Introduction
The binding of the epidermal growth factor (EGF) to
its receptor (EGFR) triggers a large set of downstream processes,
ultimately causing cell growth, differentiation and proliferation
(1,2). The receptor interaction and
subsequent events are essential for the cell, although they may
also be a threat to it. The overexpression and increased activity
of EGFR have been linked to a variety of tumor types (3), rendering EGFR an important target for
cancer therapy. A full understanding of the biology behind the
binding of EGF and its link to cancer development is thus crucial
for future drug development.
Besides EGFR, the EGFR family consists of three
other members: HER2, HER3 and HER4. The receptors are known to
dimerize, as either homodimers or as heterodimers, with other
members of the family. The extent to which the dimerization occurs
and its correlation to ligand binding and further signaling have
been discussed for several years, however, are yet to be fully
understood (2). The binding of EGF
to EGFR generates a shift in conformation, enabling EGFR to
dimerize (4–6), which in turn activates the
cytoplasmic tyrosine kinase domain to become activated through
phosphorylation (7). Studies have
also demonstrated signs of preformed EGFR dimers on the cell
surface in the absence of bound EGF (8,9). For
example, HER2 overexpression has been associated with growth
factor-independent induction of EGFR-HER2 formation (10), as well as subsequent EGFR tyrosine
phosporylation (11,12). HER2 is consistently and
ligand-independently activated and is the preferred binding partner
of EGFR (13).
We have previously shown that the kinetic behavior
of the EGF-EGFR interaction may vary greatly among cell lines
(14), as demonstrated by
real-time interaction measurements performed using
LigandTracer® instruments. The binding curves showed at
least two parallel interaction events, one fast-on-fast-off
interaction and one higher affinity interaction, in which the
association and dissociation rates of EGF were much slower.
Thorough investigations have led to the hypothesis that the high
affinity contribution of the binding curve represents EGF binding
to either EGFR homodimers (EGFR-EGFR) or heterodimers (EGFR-HER2),
while the weaker interaction is EGF binding to EGFR monomers
(10). The monomeric and dimeric
EGFR forms could correspond to the previously discussed high- and
low-affinity EGFR receptor populations (15–17).
Furthermore, there is a difference between the rates of EGF
association and dissociation to EGFR homodimers and heterodimers
(10,12).
Tyrosine kinase inhibitors (TKI) are developed to
prevent downstream signaling from EGFR. Several TKI anticancer
drugs are currently available that focus on disrupting the kinase
activity of EGFR (18,19), including gefitinib (Iressa™),
lapatinib (Tykerb™) and erlotinib (Tarceva™) (20–22).
In theory, this causes a decrease in tumor growth, although the
response varies largely between patients (23,24).
Certain mutations in the receptor have been shown to be predictive
markers for either sensitivity or resistance (24,25),
although the mechanisms underlying variation between patient
responses have not been completely established (26,27).
In addition to growth rate inhibition, gefitinib,
erlotinib and the TKI AG1478 have been shown to promote EGFR
dimerization (10,28–30).
These dimers have been revealed to be non-active and
conformationally different from ligand-dependent dimer forms
(10,28,30).
By contrast, lapatinib has been shown to bind the inactive EGFR
conformation and does not induce dimer formation (31,32).
Previous studies have demonstrated that the presence
of gefitinib affects the kinetic properties (association and
dissociation rates) of the EGF-EGFR interaction in certain cell
lines, observed as an increase in affinity (10,14).
This observation may be the result of the larger number of EGFR
dimers present on the cell surface upon gefitinib treatment,
regardless of their kinase activity. Lapatinib, which in contrast
to gefitinib, stabilizes the inactive form of EGFR, was observed to
reduce the affinity of the EGF-EGFR interaction (14).
For cells growing in physiological conditions, EGF
binding and the formation of EGFR dimers induce a rapid
internalization of the occupied receptors through endocytosis.
Internalized ligand-receptor complexes are separated, inactivating
the EGFR. Through sorting endosomes, the unphosphorylated receptors
are either recycled back to the surface, or are transferred to
lysosomes for degradation (12,33,34).
The destiny of EGFR may depend on its dimerization partner.
Previous studies indicated slow or even completely disrupted EGFR
internalization in case of HER2 heterodimerization (35–37),
while findings of other studies (12,37)
claim that HER2 as a binding partner does not affect the
internalization rate per se, instead the subsequent
degradation processes. While EGFR homodimers are destined to a
rapid lysosomal degradation, EGFR-HER2 heterodimers have been
observed to be more prone to dissociate in early endosomes
(37), shown as a larger fraction
of recycled activated EGFRs in HER2 overexpressing cells (12).
The binding of TKIs to EGFR may also affect its
internalization and degradation. In their study, Nishimura et
al (27) detected a reduction
of the EGFR internalization rate and a retarded transition from
early to late endocytosis upon gefitinib treatment.
In interaction studies cell measurements are usually
performed at either room temperature or in a cool environment,
using manual end-point assays. Replacing end-point binding assays
with time-resolved assays may increase the information content of
the measurements, as shown in a previous study (14). The purpose of non-physiological
temperature is to slow other cell processes, such as growth,
internalization, recycling and formation of new receptors, i.e.,
events that may influence the interaction data, thus rendering them
more difficult to interpret. A drawback of restricting typical cell
behavior is that the measured interaction data are obtained from
artificial circumstances and may thus not always be an adequate
representation of the interaction under physiological
conditions.
The primary aim of this study was to investigate the
molecular interactions within the EGFR system in human cancer
cells, by introducing perturbations, such as temperature changes
and TKIs on cell lines expressing different proportions of EGFR and
HER2. A secondary goal was to develop new generic tools to
investigate cell processes associated with receptor interactions,
using real-time interaction measurement technology. Real-time data
of the EGF-EGFR interaction from measurements conducted at cold or
room temperatures, as well as in an incubator environment at 37°C
using the human tumor cell lines A431, U343 and SKOV3 and the four
TKIs gefitinib, lapatinib, AG1478 and erlotinib were demonstrated.
When combining interaction data with the degree of internalization
of 125I-EGF at different time points and temperature
settings, we were able to hypothesize as to how EGF is bound,
internalized, recycled and degraded depending on EGFR and HER2
expression, as well as treatment with TKIs.
Materials and methods
Cell culture
The human squamous carcinoma cell line A431 (CLR
1555; ATCC, Rocksville, MD, USA), the human ovarian carcinoma cell
line SKOV3 (HTB-77; ATCC), the human glioma cell line U343MGaCl2:6
[a subclone of U343MG (38)],
denoted U343, were used in this study. Cells were selected to
represent a range of EGFR and HER2 expression: 2E6 EGFR/cell and
2E5 HER2/cell for A431, 6E5 EGFR/cell and 3E4 HER2/cell for U343
and 3E5 EGFR/cell and 2E7 HER2/cell for SKOV3 (10). HER3 and HER4 populations are
considered small enough on the cell surface to be neglected
(30,39). EGF has previously been confirmed to
bind specifically to EGFR using different assays (40–42).
The cells were cultured in Ham’s F10 (A431 and U343)
or RPMI (SKOV3) cell culture medium (Biochrom AG, Berlin, Germany),
supplemented with 10% fetal calf serum (FCS) (Sigma-Aldrich, St.
Louis, MO, USA), L-glutamine (2 mM) and PEST (penicillin 100 IU/ml
and streptomycin 100 μg/ml, Biochrom AG) in a humified
incubator at 37°C, equilibrated with 5% CO2. The cells
were seeded on a local area of a cell culture dish (Nunclon™, size
100×20; NUNC A/S, Roskilde, Denmark) for the LigandTracer
measurements, as described previously (43).
Environments
The measurements described below were conducted i)
at room temperature, ∼22°C with normal CO2 levels; ii)
in a humified incubator, at 37°C with 5% CO2; or iii) in
a cold room, at ∼7°C.
Radiolabeling
Human EGF (Chemicon International, Temecula, CA,
USA) was labeled with 125I (Perkin-Elmer, Wellesley, MA,
USA), using the chloramine-T protocol (44). Chloramine-T (Sigma-Aldrich, St.
Louis, MO, USA) and sodium metabisulfite (Sigma-Aldrich, Stockholm,
Sweden) were used for the labeling reactions. Excess of
125I and reagents were separated from the labeled EGF
solution using a NAP-5 column (GE Healthcare, Waukesha, WI, USA)
equilibrated with phosphate-buffered saline (PBS) (10 mM, pH 7.4,
140 mM NaCl).
Treatment of cells with TKIs
Cells were treated for 48 h in 1 μM of either
gefitinib (Biaffin GmbH & Co KG, Kassel, Germany), lapatinib,
AG1478 or erlotinib (all three from LC Laboratories, Woburn, MA,
USA) prior to binding studies in LigandTracer, acid wash
measurements or cell counting. After the first 24 h of treatment,
fresh medium containing TKI was added to ensure a continuous
supply.
Real-time measurements of the
125I-EGF-EGFR interaction at room temperature and in an
incubator
The 125I-EGF-EGFR interaction was
detected in real-time on cultured tumor cells, using two
LigandTracer Grey instruments in parallel, one of which was placed
at room temperature, and the other in an incubator at 37°C. The
incubator instrument was temperature-equilibrated 1 h prior to
measurement. The technology behind and the validation of
LigandTracer has been described previously (43,45).
The uptake of 2.7 (A431), 1.5 (U343) and 2 nM (SKOV3) was measured
for either 2 or 4 h. The concentrations were sufficient to ensure
<10% loss of free 125I-EGF in solution due to
binding, reducing the risk of depletion affecting the results. The
shorter uptake phase (2 h) was followed by a 2 h retention
measurement. This procedure was performed using either normally
treated cells grown in complete culture medium or cells pre-treated
with TKI. In the latter case, the same TKI was included in the
medium during the measurement. Note that the term ‘uptake’ refers
to the sum of the events occurring during incubation, i.e., when
125I-EGF is present, while ‘retention’ is the
combination of events taking place once the 125I-EGF
solution has been removed and replaced with fresh medium. Examples
of biological events that affect measured 125I signal
are EGF association and dissociation to/from EGFR and the excretion
of 125I (or 125I-EGF) from the cell, once the
ligand has been internalized.
Each measurement was repeated at least twice,
essentially under the same experimental conditions. Obtained curves
were evaluated using TraceDrawer 1.4 (Ridgeview Instruments AB,
Uppsala, Sweden).
Acid wash measurements used to estimate
membrane bound and internalized 125I-EGF
Cultured A431, U343 and SKOV3 cells were seeded in
12-well plates (Nunclon, size 100×20; NUNC A/S). Forty-eight hours
prior to measurements, the cells were grown in a normal, TKI-free
environment or in complete cell culture medium containing 1
μM of gefitinib, lapatinib, AG1478 or erlotininb. Cells were
incubated with 125I-EGF for 40, 90 or 240 min at either
room temperature or 37°C, using the same concentrations as in the
LigandTracer incubator measurements. After a washing step, the
cells were treated with acid in 0.2 M Gly-HCl pH 2.5 containing
0.15 M NaCl and 4 M Urea for 5 min on ice to strip the cell
surface-associated 125I-EGF from the membrane. Then, 1 M
NaOH was added and incubated at 37°C for at least 1 h to fully
disrupt the structure of the cells, thus enabling collection of
internalized 125I-EGF. The activity from the solutions
was measured using an automatic gamma counter (1480 WIZARD™ 3″,
PerkinElmer, Waltham, MA, USA).
To obtain further information concerning the
EGF-EGFR interaction, a few combinations of cells and treatments
were studied after 90 min incubation on ice or at 7°C in a cold
room.
Effects of TKIs on the kinetics of the
125I-EGF-EGF interaction at room temperature and at
7°C
A stepwise increase of 125I-EGF
concentration was monitored at room temperature or inside a cold
room (7°C) using LigandTracer Grey, as described previously
(14). Instruments and liquids
were temperature equilibrated before measurements were conducted in
the cold room. The set of studied concentrations were cell
type-specific and selected to match the previously estimated
affinity of the EGF-EGFR interaction at room temperature in the
three hosting cell lines (2.7, 9 and 27 nM for A431; 0.5, 1.5 and
4.5 nM for U343 and 0.7, 2 and 6 nM for SKOV3) (14). As cell detachment may be a problem
for cells exposed to low temperatures for a longer time period,
only the two highest concentrations were used in cold room
measurements to shorten the uptake phase. The binding of
125I-EGF was followed for 2.5 h for each concentration,
followed by a retention measurement overnight using fresh medium.
Cell detachment in cold room was estimated after 5 h of retention
measurement by counting cells in the media. TKI pre-treated cells
had the same inhibitor present in the medium throughout the
run.
Interaction map analysis
Data produced in LigandTracer were analyzed using
the Interaction Map® (IM) method (Ridgeview Diagnostics
AB, Uppsala, Sweden) to investigate the heterogeneity of the
EGF-EGFR interaction, as described previously (10,46,47).
IM deciphers the different components of a measured interaction
from the shape of its real-time binding curve. Each component is a
contributing interaction describing one type of ligand binding to
one type of target. The components are presented as areas in an
on-off map (48) with darkness
relative to their weighing factors, i.e., degree of contribution to
the measured binding curve.
TKI effects in cell count
Tumor cells were cultivated in 12-well plates and
treated with TKIs for 48 h prior to the addition of a trypsin-EDTA
solution (Biochrom AG) at 37°C to cause cell detachment. Cells were
counted to estimate growth-disturbing effects caused by TKIs in
A431, U343 and SKOV3 cells.
Results
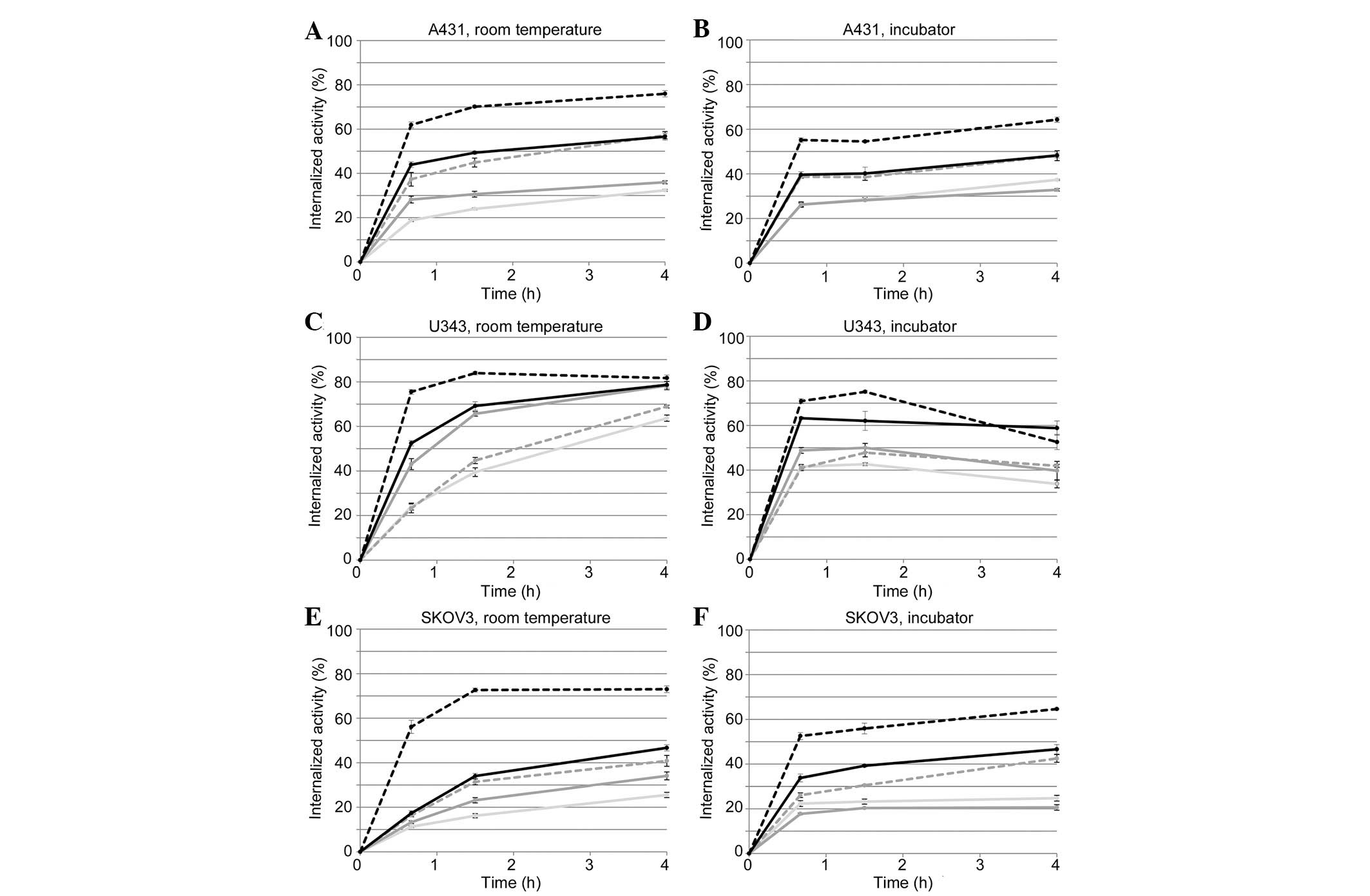
Real-time measurement data of the
125I-EGF-EGFR interaction in incubator shows signs of
internalization and processing of ligand
Data from real-time measurements of the binding of
125I-EGF are shown in Fig.
1. The interaction between 125I-EGF and A431
(Fig. 1A and B), U343 (Fig. 1C and D) or SKOV3 (Fig. 1E and F) cells was followed at room
temperature (black curves) or inside an incubator at 37°C (grey
curves), for 4 h (solid line) or 2 h, followed by a 2 h retention
measurement (dotted line; mark indicates the start of this
retention phase). Curves have been normalized at t=10 min to
minimize the impact on signal level due to variations in the
receptor number caused by differences in cell counts. The
measurements were performed under either normal conditions, with
control cells grown in complete cell culture medium, or in the
presence of 1 μM of gefitinib, lapatinib, AG1478 or
erlotinib. The effects of TKI on the cell uptake of
125I-EGF at 37°C compared to room temperature were
similar between the four TKIs and are represented by data from the
gefitinib effect measurements (Fig.
1B, D and F).
When 125I-EGF was added to A431 or U343
cells at 37°C, the signal initially increased rapidly due to
binding of 125I-EGF to membrane bound EGFR. After ∼40
min the increase was reduced and the signal started to decrease as
EGFR with bound 125I-EGF likely was internalized and
degraded, inducing an excretion of the nuclide. In SKOV3 cells no
clear decrease was visible in signal, suggesting that some (or
several) of the cell processes that evenutally led to excretion of
125I were considerably slower in the SKOV3 compared to
the A431 or U343 cells.
Temperature changes may affect the kinetics of an
interaction. This was clearly observed for the dissociation of
125I from SKOV3 cells, which accelerated with increased
temperature.
When treated with gefitinib, lapatinib, AG1478 or
erlotinib, the differences between room temperature and incubator
measurements were reduced. The clear decrease in signal observed in
TKI untreated A431 and U343 cells was small or non-existent in the
presence of any of the four TKIs (Fig.
1B and D). This finding indicates that either the
internalization rate or intracellular degradation was affected by
the TKIs.
In SKOV3 cells, the effect of TKIs (Fig. 1F) was not as clear as in A431 or
U343 cells. The main difference was observed in the dissociation
rate, which was slower when cells had been treated with lapatinib
(data not shown) and notably slower with gefitinib, AG1478 and
erlotinib.
Acid wash measurements demonstrated a
decrease in the amount of internalized 125I-EGF in A431,
U343 and SKOV3 cells induced by TKIs
Data from acid- and subsequently base-treated cells
show the amount of membrane-bound and internalized
125I-EGF, respectively. The average fraction of the
internalized activity of total cell-associated activity (membrane
bound + internalized) at room temperature or at 37°C (inside an
incubator) at different time points is shown in Fig. 2. Error bars show the distribution
of the triplicates, (i.e., maximum value-minimum value)/2. For the
three cell lines, the TKIs significantly decreased the fraction of
internalized 125I activity at both room temperature and
at 37°C, although to a lesser extent by erlotinib. AG1478 in A431
and SKOV3 cells, and lapatinib in U343 cells, also showed a lower
impact on the internalization degree.
 | Figure 2Internalized activity of
125I-EGF as percentage of total cell-associated activity
for cultured (A and B) A431, (C and D) U343 and (E and F) SKOV3
cells, grown either in normal cell culture medium (black, dashed
line) or pre-treated with 1 μM of gefitinib (light grey,
solid line), lapatinib (grey, solid line), AG1478 (grey, dashed
line) or erlotinib (black, solid line). The tyrosine kinase
inhibitors (TKIs) reduced the degree of internalized activity, most
notably observed for gefitinib, lapatinib and AG1478. Error bars
show the distribution of triplicates, (maximum value-minimum
value)/2. |
The total cell-associated activity was plotted over
time (data not shown) and matched the real-time binding curves from
LigandTracer measurements at room temperature and 37°C well
(Fig. 1), which serves as a
quality control for the two methods. The degree of internalization
was similar at room temperature and at 37°C, for the three cell
lines, or even higher at room temperature.
A relatively large amount of 125I-EGF was
internalized in SKOV3 cells (Fig.
2E–F) at both temperatures (∼70% of total cell-associated
activity), similar to what was observed in A431 cells (Fig. 2A and B).
The fraction of internalized 125I-EGF of
the cell associated 125I-EGF was constant or increased
slightly over time for A431 and SKOV3 cells at 37°C, for the
treatments (Fig. 2B and F). For
U343 cells the degree of internalized activity was constant or
decreased (Fig. 2D). The decrease
indicates that the degradation and excretion of 125I-EGF
was faster than the internalization.
When incubated inside a cold room (7°C) or on ice,
the percentage of cell-associated 125I-EGF inside the
cell was ∼5 and 2%, respectively (Table I).
 | Table IDegree of internalization at 7°C and
on ice. |
Table I
Degree of internalization at 7°C and
on ice.
| Cell line | 7°C (%) | On ice (%) |
|---|
| A431 | 6.0±0.3 | 2.2±0.3 |
| A431+gef | 5.1±0.4 | 2.8±0.1 |
| U343 | 4.5±0.2 | 1.7±0.3 |
| SKOV3 | 4.2±0.1 | 1.5±0.1 |
TKIs affect the kinetics of the
125I-EGF-EGFR interaction albeit differently, depending
on the inhibitor and cell line
The uptake and retention of 125I-EGF were
detected using a stepwise increase of the 125I-EGF
concentration, three concentrations at room temperature and two for
the measurements performed at 7°C in a cold room (Fig. 3). The curves were normalized at the
beginning of the final incubation to remove any signal magnitude
differences originating from total receptor number related to cell
count. When comparing the binding of 125I-EGF to the
differently treated cells, the shape of the curve at each uptake
and retention phase is important, as well as the manner in which
the signal increases at each addition of 125I-EGF.
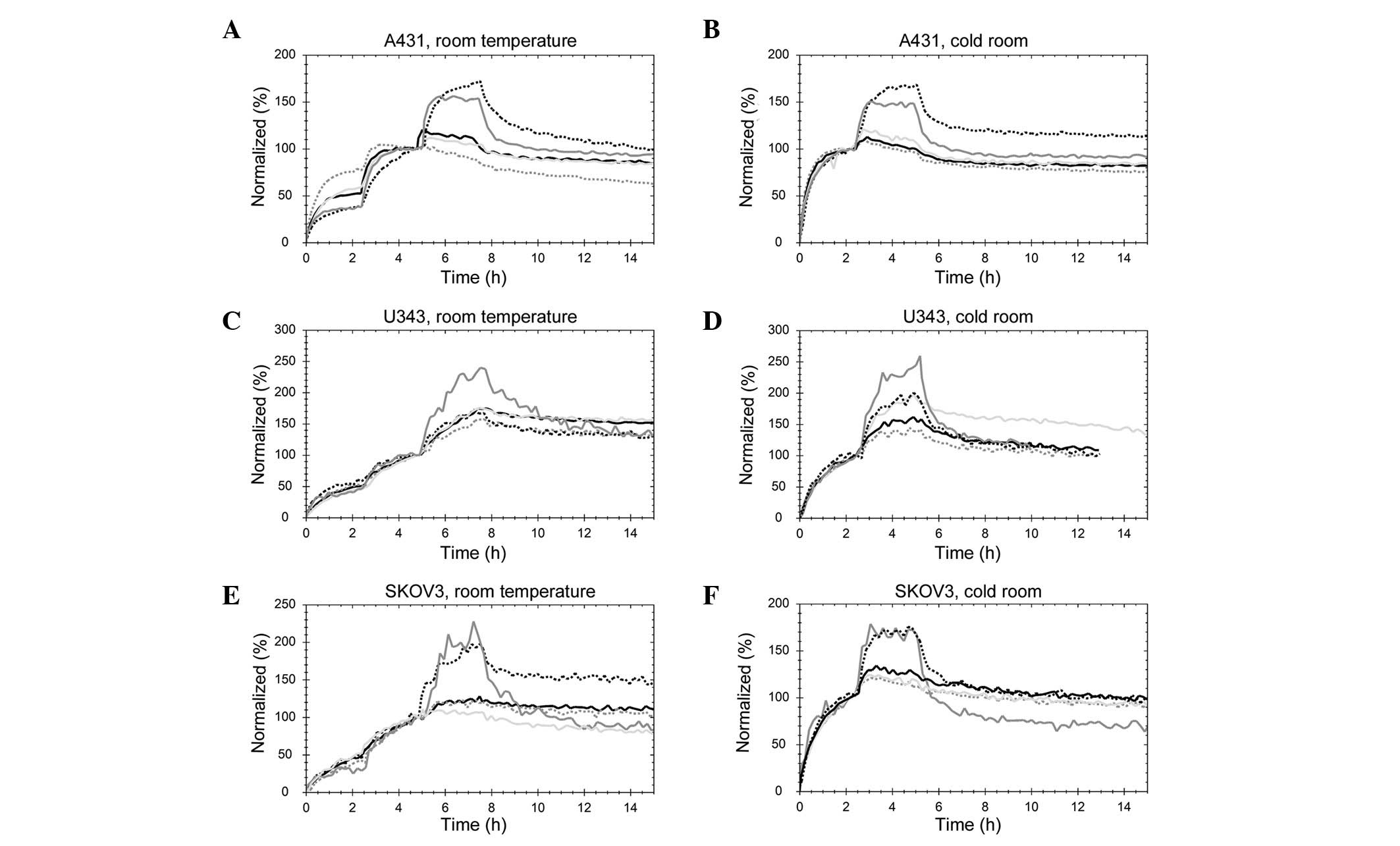 | Figure 3Real-time binding curves of the
125I-EGF-EGFR interaction measured in (A) A431 at room
temperature, (B) A431 at 7°C, (C) U343 at room temperature, (D)
U343 at 7°C, (E) SKOV3 at room temperature or (F) SKOV3 cells at
7°C. 125I-EGF was detected using stepwise increase to
match previously estimated overall affinity: 2.7, 9 and 27 nM for
A431 cells (9 and 27 nM at 7°C); 0.5, 1.5 and 4.5 nM for U343 cells
(1.5 and 4.5 nM at 7°C) and 0.7, 2 and 6 nM for SKOV3 cells (2 and
6 nM at 7°C). The cells were grown either in normal cell culture
medium (black, dotted line) or pre-treated with 1 μM of
gefitinib (light grey, solid line), lapatinib (grey, solid line),
AG1478 (grey, dotted line) or erlotinib (black, solid line). |
Gefitinib (light grey, solid line), AG1478 (grey,
dotted line) and erlotinib (black, solid line) clearly reduced the
dissociation rate, or possibly disrupted the fast-off component of
the interaction in A431 cells at both room temperature and 7°C
(Fig. 3A and B). Furthermore, the
receptors were almost fully saturated already after the second
concentration (9 nM), as low or no increase was observed in formed
complexes for the third concentration. This finding indicates that
9 nM is considerably higher compared to the apparent KD
of the 125I-EGF-EGFR interaction, when A431 cells are
treated with gefitinib, AG1478 or erlotinib. AG1478 increased the
affinity the most, observed as a low increase in signal when adding
the second concentration at room temperature. This finding
indicates that as little as 2.7 nM of 125I-EGF is enough
to make most of the receptors have an EGF bound, when A431 cells
are treated with AG1478. Lapatinib modified the interaction in a
completely different manner. The increases in signal following the
addition of 125I-EGF were similar to the control cells,
although the time to reach equilibrium was considerably shorter.
The TKI made the faster contributing interaction even faster, and
increased the stability of the slower interaction (resulting in a
slower dissociation rate).
In U343 cells, small effects of gefitinib, AG1478
and erlotinib on the 125I-EGF-EGFR interaction were
visible at room temperature (Fig.
3C). Lapatinib caused a faster dissociation of
125I-EGF and amplified the degree of signal increase at
each concentration, both of which indicate a lower affinity in the
interaction between ligand and receptor. At 7°C, gefitinib, AG1478
and erlotinib increased the affinity of the
125I-EGF-EGFR interaction, observed as a slower
dissociation and (for AG1478 and erlotinib) a larger fraction of
EGFR bound by low EGF concentrations (Fig. 3D).
In SKOV3 cells treated with gefitinib, AG1478 and
erlotinib, the 125I-EGF-EGFR interaction had a higher
affinity compared to control cells at both room temperature and
7°C, most clearly visible as a lower increase in signal at each
addition of 125I-EGF. Lapatinib treatment made the
fast-on-fast-off contribution of the binding of 125I-EGF
more abundant, shifting the overall interaction into a lower
affinity state (Fig. 3E and
F).
The fact that the binding curves obtained at room
temperature and at 7°C are similar and the fact that
125I-EGF is internalized to a larger degree at room
temperature compared to 7°C suggests that there is a recycling of
125I-EGF-EGFR complexes and that this process is more
rapid than the dissociation rate of the 125I-EGF-EGFR
complexes.
In general, TKIs are capable of modifying the
interaction between EGF and its receptor. Gefitinib, AG1478 and
erlotinib sometimes increased the affinity of the interaction,
although the magnitiude of the affinity increase varied among the
host cell lines. Lapatinib behaved in a markedly different way,
decreasing the overall affinity and sometimes increasing the rate
of 125I-EGF association and dissociation.
Real-time interaction data produced at room
temperature were further analyzed using IM (Fig. 4). This method analyzes the shape of
a binding curve and deciphers the mixture of contributing
interactions possibly resulting in such a curve shape. Each
interaction is presented as an area in an on-off map, where the
coordinate represents the kinetic properties ka and kd,
and the darkness the extent to which the interaction contributes to
the measured real-time binding curve. The effects of gefitinib,
AG1478 and erlotinib on the EGF-EGFR interaction were almost
identical and are represented by the erlotinib treatment.
Similarly, TKI effects were comparable in the three cell lines and
depicted by the binding of EGF to SKOV3 cells. The contributions of
the interactions were calculated by integration of the respective
areas in the map.
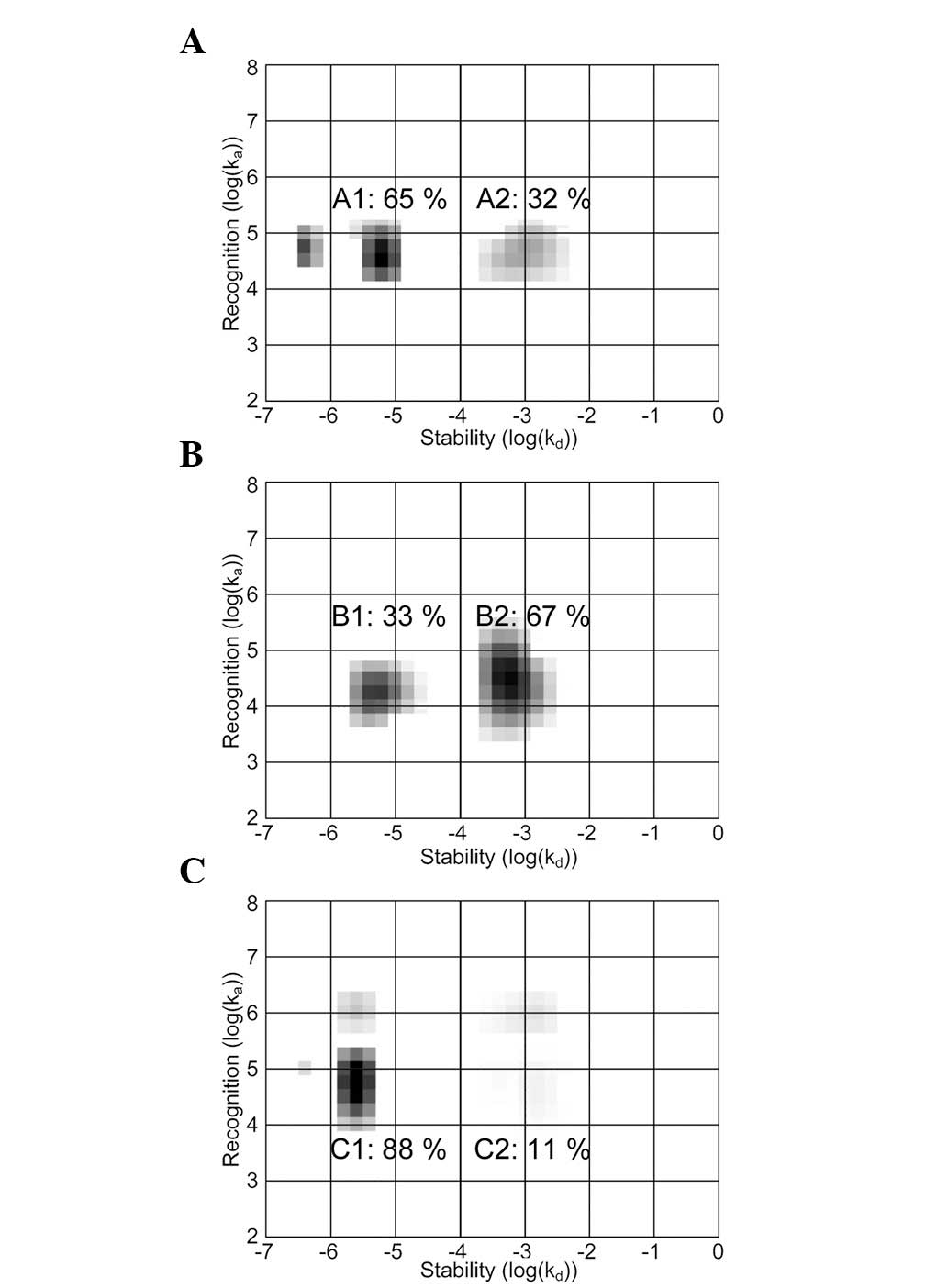
The EGF-EGFR interaction measured in A431, U343 and
SKOV3 cells was heterogeneous, observed as two major areas in the
maps. The areas, corresponding to parallel EGF-EGFR interactions,
represent various dissociation rate constants of kd. The
contribution of the lesser stable interaction to the measured curve
increased from 32 to 67% when SKOV3 cells were treated with
lapatinib (B2). The presence of gefitinib, AG1478 and erlotinib
induced the reverse effect, making the higher-affinity interaction
(C1) more common.
Gefitinib, lapatinib, AG1478 and
erlotininb decrease cell growth differently depending on cell
line
Cells grown in 12-well plates were treated with
gefitinib, lapatinib, AG1478 or erlotininb for 48 h. The number of
cells were counted in triplicates and compared with control cells
grown in complete medium, for a rough estimation of growth effects
caused by the TKIs. Table II shows
the average cell count for each treatment, in relation to cell
count for the control (set to 100%). Errors are presented as
(maximum value-minimum value)/2 of the cell count, with quadratic
propagation of error. In this experiment, the four TKIs decreased
cell growth to various extents, depending on the inhibitor and cell
line. SKOV3 shows a similar sensitivity to the four TKIs, reducing
the growth rate to in average 55–68%. In A431 and U343, however,
the effect varied markedly, with some extreme cases where the cell
count was only 25–30% of the control.
 | Table IIEffect of gefitinib, lapatinib,
AG1478 and erlotinib on cell growth. |
Table II
Effect of gefitinib, lapatinib,
AG1478 and erlotinib on cell growth.
| Cell number
|
|---|
| A431 (%) | U343 (%) | SKOV3 (%) |
|---|
| Gefitinib | 52.7±7.2 | 70.4±6.4 | 67.7±12.9 |
| Lapatinib | 36.8±3.1 | 27.0±4.2 | 67.7±6.9 |
| AG1478 | 30.2±3.3 | 28.2±4.8 | 63.6±3.9 |
| Erlotinib | 23.3±4.3 | 66.6±13.1 | 55.5±5.6 |
Discussion
The aim of this study was to further investigate the
interaction between EGF and its receptor and to use interaction
analysis at different temperatures as a new method for resolving
parts of the receptor biology. Although a temperature shift towards
37°C renders interaction data on cells markedly more complicated,
this was deemed an important development for a proper understanding
of the processes involving the EGFR system. The data described in
this study are complex and demonstrate several underlying
processes: the association and dissociation rates of the EGF-EGFR
interaction itself, as well as related events, such as
dimerization, internalization, recycling and degradation of the
receptor and its ligand. Thus, this paper describes a first attempt
to separate the velocities of these processes, at normal conditions
and in the presence of TKIs to identify the rate-limiting steps of
EGFR-related biology.
The detection technology used in this study relies
on quantification of a label, in this case 125I. Thus,
the measurements follow the path of the label only. Several of the
mechanistic steps in the EGF-EGFR interaction are conducted in a
manner that cannot easily be resolved. The interaction analysis
method used will not distinguish between surface-bound and
internalized ligands, and the only way to separate the signal from
such events is through perturbation with agents and conditions (in
the present case TKIs and temperature) known to impact the
processes in different ways, as well as cell lines with different
proportions of the receptor components. Furthermore, when several
events are mixed into one measurement step, the rate-limiting
process dictates the detected output. Thus, the measurements
presented in this study constitute a complex pattern of
complementing experiments, aiming at deconvoluting the underlying
processes.
For A431 and U343 cells grown in the absence of
TKIs, the signal from 125I-EGF associated with cells
started to decrease after ∼40min. Consequently, this indicated
125I-EGF to be associated with EGFR, further receptor
dimerization, internalization and at a later stage the degradation
and excretion of EGF and its nuclide. The combined results from
LigandTracer and acid-wash measurements at room temperature and
37°C indicated that the main difference between the two
temperatures (in the absence of TKIs) was not the internalization
itself, but the rate of subsequent degradation and excretion of
125I-EGF once inside the cells. These degradation
processes become much slower when going from 37°C to room
temperature. For HER2 overexpressing SKOV3 cells, the
internalization mechanisms were active, although the degradation
processes were slow or even disrupted at 37°C. This clarifies which
of the cell processes are affected when HER2 serves as a dimer
partner, as discussed previously (12,35,36).
When comparing cold conditions to room temperature
the interaction data is similar, however, the internalization is
turned off at 7°C. We thus conclude that the internalization and
recycling processes are more rapid compared to the dissociation
rate of the EGF-EGFR complex, rendering internalization and
recycling difficult to detect by interaction analysis alone at room
temperature.
When cells were treated with any of the four TKIs
gefitinib, lapatinib, AG1478 or erlotinib, the LigandTracer-binding
curves from the incubator measurements became more similar to data
from the room temperature measurements, indicating that the degree
of either nuclide excretion or internalization was reduced
(Fig. 1). Acid wash measurements
provided additional information by showing that the degree of
internalization was significantly lower in the three cell lines in
the presence of TKIs, although most likely not low enough to cause
the similarity of 37°C to room temperature. Thus, the TKIs are
believed to shift the balance towards receptor complexes on the
cell surface, as well as to reduce the speed of the excretion.
Measurements of the impact of TKIs on interaction
kinetics revealed that gefitinib, AG1478 and erlotinib increased
the affinity in A431 and SKOV3 cells, while lapatinib decreased it
in the three cell lines. Gefitinib, AG1478 and erlotinib have been
shown to cause growth factor-independent formation of EGFR dimers,
while lapatinib strengthens the inactive monomeric form of EGFR
(10,28–32).
The correlation between affinity increases and dimerization
induction strengthens previous hypothesis that EGF binds to dimeric
EGFR forms with a higher affinity compared to the EGFR monomer
(10,17). Notably, this is not associated with
internalization and degradation perturbations, as all four TKIs had
an effect on these events (Fig. 1B and
D and 2).
Translating real-time binding data into IMs
demonstrated the heterogeneity of the EGF-EGFR interaction further.
Through experiments with dimerization-preventing agents, the
authors previously hypothesized that the two main areas in the maps
correspond to EGF interacting with either EGFR monomers or dimers,
where the dimer interaction was assumed to be the more stable one
(Fig. 4; A1, B1 and C1,) (10). TKI effects investigated in this
study confirm the identification of these two map areas. The IM
method provides means to quantify the effects on dimerization
caused by TKIs. The quantification is fast and straightforward and
may be considered to be a convenient alternative to more
time-consuming manual dimerization determining methods, such as
immunoblotting. Additionally, this quantification is based entirely
on the function of the receptor forms (i.e., binding abilities) and
not their structures per se, in contrast to several manual
alternatives.
From the findings described above, we formed a
hypothesis on the pattern of EGF-binding kinetics, internalization,
recycling, degradation and excretion in the presence of TKIs at
different temperatures (Fig. 5).
EGF may interact with EGFR monomers present on the cell surface
(black, dashed line), enabling EGFR to dimerize, likely as
homodimers on EGFR-rich A431 and U343 cells (black, solid line) and
HER2 heterodimers on SKOV3 cells, which have a high HER2 expression
(black, dotted line). EGF was suggested to possibly associate to as
well as dissociate from the catalytically inactive pre-formed
gefitinib-, AG1478- and erlotinib-induced EGFR dimers (grey solid
and dotted lines for EGFR homo- and EGFR-HER2 heterodimers,
respectively). IM data show that the dissociation of EGF from EGFR
dimers is slower compared to monomeric EGFR. The TKI induction of
dimers reduces the overall dissociation rate, thus increasing the
apparent affinity. At room temperature and 37°C, but not at 7°C,
the two EGF dependently formed homo- and heterodimers (black solid
and dotted line) are rapidly internalized. EGF is only visibly
excreted from A431 and U343 cells at 37°C, indicating that full
degradation requires a physiological temperature and that EGFR-HER2
heterodimers (black, dotted line) are degraded to a lesser extent
compared to EGFR homodimers (black, solid line). Gefitinib-,
AG1478- and erlotinib-induced dimers have a perturbed
internalization and may also be degraded more slowly. Lapatinib
strengthens the monomeric EGFR form, thus reducing the overall
number of dimers, which may have caused the lower degree of
internalization (Fig. 2, grey solid
line) as EGFR monomers are not likely to internalize. We
strongly believe that a subset of internalized, TKI-induced or
naturally formed EGFR dimers may recycle to the surface, although
this remains to be investigated. Recycling is likely to be less in
cells with a high degradation and excretion level.
 | Figure 5Proposed mechanism of EGF-GFR
kinetics, internalization, recycling, degradation and excretion
with regard to temperature and tyrosine kinase inhibitors (TKIs)
treatment. EGFR monomers (black square, dashed line) mainly forms
homodimers (black square, solid line) on A431 and U343 cells and
EGFR-HER2 heterodimers (black square, dotted line) on SKOV3 cells
upon EGF binding, where the choice of dimer partner depends on
overall EGFR/HER2 expression. Gefitinib, AG1478 and erlotinib
induce dimers (grey squares, solid and dotted lines) that are
catalytically inactive. EGF dissociates slower from EGFR dimers
compared to monomers. Internalization occurs at room temperature
and 37°C, but not at 7°C. EGFR homodimers are degraded and EGF
residues and 125I excreted at 37°C, but not at room
temperature (RT). No or little excretion was observed on SKOV3
cells, indicating that EGFR-HER2 heterodimers are degraded to a
lesser extent. The interaction between gefitinib, AG1478 or
erlotinib and EGFR perturbs internalization and possibly also
degradation. Lapatinib (not shown) stabilizes the monomeric EGFR
form, which may explain the lower amount of internalized EGF as
internalization is likely restricted to EGFR dimers. EGFR recycling
is probable, but has not been investigated in detail. |
As discussed previously, measurements of
interactions in cells are often conducted at non-physiological
temperatures to reduce the effect of other cell activities. This is
a convenient strategy and often necessary to isolate information on
the interaction itself in a measurable manner. In this study, the
problem is approached from a different perspective by utilizing
cell processes such as internalization and excretion present during
measurement, thereby providing additional information on the
receptor system. The variation of temperature is a means to turn on
and off some of these processes, rendering it possible to separate
and investigate these processes a few at a time. The authors
understand that the method and approach used in this paper are
novel and different from the conventional ones in the field.
Findings reported in this paper are consistent with findings of
previous studies, strengthening the credibility of our results.
Following the labeled ligand in real time is a convenient and
information-rich approach to estimate the combined rates of
internalization, degradation and excretion to identify the
rate-limiting step with minimal labor time. The use of
complementing time-resolved assay set-ups to deconvolute complex
receptor biology processes, as exemplified for the EGF-EGFR system
in this paper, is a powerful strategy to advance the field of
molecular oncology.
In summary, this study provides additional
information on the intricate interplay of EGF and EGFR through the
real-time analysis of the cell association of 125I-EGF.
Analyses in the presence of the TKIs rendered the comparison of the
impact of the EGF-EGFR interaction on the kinetics, as well as the
evaluation of the way the cell processes subsequently leading to
125I excretion were altered, possible. By applying IM on
the continuous data additional information was obtained on the
manner in which the TKIs alter EGFR dimer levels. This study also
confirms some of the previous findings described in the literature,
providing additional information to create a broad understanding of
effects on the EGFR system induced by the TKIs gefitinib,
lapatinib, AG1478 and erlotinib, using completely different
assays.
Acknowledgements
The authors would like to thank Rick
Filonzi for encouraging scientific discussions on the need for
comparing protein interactions at different temperatures.
References
|
1
|
Scaltriti M and Baselga J: The epidermal
growth factor receptor pathway: a model for targeted therapy. Clin
Cancer Res. 12:5268–5272. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blume-Jensen P and Hunter T: Oncogenic
kinase signalling. Nature. 411:355–365. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferguson KM, Berger MB, Mendrola JM, Cho
HS, Leahy DJ and Lemmon MA: EGF activates its receptor by removing
interactions that autoinhibit ectodomain dimerization. Mol Cell.
11:507–517. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garrett TP, McKern NM, Lou M, et al:
Crystal structure of a truncated epidermal growth factor receptor
extracellular domain bound to transforming growth factor alpha.
Cell. 110:763–773. 2002. View Article : Google Scholar
|
|
6
|
Ogiso H, Ishitani R, Nureki O, et al:
Crystal structure of the complex of human epidermal growth factor
and receptor extra-cellular domains. Cell. 110:775–787. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Gureasko J, Shen K, Cole PA and
Kuriyan J: An allosteric mechanism for activation of the kinase
domain of epidermal growth factor receptor. Cell. 125:1137–1149.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu P, Sudhaharan T, Koh RM, et al:
Investigation of the dimerization of proteins from the epidermal
growth factor receptor family by single wavelength fluorescence
cross-correlation spectroscopy. Biophys J. 93:684–698. 2007.
View Article : Google Scholar
|
|
9
|
Tao RH and Maruyama IN: All EGF(ErbB)
receptors have preformed homo- and heterodimeric structures in
living cells. J Cell Sci. 121:3207–3217. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Björkelund H, Gedda L, Barta P, Malmqvist
M and Andersson K: Gefitinib induces epidermal growth factor
receptor dimers which alters the interaction characteristics with
125I-EGF. PLoS One. 6:e24739
|
|
11
|
Bollig-Fischer A, Dziubinski M, Boyer A,
Haddad R, Giroux CN and Ethier SP: HER-2 signaling, acquisition of
growth factor independence, and regulation of biological networks
associated with cell transformation. Cancer Res. 70:7862–7873.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Worthylake R, Opresko LK and Wiley HS:
ErbB-2 amplification inhibits down-regulation and induces
constitutive activation of both ErbB-2 and epidermal growth factor
receptors. J Biol Chem. 274:8865–8874. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tzahar E, Waterman H, Chen X, et al: A
hierarchical network of interreceptor interactions determines
signal transduction by Neu differentiation factor/neuregulin and
epidermal growth factor. Mol Cell Biol. 16:5276–5287. 1996.
|
|
14
|
Björkelund H, Gedda L and Andersson K:
Comparing the epidermal growth factor interaction with four
different cell lines: intriguing effects imply strong dependency of
cellular context. PLoS One. 6:e165362011.PubMed/NCBI
|
|
15
|
Lax I, Bellot F, Howk R, Ullrich A, Givol
D and Schlessinger J: Functional analysis of the ligand binding
site of EGF-receptor utilizing chimeric chicken/human receptor
molecules. EMBO J. 8:421–427. 1989.PubMed/NCBI
|
|
16
|
Ullrich A and Schlessinger J: Signal
transduction by receptors with tyrosine kinase activity. Cell.
61:203–212. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ozcan F, Klein P, Lemmon MA, Lax I and
Schlessinger J: On the nature of low- and high-affinity EGF
receptors on living cells. Proc Natl Acad Sci USA. 103:5735–5740.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baselga J: Targeting tyrosine kinases in
cancer: the second wave. Science. 312:1175–1178. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ciardiello F: Epidermal growth factor
receptor tyrosine kinase inhibitors as anticancer agents. Drugs.
60(Suppl 1): 25–32, Discussion 41–22,. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baselga J and Averbuch SD: ZD1839
(‘Iressa’) as an anticancer agent. Drugs. 60(Suppl 1): 33–40,
Discussion 41–42,. 2000.
|
|
21
|
Pollack VA, Savage DM, Baker DA, et al:
Inhibition of epidermal growth factor receptor-associated tyrosine
phosphorylation in human carcinomas with CP-358,774: dynamics of
receptor inhibition in situ and antitumor effects in athymic
mice. J Pharmacol Exp Ther. 291:739–748. 1999.
|
|
22
|
Xia W, Liu LH, Ho P and Spector NL:
Truncated ErbB2 receptor (p95ErbB2) is regulated by heregulin
through heterodimer formation with ErbB3 yet remains sensitive to
the dual EGFR/ErbB2 kinase inhibitor GW572016. Oncogene.
23:646–653. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kris MG, Natale RB, Herbst RS, et al:
Efficacy of gefitinib, an inhibitor of the epidermal growth factor
receptor tyrosine kinase, in symptomatic patients with non-small
cell lung cancer: a randomized trial. JAMA. 290:2149–2158. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Petrelli F, Borgonovo K, Cabiddu M and
Barni S: Efficacy of EGFR tyrosine kinase inhibitors in patients
with EGFR-mutated non-small cell lung cancer: a meta-analysis of 13
randomized trials. Clin Lung Cancer. 13:107–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jamal-Hanjani M and Spicer J: Epidermal
growth factor receptor tyrosine kinase inhibitors in the treatment
of epidermal growth factor receptor-mutant non-small cell lung
cancer metastatic to the brain. Clin Cancer Res. 18:938–944. 2012.
View Article : Google Scholar
|
|
26
|
Kwak EL, Sordella R, Bell DW, et al:
Irreversible inhibitors of the EGF receptor may circumvent acquired
resistance to gefitinib. Proc Natl Acad Sci USA. 102:7665–7670.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nishimura Y, Bereczky B and Ono M: The
EGFR inhibitor gefitinib suppresses ligand-stimulated endocytosis
of EGFR via the early/late endocytic pathway in non-small cell lung
cancer cell lines. Histochem Cell Biol. 127:541–553. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bublil EM and Yarden Y: The EGF receptor
family: spearheading a merger of signaling and therapeutics. Curr
Opin Cell Biol. 19:124–134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lichtner RB, Menrad A, Sommer A, Klar U
and Schneider MR: Signaling-inactive epidermal growth factor
receptor/ligand complexes in intact carcinoma cells by quinazoline
tyrosine kinase inhibitors. Cancer Res. 61:5790–5795. 2001.
|
|
30
|
Arteaga CL, Ramsey TT, Shawver LK and
Guyer CA: Unliganded epidermal growth factor receptor dimerization
induced by direct interaction of quinazolines with the ATP binding
site. J Biol Chem. 272:23247–23254. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Stamos J, Sliwkowski MX and Eigenbrot C:
Structure of the epidermal growth factor receptor kinase domain
alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol
Chem. 277:46265–46272. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wood ER, Truesdale AT, McDonald OB, et al:
A unique structure for epidermal growth factor receptor bound to
GW572016 (Lapatinib): relationships among protein conformation,
inhibitor off-rate, and receptor activity in tumor cells. Cancer
Res. 64:6652–6659. 2004. View Article : Google Scholar
|
|
33
|
Barros FF, Powe DG, Ellis IO and Green AR:
Understanding the HER family in breast cancer: interaction with
ligands, dimerization and treatments. Histopathology. 56:560–572.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wells A: EGF receptor. Int J Biochem Cell
Biol. 31:637–643. 1999. View Article : Google Scholar
|
|
35
|
Haslekas C, Breen K, Pedersen KW,
Johannessen LE, Stang E and Madshus IH: The inhibitory effect of
ErbB2 on epidermal growth factor-induced formation of
clathrin-coated pits correlates with retention of epidermal growth
factor receptor-ErbB2 oligomeric complexes at the plasma membrane.
Mol Biol Cell. 16:5832–5842. 2005. View Article : Google Scholar
|
|
36
|
Hommelgaard AM, Lerdrup M and van Deurs B:
Association with membrane protrusions makes ErbB2 an
internalization-resistant receptor. Mol Biol Cell. 15:1557–1567.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lenferink AE, Pinkas-Kramarski R, van de
Poll ML, et al: Differential endocytic routing of homo- and
hetero-dimeric ErbB tyrosine kinases confers signaling superiority
to receptor heterodimers. EMBO J. 17:3385–3397. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Westermark B, Magnusson A and Heldin CH:
Effect of epidermal growth factor on membrane motility and cell
locomotion in cultures of human clonal glioma cells. J Neurosci
Res. 8:491–507. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Carlsson J: Potential for clinical
radionuclide-based imaging and therapy of common cancers expressing
EGFR-family receptors. Tumour Biol. 33:653–659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Björkelund H, Gedda L and Andersson K:
Avoiding false negative results in specificity analysis of
protein-protein interactions. J Mol Recognit. 24:81–89.
2010.PubMed/NCBI
|
|
41
|
Sundberg AL, Almqvist Y, Orlova A, et al:
Combined effect of gefitinib (‘Iressa’, ZD1839) and targeted
radiotherapy with 211At-EGF. Eur J Nucl Med Mol Imaging.
30:1348–1356. 2003.
|
|
42
|
Nordberg E, Friedman M, Gostring L, et al:
Cellular studies of binding, internalization and retention of a
radiolabeled EGFR-binding affibody molecule. Nucl Med Biol.
34:609–618. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bjorke H and Andersson K: Automated,
high-resolution cellular retention and uptake studies in vitro.
Appl Radiat Isot. 64:901–905. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hunter WM and Greenwood FC: Preparation of
iodine-131 labelled human growth hormone of high specific activity.
Nature. 194:495–496. 1962. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bjorke H and Andersson K: Measuring the
affinity of a radioligand with its receptor using a rotating cell
dish with in situ reference area. Appl Radiat Isot. 64:32–37. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Svitel J, Balbo A, Mariuzza RA, Gonzales
NR and Schuck P: Combined affinity and rate constant distributions
of ligand populations from experimental surface binding kinetics
and equilibria. Biophys J. 84:4062–4077. 2003. View Article : Google Scholar
|
|
47
|
Barta P, Malmberg J, Melicharova L, et al:
Protein interactions with HER-family receptors can have different
characteristics depending on the hosting cell line. Int J Oncol.
40:1677–1682. 2012.PubMed/NCBI
|
|
48
|
Huber W: A new strategy for improved
secondary screening and lead optimization using high-resolution SPR
characterization of compound-target interactions. J Mol Recognit.
18:273–281. 2005. View Article : Google Scholar : PubMed/NCBI
|