Introduction
Colorectal cancer (CRC) is the third most common
cause of cancer mortality worldwide (1). Despite the fact that recent advances
in chemotherapeutic regimens and combination with radiotherapy have
improved survival of advanced-stage CRC patients, an increased risk
of recurrence and metastasis, and thus high mortality rates, are
associated with advanced-stages of the disease (2). Therefore, identification of novel
prognostic biomarkers to improve patient outcome and to assess
individual prognosis is required.
MicroRNAs (miRNAs) are a class of small, mature
non-coding 21–25 nucleotides that participate in the regulation of
cell differentiation, cell cycle progression and apoptosis
(3–5). miRNAs target protein-coding mRNAs at
the post-transcriptional level by direct cleavage of the mRNA or by
inhibition of protein synthesis (6). It has been hypothesized that miRNAs
are highly involved in cancer development (4). The synthesis and maturation of miRNAs
requires a set of proteins known collectively as miRNA-processing
machinery (7). miRNAs are
transcribed by polymerase II as long primary transcripts
(pri-miRNAs). Pri-miRNAs are spliced in the nuceleus by the enzyme
Drosha to form 70–100 nucleotide hairpin precursors of miRNA
(pre-miRNA) (8). The pre-miRNAs
are exported to the cytoplasm by Exportin-5, where they are further
processed by the RNAse III endonuclease enzyme DICER 1, resulting
in short double-stranded miRNA (miRNA douplex) of 19–24 nucleotides
(9,10).
Cytoplasmic ribonuclease type III DICER1 is a key
enzyme involved in the miRNA processing pathway that regulates
RNA-based gene silencing by the cleavage of miRNA precursor
(11–16). Mounting evidence suggests that
DICER1 expression levels are associated with clinical outcomes in
lung, ovarian, breast and prostate cancers (17–21).
Therefore, it is of interest to determine whether or not DICER1 may
be used as a prognostic marker to predict an individual patient’s
risk. Using the immunohistochemical method (IHC), only a few
clinical studies have investigated the usefulness of DICER1 protein
levels as a prognostic factor in CRC patients (22,23).
However, the prognostic values of DICER1 protein levels in these
studies were contradictory. Furthermore, the prognostic
significance of DICER1 mRNA level in CRC patients has yet to be
elucidated.
In this study, we investigated the association
between the clinicopathological characteristics and prognostic
value of DICER1 mRNA in 260 CRC patients.
Patients and methods
Patients and tissue samples
A total of 260 CRC patients were studied between
September, 2000 and Apri l, 2006 at the Teikyo University Hospital
(Tokyo, Japan). The median follow-up period was 45 months (range,
24–70). The samples were obtained from patients who did not receive
any chemotherapy or radiotherapy prior to surgery. Immediately
following surgical resection, primary CRC and normal adjacent tumor
tissues (normal tissue) were mounted using Tissue-Tek O.C.T
Compound (Sakura Finetechnical Co., Ltd., Tokyo, Japan) and frozen
in liquid nitrogen. The tissues were then stored at −80°C until
laser-capture micro-dissection (LCM). The study protocol conformed
to the guidelines of the ethics committee, and was approved by the
review board of the Teikyo University, while written informed
consent was obtained from the patients.
Follow-up of patients
Post-operative follow-up was performed along the
guidelines published by the Japanese Society for Cancer of the
Colon and Rectum. Confirmation of recurrence in the patients was
required to evaluate imaging or pathological diagnosis. Physical
examination and tumor marker (CEA and CA19-9) testing was conducted
every 3 months for 3 years and then every 6 months for 5 years.
Computed tomography (CT) or magnetic resonance imaging (MRI) scans
were repeated every 3 months for 3 years and then every 6 months
for up to 5 years, following surgery. Colon evaluation, including
colonoscopy or colon radiography was performed every 2 years or
annually for 3 years.
LCM and RNA isolation
Frozen sections (10 μm) of CRC and normal
tissues were prepared using a Leica CM 1900 cryostat (Leica,
Wetzlar, Germany) at −25°C. The sections were placed on
membrane-coated glass slides (Leica), fixed in 75% alcohol for 30
sec and stained with 0.5% violet-free methyl green (Sigma-Aldrich,
St. Louis, MO, USA). After staining, the sections were air-dried
and micro-dissected using a Leica AS LMD system (Leica). LCM caps
were stored at −80°C until RNA isolation.
Total RNAs were extracted using a miRNeasy Mini Kit
(Qiagen, Inc., Valencia, CA, USA) and were treated with DNase I,
according to the manufacturer’s instructions (Qiagen). Total RNA
was reverse-transcribed to complementary DNA (cDNA), using the
SuperScript II reverse transcriptase system with random hexamer
primers, according to the manufacturer’s instructions (Invitrogen
Corporation, Carlsbad, CA, USA).
Quantitative real-time reverse
transcription polymerase chain reaction (RT-PCR) for DICER1
mRNA
The relative expression levels of DICER1 and GAPDH
mRNA (internal control) were determined by quantitative real-time
PCR amplification (qRT-PCR) using a LightCycler 480 (Roche
Diagnostics Corp., Indianapolis, IN, USA). The amplifications of
these genes were performed using the LightCycler 480 Probe Master
(Roche Diagnostics Corp.) and TaqMan Gene Expression Assays for
DICER1 (Hs00229023_m1) and GAPDH (Hs02758991_g1) (Applied
Biosystems, Inc., Carlsbad, CA, USA). The PCR conditions of these
genes are 95°C for 5 min, followed by 40 cycles 95°C for 1 sec,
60°C for 20 sec. All the samples were performed in triplicates. The
expression levels of DICER1 mRNA were normalized to GAPDH mRNA
expression.
Statistical analysis
Data were shown as the mean ± standard error. The
correlations were analyzed using the Student’s t-test, the
Chi-square test and analysis of variance (ANOVA). The cut-off value
of DICER1 mRNA was determined by receiver operating characteristic
(ROC) curves, which used the JMP 9.0 software (SAS Inst. Inc.,
Cary, NC, USA). Overall survival (OS) and disease-free survival
(DFS) curves were analyzed using the Kaplan-Meier method and the
differences were examined using log-rank tests. Cox
proportional-hazards regression analysis was used to estimate
univariate and multivariate hazard ratios for OS and DFS. P values
were two-sided, and P<0.05 was considered to indicate a
statistically significant difference. Statistical analyses were
performed using the JMP 9.0 software (SAS Inst. Inc.).
Results
Expression of DICER1 mRNA in CRC and
normal tissues
A comparison of DICER1 mRNA expression levels of the
primary CRC and normal adjacent tumor tissues (normal tissues) were
compared (Fig. 1). The samples
were collected from 260 CRC patients. DICER1 mRNA levels were
normalized by GAPDH mRNA levels. In this study, DICER1 mRNA levels
of CRC tissues showed a significant decrease as compared to normal
tissues (P=0.039).
Correlation between the expression of
clinicopathological factors and DICER 1 mRNA in tumor tissues
This study comprised 260 CRC patients (153 men and
107 women), with a mean age of 67 years (range, 27–88). To evaluate
the correlation between the DICER1 mRNA levels and the
clinicopathological characteristics, patients were divided into the
high- and low-level groups. The cut-off level for DICER1 mRNA was
set at 0.275 based on analysis of the ROC curve. As shown in
Table I, a statistically
significant association was observed between DICER1 mRNA expression
and tumor size, depth of invasion, lymph node metastasis, lymphatic
invasion and Dukes’ stage.
 | Table IClinicopathological data and DICER1
mRNA expression in 260 CRC patients. |
Table I
Clinicopathological data and DICER1
mRNA expression in 260 CRC patients.
| | DICER1 mRNA
expression, no. of patients (%)
| |
|---|
| Variables | Total no. of
patients | High | Low | P-value |
|---|
| Gender | | | | 0.909 |
| Male | 153 | 64 (41.83) | 89 (58.17) | |
| Female | 107 | 44 (41.12) | 63 (58.88) | |
| Tumor size (cm) | | | | 0.001a |
| <5 | 147 | 74 (50.34) | 73 (49.66) | |
| ≥5 | 113 | 34 (30.09) | 79 (69.91) | |
| Depth of
invasion | | | | 0.002a |
| ≤pT2 | 24 | 17 (70.83) | 7 (29.17) | |
| ≥pT3 | 236 | 91 (38.56) | 145 (61.44) | |
| Localization | | | | 0.860 |
| Colon | 150 | 63 (42.00) | 87 (58.00) | |
| Rectum | 110 | 45 (40.91) | 65 (59.09) | |
| Histological
type | | | | 0.319 |
| Well | 174 | 76 (43.68) | 98 (56.32) | |
| Unwell | 86 | 32 (37.21) | 54 (62.79) | |
| Lymph node
metastasis | | | | 0.008a |
| Negative | 124 | 62 (50.00) | 62 (50.00) | |
| Positive | 136 | 46 (33.82) | 90 (66.18) | |
| Lymphatic
invasion | | | | 0.014a |
| Negative | 150 | 72 (48.00) | 78 (52.00) | |
| Positive | 110 | 36 (32.73) | 74 (67.27) | |
| Venous
invasion | | | | 0.804 |
| Negative | 106 | 45 (42.45) | 61 (57.55) | |
| Positive | 154 | 63 (40.91) | 91 (59.09) | |
| Liver
metastasis | | | | 0.178 |
| Negative | 222 | 96 (43.24) | 126 (56.76) | |
| Positive | 38 | 12 (31.58) | 26 (68.42) | |
| Peritoneum
dissemination | | | | 0.116 |
| Negative | 243 | 104 (42.80) | 139 (57.20) | |
| Positive | 17 | 4 (23.53) | 13 (76.47) | |
| Dukes’ stage | | | | 0.015a |
| A | 40 | 25 (62.50) | 15 (37.50) | |
| B | 68 | 30 (44.12) | 38 (55.88) | |
| C | 88 | 32 (36.36) | 56 (63.64) | |
| D | 64 | 21 (32.81) | 43 (67.19) | |
Correlation between DICER1 mRNA levels
and OS and DFS
The prognostic significances of DICER1 mRNA levels
was evaluated for the OS in all 260 patients and DFS in the 196
patients who underwent curative surgery. The average follow-up
period for OS was 39.4±25.8 months and that of DFS was 38.8±21.5
months. In each analysis, patients were divided into the high and
low DICER1 mRNA expression groups, as described above.
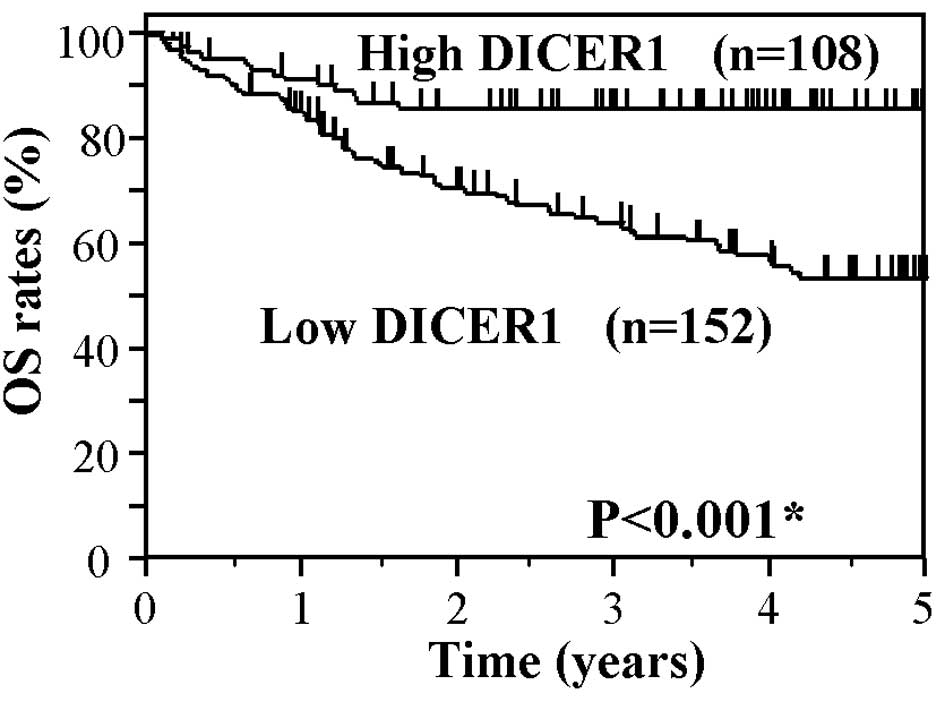
Fig. 2 shows the
Kaplan-Meier OS curve of the CRC patients based on the status of
DICER1 mRNA levels. The OS of patients in the low DICER1 group
showed significantly worse survival rates as compared to the high
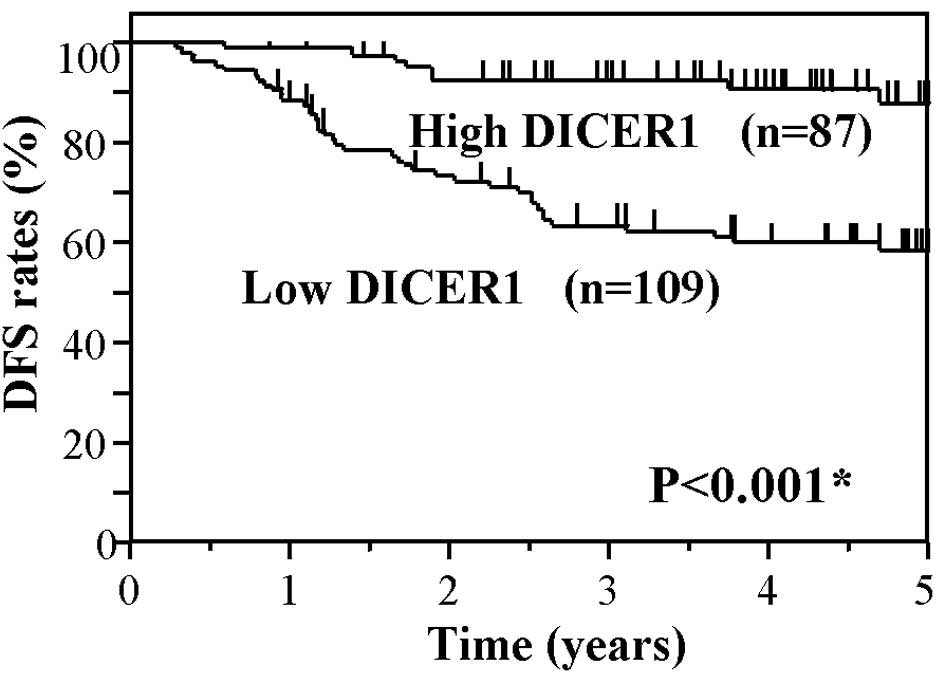
DICER1 group (P<0.001). Fig. 3
shows the Kaplan-Meier DFS curves of the CRC patients, based on the
status of DICER1 mRNA levels. The DFS of patients in the low DICER1
group also showed significantly worse survival rates as compared to
the high DICER1 group (P<0.001). These findings suggest that a
low expression of DICER1 mRNA is associated with worse OS and DFS
in CRC patients.
Univariate and multivariate Cox analyses
for OS
Table II shows the
results of the univariate and multivariate Cox proportional hazard
regression analyses for OS in the CRC patients. Multivariate
analysis was performed for factors exhibiting statistical
significance in the univariate analysis. In the univariate
analysis, tumor size, depth of invasion, lymph node metastasis,
lymphatic invasion, venous invasion, histological type, liver
metastasis, peritoneal dissemination, serum CEA, serum CA19-9,
Dukes’ stage and DICER1 mRNA level, while in the multivariate
analysis, Dukes’ stage and DICER1 mRNA showed statistical
significance for OS. Table III
shows the results of univariate and multivariate Cox analyses for
DFS in CRC patients who underwent curative surgery (n=196). In the
univariate analysis, depth of invasion, venous invasion, serum CEA,
Dukes’ stage and DICER1 mRNA, while in the multivariate analysis,
venous invasion, Dukes’ stage and DICER1 mRNA showed statistical
significance for DFS.
 | Table IIUnivariate and multivariate analysis
of prognostic factors for OS. |
Table II
Univariate and multivariate analysis
of prognostic factors for OS.
| Univariate analysis
| Multivariate
analysis
|
|---|
| Variables | Regression
coefficient | Hazard ratio (95%
CI) | P-value | Regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| Tumor size | 0.65 | 1.92
(1.22–3.04) | 0.005a | −0.23 | 0.79
(0.43–1.47) | 0.467 |
| Depth of
invasion | 2.23 | 9.34
(2.07–164.75) | 0.008a | 0.13 | 1.10
(0.19–20.85) | 0.907 |
| Lymph node
metastasis | 1.25 | 3.50
(2.12–6.05) | <0.001a | 0.03 | 1.03
(0.51–2.13) | 0.945 |
| Lymphatic
invasion | 0.99 | 2.70
(1.71–4.33) | <0.001a | 0.13 | 1.14
(0.61–2.16) | 0.683 |
| Venous
invasion | 1.00 | 2.71
(1.64–4.69) | <0.001a | 0.27 | 1.31
(0.66–2.76) | 0.444 |
| Histological
type | 0.65 | 1.92
(1.22–3.04) | 0.005a | 0.52 | 1.67
(0.89–3.15) | 0.108 |
| Liver
metastasis | 1.61 | 4.98
(2.94–8.23) | <0.001a | −0.23 | 0.79
(0.31–1.96) | 0.618 |
| Peritoneum
dissemination | 1.94 | 6.95
(3.60–12.46) | <0.001a | 0.11 | 1.11
(0.38–3.03) | 0.840 |
| Serum CEA | 1.17 | 3.23
(1.93–5.60) | <0.001a | 0.15 | 1.16
(0.57–2.39) | 0.686 |
| Serum CA19-9 | 0.63 | 1.87
(1.07–3.20) | 0.027a | 0.07 | 1.07
(0.52–2.14) | 0.855 |
| Dukes’ stage | 1.47 | 4.33
(3.11–6.21) | <0.001a | 1.18 | 3.27
(1.75–6.22) | 0.001a |
| DICER1 mRNA | −1.93 | 0.15
(0.02–0.46) | 0.002a | −1.19 | 0.30
(0.13–0.64) | 0.001a |
 | Table IIIUnivariate and multivariate analysis
of prognostic factors for DFS. |
Table III
Univariate and multivariate analysis
of prognostic factors for DFS.
| Univariate analysis
| Multivariate
analysis
|
|---|
| Variables | Regression
coefficient | Hazard ratio (95%
CI) | P-value | Regression
coefficient | Hazard ratio (95%
CI) | P-value |
|---|
| Tumor size | 0.27 | 1.31
(1.31–2.31) | 0.369 | | _ | |
| Depth of
invasion | 1.26 | 3.51
(1.09–6.48) | 0.034a | −0.44 | 0.66
(0.16–4.39) | 0.604 |
| Lymphatic
invasion | 0.52 | 1.69
(0.94–2.97) | 0.077 | | _ | |
| Venous
invasion | 0.70 | 2.00
(1.13–3.67) | 0.018a | 0.54 | 1.73
(0.95–3.21) | 0072 |
| Histological
type | 0.40 | 1.49
(0.81–2.66) | 0.195 | | _ | |
| Serum CEA | 0.63 | 1.87
(1.05–3.30) | 0.035a | 0.39 | 1.48
(0.82–2.62) | 0.189 |
| Serum CA19-9 | 0.60 | 1.82
(0.91–3.46) | 0.089 | | _ | |
| Dukes’ stage | 0.87 | 2.39
(1.54–3.91) | <0.001a | 0.73 | 2.07
(1.28–3.53) | 0.003a |
| DICER1 mRNA | −1.54 | 0.21
(0.09–0.43) | <0.001a | −1.46 | 0.23
(0.10–0.48) | 0.001a |
These results suggest that DICER1 mRNA levels of
tumor tissues have an independent prognostic value for OS and DFS
in CRC patients.
Discussion
In the present study, we aimed to examine the
association of clinicopathological variables and the prognostic
value of DICER1 mRNA in 260 CRC patients. Our findings demonstrate
that the expression of DICER1 mRNA of CRC tissues showed a
significant correlation between the tumor size, depth of invasion,
lymph node metastasis, lymphatic invasion and Dukes’ stage.
Furthermore, the low expression of DICER1 mRNA in CRC tissue showed
a markedly poor prognosis for OS and DFS in CRC patients.
DICER1 is a key enzyme responsible for the cleavage
of miRNA precursors that is necessary for the production of mature
miRNAs (11–16). DICER1 is capable of splicing the
hairpin-like structure RNA and double-stranded RNA into mature
miRNA or siRNA. MiRNAs are highly involved in several developmental
and biological cell processes including timing of cell development,
haematopoiesis, organogenesis, apoptosis, cell differentiation and
proliferation (12,25). Involvement of miRNA-base regulatory
mechanisms is important in several diseases, including cancer
(25–27). A general deregulation of miRNAs has
been described as a key feature of several cancer types. Therefore,
alternation of DICER1 expression may affect the development and
progression of cancer via the loss of miRNA-mediated gene
regulation.
DICER1 expression in cancer and normal tissues has
shown inconsistent results in various cancer types. In this study,
DICER1 mRNA levels of CRC showed a significant decrease compared to
normal tissues. Similar to our findings, a decreased DICER1
expression has been shown in CRC, lung, gastric and ovarian cancers
(17,20,28,29).
In their study, Papachristou et al (30) reported that the mRNA levels of
DICER did not exhibit significant differences in normal and CRC
tissues. Conversely, the overexpression of DICER1 has been reported
in prostate adenocarcinoma, ovarian cancer and acute myeloid
leukemia (19,31,32).
The correlation between the DICER1 levels in CRC tissues and their
clinicopathological factors are also noteworthy. We have
demonstrated significant correlations between the DICER1 mRNA and
tumor size, depth of invasion, lymph node metastasis, lymphatic
invasion and Dukes’ stage. Using the IHC method, Faggad et
al (23) reported that the
DICER1 protein expression in CRC tissues showed a significant
correlation with tumor grade, lymph node metastasis, localization
of tumor and tumor stage, thereby partially supporting our
findings.
We also evaluated the prognostic value of DICER1
mRNA in CRC tissues. The effect of expression on prognosis has been
studied in several cancers with controversial outcomes in various
cancer types. In patients with ovarian, lung and breast cancers,
decreased DICER1 levels in the tumor tissues showed a poor
prognosis (17,18,21).
Similarly, in patients with myeloma, nasopharyngeal carcinoma and
chronic lymphocytic leukemia, decreased DICER1 levels were also a
marker of poor prognosis (33–35).
Conversely, the overexpression of DICER1 expression has been
reported as a prognostic factor in prostate adenocarcinoma
(19). Regarding CRC, the results
of prognostic values of DICER1 in tumor tissues are also
controversial.
Recently, Faber et al (24) reported that the overexpression of
DICER1 predicts poor survival in CRC patients with pT2 or pT3
stages and without metastatic disease (pN0 and pM0). By contrast,
Faggad et al (23) reported
that the downregulation of DICER1 is a prognostic factor in CRC
patients with WHO stage I, II, III and IV. These studies examined
the DICER1 protein levels of tissue microarrays (TMA) using IHC
staining. In this study, we selected the DICER1 mRNA detection
using the Taqman RT-PCR method, with the aim of obtaining high
sensitivity and objective analysis, since a small sample size of
TMA may limit the value of TMA due to tumor heterogeneity.
Furthermore, Grelier et al (21) compared the prognostic value of
DICER1 of breast cancer in the two measuring methods: real time
RT-PCR and IHC. They reported that DICER1 mRNA levels were
predictive for metastatic-free survival. However, the protein
expression was not informative for survival. Of note, our data
showed that low expression levels of DICER1 mRNA in CRC tissues
significantly correlate with poorer OS and DFS, thereby supporting
the findings of Faggad et al (36). The expression level of DICER1
directly influences the biosynthesis of miRNA. In ovarian cancer, a
link between reduced DICER1 expression and a global down-regulation
of miRNA are reported.
The aggressive tumors are thought to have decreased
total microRNA levels, contributing to their poor differentiation.
The downregulation of miRNA may have an impact on the development
of tumor cells leading to poor prognosis of patients. However, the
reason for the discrepancy with the survival results reported by
Faber et al (24) is to be
studied in future large-scale studies of each CRC stage. To the
best of our knowledge, this is the first study to demonstrate the
independent prognostic value of a decreased expression of DICER1
mRNA in CRC.
In conclusion, our study has demonstrated that
reduced DICER1 mRNA expression of tumor tissues shows a prognostic
significance in CRC patients. Our finding of DICER1 mRNA expression
being an independent marker capable of predicting high-risk
patients is potentially useful in the individualized management and
monitoring of CRC patients. In the future, these molecules may
serve as a novel target with beneficial therapeutic
applications.
Acknowledgements
The author thanks Professor Y.
Hashiguchi and Dr Iinuma for their helpful suggestions, as well as
Dr K. Matsuda, Miss J. Tamura and all members of the colorectal
group for their help. This study was supported by the JSPS KAKENHI,
grant no. 24591984.
References
|
1
|
Gill S, Thomas RR and Goldberg RM: Review
article: colorectal cancer chemotherapy. Aliment Pharmacol Ther.
18:683–692. 2003. View Article : Google Scholar
|
|
2
|
Aggarwal S and Chu E: Current therapies
for advanced colorectal cancer. Oncology. 19:589–595.
2005.PubMed/NCBI
|
|
3
|
Caldas C and Brenton JD: Sizing up miRNAs
as cancer genes. Nat Med. 11:712–714. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar
|
|
5
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mott JL: MicroRNAs involved in tumor
suppressor and oncogene pathways: implications for hepatobiliary
neoplasia. Hepatology. 50:630–637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee Y, Ahn C, Han J, et al: The nuclear
RNase III Drosha initiates microRNA processing. Nature.
6956:415–419. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lund E, Güttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
5654:95–98. 2004. View Article : Google Scholar
|
|
10
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Genes Dev. 17:3011–3016. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hutvágner G, McLachlan J, Pasquinelli AE,
Bálint E, Tuschl T and Zamore PD: A cellular function for the
RNA-interference enzyme Dicer in the maturation of the let-7 small
temporal RNA. Science. 5531:834–838. 2001.PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cullen BR: Transcription and processing of
human microRNA precursors. Mol Cell. 16:861–865. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carmell MA and Hannon GJ: RNase III
enzymes and the initiation of gene silencing. Nat Struct Mol Biol.
11:214–218. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cummins JM, He Y, Leary RJ, et al: The
colorectal microRNAome. Proc Natl Acad Sci USA. 103:3687–3692.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ambros V, Bartel B, Bartel DP, et al: A
uniform system for microRNA annotation. RNA. 9:277–279. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karube Y, Tanaka H, Osada H, et al:
Reduced expression of Dicer associated with poor prognosis in lung
cancer patients. Cancer Sci. 96:111–115. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Merritt WM, Lin YG, Han LY, et al: Dicer,
Drosha, and outcomes in patients with ovarian cancer. N Engl J Med.
359:2641–2650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiosea S, Jelezcova E, Chandran U,
Acquafondata M, McHale T, Sobol RW and Dhir R: Up-regulation of
dicer, a component of the MicroRNA machinery, in prostate
adenocarcinoma. Am J Pathol. 169:1812–1820. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chiosea S, Jelezcova E, Chandran U, Luo J,
Mantha G, Sobol RW and Dacic S: Overexpression of Dicer in
precursor lesions of lung adenocarcinoma. Cancer Res. 67:2345–2350.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grelier G, Voirin N, Ay AS, et al:
Prognostic value of Dicer expression in human breast cancers and
association with the mesenchymal phenotype. Br J Cancer.
101:673–683. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stratmann J, Wang CJ, Gnosa S, Wallin A,
Hinselwood D, Sun XF and Zhang H: Dicer and miRNA in relation to
clinicopathological variables in colorectal cancer patients. BMC
Cancer. 11:3452011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Faggad A, Kasajima A, Weichert W,
Stenzinger A, Elwali NE, Dietel M and Denkert C: Down-regulation of
the microRNA processing enzyme Dicer is a prognostic factor in
human colorectal cancer. Histopathology. 20: View Article : Google Scholar : 2012.PubMed/NCBI
|
|
24
|
Faber C, Horst D, Hlubek F and Kirchner T:
Overexpression of Dicer predicts poor survival in colorectal
cancer. Eur J Cancer. 47:1414–1419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wiemer EA: The role of microRNAs in caner:
no small matter. Eur J Cancer. 43:1529–1544. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nelson P, Kiriakidou M, Sharma A,
Maniataki E and Mourelatos Z: The microRNA world: small is mighty.
Trends Biochem Sci. 28:534–540. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Calin GA and Croce CM: MicroRNA-cancer
connection: the beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng ZH, Sun XJ, Fu WN, Guan Y, Gao F,
Wang Y and Sun KL: Decreased expression of DICER1 in gastric
cancer. Chin Med J. 120:2099–2104. 2007.PubMed/NCBI
|
|
29
|
Pampalakis G, Diamandis EP, Katsaros D and
Sotiropoulou G: Down-regulation of dicer expression in ovarian
cancer tissues. Clin Biochem. 43:324–327. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Papachristou DJ, Korpetinou A,
Giannopoulou E, et al: Expression of the ribonucleases Drosha,
Dicer, and Ago2 in colorectal carcinomas. Virchows Arch.
459:431–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Flavin RJ, Smyth PC, Finn SP, et al:
Altered eIF6 and Dicer expression is associated with
clinicopathological features in ovarian serous carcinoma patients.
Mod Pathol. 21:676–684. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Martin MG, Payton JE and Link DC: Dicer
and outcomes in patients with acute myeloid leukemia (AML). Leuk.
Res. 33:e1272009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo X, Liao Q, Chen P, et al: The
microRNA-processing enzymes: Drosha and Dicer can predict prognosis
of nasoparyngeal carcinoma. J Cancer Res Clin Oncol. 138:49–56.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu DX, Fan L, Lu RN, et al:
Downregulation Dicer expression predicts poor prognosis in chronic
lymphocytic leukemia. Cancer Sci. 103:875–881. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sarasquete M, Gutierrez NC,
Misiewicz-Krzeminska I, et al: Upregulation of Dicer is more
frequent in monoclonal gammopathies of undetermined significance
than in multiple myeloma patients and is associated with longer
survival in symptomatic myeloma patients. Haematologica.
96:468–471. 2011. View Article : Google Scholar
|
|
36
|
Faggad A, Budczites J, Tchemitsa O, et al:
Prognostic significance of Dicer expression in ovarian cancer-link
to global microRNA changes and oestrogen receptor expression. J
Pathol. 220:382–391. 2010.PubMed/NCBI
|