resorption. Nat Med. 22:539–546. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Wang H, Guo J, Gao J, Wang M, Xia
M, Wen Y, Su P, Yang M, Liu M, et al: LGR4, Not LGR5, enhances hPSC
hematopoiesis by facilitating mesoderm induction via TGF-Beta
signaling activation. Cell Rep. 31:1076002020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weng J, Luo J, Cheng X, Jin C, Zhou X, Qu
J, Tu L, Ai D, Li D, Wang J, et al: Deletion of G protein-coupled
receptor 48 leads to ocular anterior segment dysgenesis (ASD)
through down-regulation of Pitx2. Proc Natl Acad Sci USA.
105:6081–6086. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo W, Rodriguez M, Valdez JM, Zhu X, Tan
K, Li D, Siwko S, Xin L and Liu M: Lgr4 is a key regulator of
prostate development and prostate stem cell differentiation. Stem
Cells. 31:2492–2505. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Liu R, Wang F, Hong J, Li X, Chen
M, Ke Y, Zhang X, Ma Q, Wang R, et al: Ablation of LGR4 promotes
energy expenditure by driving white-to-brown fat switch. Nat Cell
Biol. 15:1455–1463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu J, Xie N, Xie K, Zeng J, Cheng L, Lei
Y, Liu Y, Song L, Dong D, Chen Y, et al: GPR48, a poor prognostic
factor, promotes tumor metastasis and activates β-catenin/TCF
signaling in colorectal cancer. Carcinogenesis. 34:2861–2869. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zheng H, Liu J, Cheng Q, Zhang Q, Zhang Y,
Jiang L, Huang Y, Li W, Zhao Y, Chen G, et al: Targeted activation
of ferroptosis in colorectal cancer via LGR4 targeting overcomes
acquired drug resistance. Nat Cancer. 5:572–589. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yue Z, Niu X, Yuan Z, Qin Q, Jiang W, He
L, Gao J, Ding Y, Liu Y, Xu Z, et al: RSPO2 and RANKL signal
through LGR4 to regulate osteoclastic premetastatic niche formation
and bone metastasis. J Clin Invest. 132:e1445792022. View Article : Google Scholar :
|
|
16
|
van Andel H, Ren Z, Koopmans I, Joosten
SP, Kocemba KA, de Lau W, Kersten MJ, de Bruin AM, Guikema JE,
Clevers H, et al: Aberrantly expressed LGR4 empowers Wnt signaling
in multiple myeloma by hijacking osteoblast-derived R-spondins.
Proc Natl Acad Sci USA. 114:376–381. 2017. View Article : Google Scholar :
|
|
17
|
Yi Z, Ma T, Liu J, Tie W, Li Y, Bai J, Li
L and Zhang L: LGR4 promotes tumorigenesis by activating
TGF-β1/Smad signaling pathway in multiple myeloma. Cell Signal.
110:1108142023. View Article : Google Scholar
|
|
18
|
Huang X, Guo B, Liu S, Wan J and Broxmeyer
HE: Neutralizing negative epigenetic regulation by HDAC5 enhances
human haematopoietic stem cell homing and engraftment. Nat Commun.
9:27412018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rajkumar SV, Dimopoulos MA, Palumbo A,
Blade J, Merlini G, Mateos MV, Kumar S, Hillengass J, Kastritis E,
Richardson P, et al: International Myeloma Working Group updated
criteria for the diagnosis of multiple myeloma. Lancet Oncol.
15:e538–e548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang J, Shi F, Liu X, Wu X, Hu C, Guo J,
Yang Q, Xia J, He Y, An G, et al: Proline promotes proliferation
and drug resistance of multiple myeloma by downregulation of
proline dehydrogenase. Br J Haematol. 201:704–717. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Noborio-Hatano K, Kikuchi J, Takatoku M,
Shimizu R, Wada T, Ueda M, Nobuyoshi M, Oh I, Sato K, Suzuki T, et
al: Bortezomib overcomes cell-adhesion-mediated drug resistance
through downregulation of VLA-4 expression in multiple myeloma.
Oncogene. 28:231–242. 2009. View Article : Google Scholar
|
|
22
|
Xia J, Zhang J, Wu X, Du W, Zhu Y, Liu X,
Liu Z, Meng B, Guo J, Yang Q, et al: Blocking glycine utilization
inhibits multiple myeloma progression by disrupting glutathione
balance. Nat Commun. 13:40072022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tu Q, Liu C, Wu D, Wen Y, Wang H, Su P,
Liu Y Ma F, Shi L and Zhou J: Establishment of the screening model
for highly efficient generation of megakaryocytes and platelets
from human pluripotent stem cells (in Chinese). Sci Sin Vitae.
47:1363–1374. 2017. View Article : Google Scholar
|
|
24
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2:1001412021.PubMed/NCBI
|
|
25
|
Zhou W, Yang Y, Xia J, Wang H, Salama ME,
Xiong W, Xu H, Shetty S, Chen T, Zeng Z, et al: NEK2 induces drug
resistance mainly through activation of efflux drug pumps and is
associated with poor prognosis in myeloma and other cancers. Cancer
Cell. 23:48–62. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takeyama Y, Sato M, Horio M, Hase T,
Yoshida K, Yokoyama T, Nakashima H, Hashimoto N, Sekido Y, Gazdar
AF, et al: Knockdown of ZEB1, a master epithelial-to-mesenchymal
transition (EMT) gene, suppresses anchorage-independent cell growth
of lung cancer cells. Cancer Lett. 296:216–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cui H, Li Z, Chen S, Li X, Chen D, Wang J,
Li Z, Hao W, Zhong F, Zhang K, et al: CXCL12/CXCR4-Rac1-mediated
migration of osteogenic precursor cells contributes to pathological
new bone formation in ankylosing spondylitis. Sci Adv.
8:eabl80542022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hu Y, Zhang Y, Ni CY, Chen CY, Rao SS, Yin
H, Huang J, Tan YJ, Wang ZX, Cao J, et al: Human umbilical cord
mesenchymal stromal cells-derived extracellular vesicles exert
potent bone protective effects by CLEC11A-mediated regulation of
bone metabolism. Theranostics. 10:2293–2308. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Holt RU, Baykov V, Rø TB, Brabrand S,
Waage A, Sundan A and Børset M: Human myeloma cells adhere to
fibronectin in response to hepatocyte growth factor. Haematologica.
90:479–488. 2005.PubMed/NCBI
|
|
30
|
Gazitt Y, Fey V, Thomas C and Alvarez R:
Bcl-2 overexpression is associated with resistance to
dexamethasone, but not melphalan, in multiple myeloma cells. Int J
Oncol. 13:397–405. 1998.PubMed/NCBI
|
|
31
|
Bianchi G, Czarnecki PG, Ho M, Roccaro AM,
Sacco A, Kawano Y, Gullà A, Samur AA, Chen T, Wen K, et al: ROBO1
promotes homing, dissemination, and survival of multiple myeloma
within the bone marrow microenvironment. Blood Cancer Discov.
2:338–353. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lai S, Cheng R, Gao D, Chen YG and Deng C:
LGR5 constitutively activates NF-κB signaling to regulate the
growth of intestinal crypts. FASEB J. 34:15605–15620. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wizenty J, Müllerke S, Kolesnichenko M,
Heuberger J, Lin M, Fischer AS, Mollenkopf HJ, Berger H, Tacke F
and Sigal M: Gastric stem cells promote inflammation and gland
remodeling in response to Helicobacter pylori via Rspo3-Lgr4 axis.
EMBO J. 41:e1099962022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Takeuchi M and Baichwal VR: Induction of
the gene encoding mucosal vascular addressin cell adhesion molecule
1 by tumor necrosis factor alpha is mediated by NF-kappa B
proteins. Proc Natl Acad Sci USA. 92:3561–3565. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
O'Shea JM and Perkins ND: Thr435
phosphorylation regulates RelA (p65) NF-kappaB subunit
transactivation. Biochem J. 426:345–354. 2010. View Article : Google Scholar
|
|
36
|
Perkins ND: The diverse and complex roles
of NF-κB subunits in cancer. Nat Rev Cancer. 12:121–132. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gong X, Yi J, Carmon KS, Crumbley CA,
Xiong W, Thomas A, Fan X, Guo S, An Z, Chang JT and Liu QJ:
Aberrant RSPO3-LGR4 signaling in Keap1-deficient lung
adenocarcinomas promotes tumor aggressiveness. Oncogene.
34:4692–4701. 2015. View Article : Google Scholar :
|
|
38
|
Yue F, Jiang W, Ku AT, Young AIJ, Zhang W,
Souto EP, Gao Y, Yu Z, Wang Y, Creighton CJ, et al: A
Wnt-independent LGR4-EGFR signaling axis in cancer metastasis.
Cancer Res. 81:4441–4454. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Salik B, Yi H, Hassan N, Santiappillai N,
Vick B, Connerty P, Duly A, Trahair T, Woo AJ, Beck D, et al:
Targeting RSPO3-LGR4 signaling for leukemia stem cell eradication
in acute myeloid leukemia. Cancer Cell. 38:263–278.e6. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mitchell JS, Li N, Weinhold N, Försti A,
Ali M, van Duin M, Thorleifsson G, Johnson DC, Chen B, Halvarsson
BM, et al: Genome-wide association study identifies multiple
susceptibility loci for multiple myeloma. Nat Commun. 7:120502016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Stevens PD and Williams BO: LGR4: Not just
for Wnt anymore? Cancer Res. 81:4397–4398. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cowan AJ, Green DJ, Kwok M, Lee S, Coffey
DG, Holmberg LA, Tuazon S, Gopal AK and Libby EN: Diagnosis and
management of multiple myeloma: A review. JAMA. 327:464–477. 2022.
View Article : Google Scholar : PubMed/NCBI
|