Introduction
Renal cell carcinoma (RCC) is the most common
neoplasm in the adult kidney and accounts for 2–3% of malignant
diseases in adults. Locally extensive or metastatic RCC, even
following complete resection, have worse prognoses compared to
organ-confined diseases. Therefore, there is a need to identify
novel biomarkers that enable prediction of early metastasis after
nephrectomy, and to develop novel targeted therapies.
Cancer develops as a result of multiple genetic and
epigenetic alterations. Better knowledge of changes in gene
expression that occur during carcinogenesis may lead to
improvements in diagnosis, treatment and prevention. Potential
biomarkers identifying high-risk patients have been reported,
however, assessment methods for these biomarkers often involve
RNA-based techniques and require fresh frozen tissues. By contrast,
formalin-fixed paraffin-embedded (FFPE) tissue samples have been
collected during decades of routine histopathological examination
and those are the most widely available materials in clinical use.
However, formaldehyde-containing fixatives cause cross-linkage
between nucleic acids and proteins, rendering subsequent extraction
and quantification of RNA challenging (1). A major obstacle to RNA expression
analysis of FFPE tissues has been the uncertainty as to whether or
not gene expression analyses from routinely archived tissues
accurately reflect the expression levels prior to fixation, since
the fixation process is likely to cause a high degree of RNA
fragmentation (2). Given that
naturally occurring small RNAs are not affected by fragmentation
and therefore do not experience loss of quality, targeting miRNAs
is more suitable for analysis of RNA extracted from FFPE
samples.
miRNAs are 18- to 25-nucleotide, non-coding RNA
molecules that regulate the translation of several genes (3). miRNA expression levels are altered in
most types of human cancers (4–6).
Several RCC studies have examined miRNAs by microarray analysis
using a relatively small amount of frozen tissue samples. Various
miRNAs that are dysregulated in RCC have been identified (7,8).
Several lines of evidence indicate that miR-486 has
oncogenic properties. Overexpression of miR-486 has been
reported in cutaneous T-cell lymphoma (9). High serum or plasma expression levels
of miR-486 have been reported in non-small cell lung
(10) and gastric cancers
(11). miR-486 has also
been reported to target PTEN and FOXO1, which
negatively affect PI3K/Akt signaling (12). The Akt-signaling pathway functions
as a potent activator of growth and survival signaling (13). However, expression of
miR-486 has not been investigated in RCC.
In the present study, we investigated the
association of miR-486 expression with RCC patient prognosis
(n=150) using quantitative reverse transcription polymerase chain
reaction (qRT-PCR) of FFPE samples. In addition, since it has been
reported that olfactomedin 4 (OLFM4), which encodes
the olfactomedin 4 protein, is one of the direct targets of
miR-486 (14),
immunohistochemical analysis of olfactomedin 4 was carried out in
RCC tissues.
Materials and methods
Tissue samples
In a retrospective study design, 150 primary tumors
were collected consecutively from patients diagnosed with RCC who
underwent surgery between 2001 and 2008 at the Hiroshima University
Hospital (Hiroshima, Japan). Only patients without preoperative
radiotherapy or chemotherapy were enrolled in the study. Of 150
patients, 129 were at stages I, II and III, while 21 were at stage
IV. The 129 patients with stage I, II and III RCC underwent
curative resection. Of the 129 patients with stage I, II and III
RCC, 20 received adjuvant therapy (interferon-α) (15). Of the 21 patients with stage IV
RCC, 17 received postoperative therapy using interferon-α (15). Postoperative follow-up was
scheduled every 1, 2 or 3 months during the first 2 years after
surgery, and every 6 months thereafter unless more frequent
follow-up was deemed necessary. Chest-abdominal computed tomography
scan and serum chemistries were performed during each 6 month visit
at least. Patients were monitored by their physician until they
succumbed to the disease or the date of the last documented
contact. The median follow-up period was 64 months (range, 2–120).
Operative mortality was defined as death within 30 days of the
patient leaving the hospital, and these patients were omitted from
the analysis.
For qRT-PCR analysis, archival FFPE tissues were
used. Histological classification was based on the World Health
Organization system. RCC cases were classified into clear cell RCC
(ccRCC) and non-ccRCC. Tumor staging was performed according to the
TNM grouping system. Since written informed consent was not
obtained, for reasons of strict privacy protection, identifying
information for the samples was removed prior to analysis. This
procedure was in accordance with the Ethical Guidelines for Human
Genome/Gene Research of the Japanese Government.
RNA extraction and qRT-PCR
FFPE samples were sectioned (10 μm),
deparaffinized and stained with hematoxylin and eosin (H&E) to
ensure that the sectioned block contained tumor cells. The tumor
areas in the adjacent sections were marked under a light microscope
without hematoxylin staining. Tumor areas were macrodissected with
sterile disposable scalpels and subjected to RNA isolation using
the Recover All™ Total Nucleic Acid Isolation kit (Ambion, Austin,
TX, USA), according to the manufacturer’s instructions. Expression
levels of miR-486 and RNU6B were measured using
TaqMan® assays for miRNA (Applied Biosystems, Austin, TX, USA) in
such a manner that the identity and clinical outcomes of samples
were blinded. Complementary DNA (cDNA) was synthesized using
miRNA-specific primers and the TaqMan® MicroRNA Reverse
Transcription kit (Applied Biosystems) according to the
manufacturer’s instructions. Briefly, 40 ng of RNA was reverse
transcribed in a 20 μl reaction with gene-specific RT
probes. qRT-PCR was performed using the ABI 7900 Version 2.3
Sequence Detection System (Applied Biosystems). RNU6B was
used as an endogenous normalization control for miR-486. The
assays were performed in triplicate. Quantification of
miR-486 relative expression was calculated using the RQ
manager 1.2 (Applied Biosystems).
Immunohistochemistry
FFPE samples were sectioned, deparaffinized and
stained with H&E to ensure that the sectioned block contained
tumor cells. Adjacent sections were then stained
immunohistochemically with the Dako EnVision+ Mouse Peroxidase
Detection System (DakoCytomation, Carpinteria, CA, USA). Antigen
retrieval was performed by microwave heating in citrate buffer (pH
6.0) for 30 min. After peroxidase activity was blocked with 3%
H2O2 methanol for 10 min, sections were
incubated with normal goat serum (DakoCytomation) for 20 min to
block non-specific antibody binding sites. Sections were incubated
with primary antibodies against olfactomedin 4 (1:50 dilution,
using an anti-olfactomedin 4 antibody raised in our laboratory)
(16) for 1 h at room temperature,
followed by incubations with EnVision+ anti-mouse peroxidase for 1
h. Staining was completed with 10-min incubation with the
substrate-chromogen solution. Sections were counterstained using
0.1% hematoxylin.
Statistical analysis
Associations between clinicopathological
characteristics and miR-486 expression were analyzed using
the Fisher’s exact test. To evaluate the associations between
clinical covariates and cancer-specific mortality univariate and
multivariate Cox regression analysis was used and conducted using
SPSS software (SPSS Inc., Chicago, IL, USA). Hazard ratio (HR) and
95% confidence interval (CI) were estimated from the Cox
proportional hazard models. For the analyses, age was treated as a
categorical variable (65 plus >65 vs. <65 years old). For the
final multivariate Cox regression models, the variables that were
moderately associated (P<0.10) with cancer-specific mortality
were included. Differences in miR-486 expression levels
between the two groups were determined by the Mann-Whitney U test
using Graphpad Prism 5.0 (Graphpad Software, Inc., San Diego, CA,
USA). Kaplan-Meier survival curves were constructed for high- and
low-miR-486 patients using the Graphpad Prism 5.0 software.
Differences between survival curves were tested for statistical
significance using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of miR-486 in RCC and
non-neoplastic kidney tissue
The expression of miR-486 and the
corresponding non-neoplastic kidney tissue samples were assessed in
86 RCC cases using qRT-PCR. The expression of miR-486 in RCC
samples was ∼2.7-fold higher compared to that in corresponding
non-neoplastic kidney samples (P<0.0001, Mann-Whitney
U-test).
Association between miR-486 expression
levels and clinicopathological characteristics
Expression of miR-486 was examined in 64
additional RCC tissue samples using qRT-PCR. We investigated the
association between the clinicopathological characteristics and
miR-486 expression levels in 150 RCC cases (Table I). These 150 RCC cases were divided
into high- and low-miR-486 cases. When low-miR-486
expression was classified according to the lowest quartile, the
number of high- and low-miR-486 cases was 112 and 38,
respectively. Expression of miR-486 was not associated with
clinicopathological characteristics such as gender, T, N and M
classifications, tumor stage or venous invasion.
 | Table IAssociation between miR-486
expression and clinicopathological characteristics. |
Table I
Association between miR-486
expression and clinicopathological characteristics.
| miR-486
expression
|
|---|
| Characteristics | High (%) | Low | P-value |
|---|
| Age (years) | | | |
| <66 | 48 (69) | 22 | 0.1084 |
| ≥66 | 64 (80) | 16 | |
| Gender | | | |
| Male | 80 (73) | 29 | 0.5552 |
| Female | 32 (78) | 9 | |
| T
classification | | | |
| 1 | 71 (77) | 21 | 0.5009 |
| 2 | 12 (71) | 5 | |
| 3 | 27 (69) | 12 | |
| 4 | 2 (100) | 0 | |
| N
classification | | | |
| 0 | 107 (76) | 34 | 0.4049 |
| 1 | 2 (50) | 2 | |
| 2 | 3 (60) | 2 | |
| M
classification | | | |
| 0 | 98 (75) | 32 | 0.6119 |
| 1 | 14 (70) | 6 | |
| Tumor stage | | | |
| I | 71 (79) | 19 | 0.1792 |
| II | 10 (71) | 4 | |
| III | 16 (67) | 8 | |
| IV | 15 (68) | 7 | |
| Histological
classification | | | |
| ccRCC | 101 (74) | 36 | 0.3649 |
| Non-ccRCC | 11 (85) | 2 | |
| Venous
invasion | | | |
| Positive | 35 (74) | 12 | 0.9699 |
| Negative | 77 (75) | 26 | |
| Interferon-α
treatment | | | |
| Received | 23 (62) | 14 | 0.0519 |
| Not received | 89 (79) | 24 | |
Association between miR-486 expression
and survival
The association between miR-486 expression
levels and cancer-specific mortality was evaluated. The
Kaplan-Meier analysis for the 150 RCC cases (stages I–IV) was
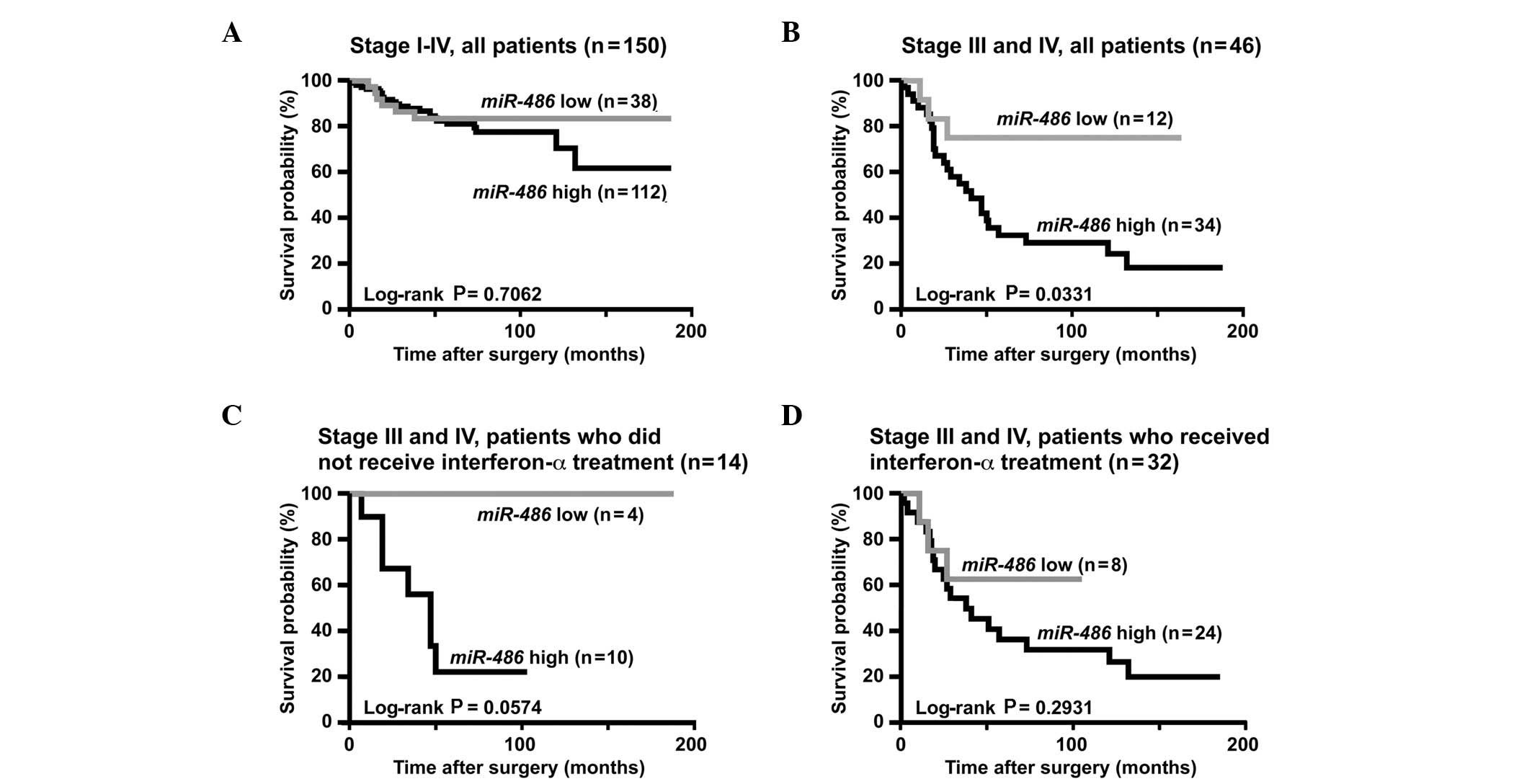
performed. As shown in Fig. 1A,
cancer-specific mortality did not statistically vary in high- and
low-miR-486 cases.
Patients with RCC at stage I and II have a good rate
of survival, whereas it is difficult to predict patient survival
with stage III and IV RCC. These patients would benefit greatly
from prognostic biomarkers. Therefore, we analyzed the prognostic
value of miR-486 expression in patients with stage III and
IV RCC (n=46). Using the Kaplan-Meier analysis, we found that RCC
cases with high-miR-486 expression had significantly worse
cancer-specific mortality compared to those with low-miR-486
expression (P=0.0331, log-rank test, Fig. 1B). Since treatment with
interferon-α for patients with stage III and IV RCC may affect
patient survival, individuals who did not receive interferon-α
treatment (n=14) were assessed. Although cancer-specific mortality
among patients with high- and low-miR-486 expression
exhibited no statistically significant difference (P=0.0574,
log-Rank test, Fig. 1C),
cancer-specific mortality of patients with high-miR-486
expression tended to be worse compared to that of patients with
low-miR-486 expression.
To evaluate the potential for miR-486
expression as a prognostic predictor in patients with stage III and
IV RCC, univariate and multivariate Cox proportional hazards
analyses were used to evaluate the association of miR-486
expression with cancer-specific mortality (Table II). Findings of the univariate
analysis showed that N (HR, 2.73; 95% CI, 1.10–6.24; P=0.0315) and
M classifications (HR, 3.16; 95% CI, 1.46–7.08; P=0.0035), tumor
stage (HR, 4.03; 95% CI, 1.82–9.54; P=0.0005) and miR-486
expression (HR, 3.38; 95% CI, 1.18–14.26; P=0.0202) were
significantly associated with cancer-specific mortality. According
to the multivariate model, which included N and M classifications,
tumor stage and miR-486 expression, expression of
miR-486 was an independent prognostic classifier of
cancer-specific mortality (HR, 4.33; 95% CI, 1.45–18.71;
P=0.0064).
 | Table IIUnivariate and multivariate Cox
regression analysis of miR-486 expression and
cancer-specific mortality in stage III and IV RCC. |
Table II
Univariate and multivariate Cox
regression analysis of miR-486 expression and
cancer-specific mortality in stage III and IV RCC.
| Univariate analysis
| Multivariate
analysis
|
|---|
|
Characteristics | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (years) | | | | |
| <66 | 1 (Ref.) | 0.5719 | | |
| ≥66 | 1.24
(0.58–2.66) | | | |
| Gender | | | | |
| Male | 1 (Ref.) | 0.5919 | | |
| Female | 0.73
(0.17–2.08) | | | |
| Histological
classification | | | | |
| Non-ccRCC | 1 (Ref.) | 0.5714 | | |
| ccRCC | 0.64
(0.18–4.00) | | | |
| Venous
invasion | | | | |
| Negative | 1 (Ref.) | 0.9318 | | |
| Positive | 1.04
(0.42–3.10) | | | |
| T
classification | | | | |
| 1/2 | 1 (Ref.) | 0.8861 | | |
| 3/4 | 1.09
(0.37–4.60) | | | |
| N
classification | | | | |
| 0 | 1 (Ref.) | 0.0315 | 1 (Ref.) | 0.9832 |
| 1/2 | 2.73 (1.10–
6.24) | | 1.01
(0.32–2.89) | |
| M
classification | | | | |
| 0 | 1 (Ref.) | 0.0035 | 1 (Ref.) | 0.2398 |
| 1 | 3.16
(1.46–7.08) | | 0.31
(0.05–2.47) | |
| Tumor stage | | | | |
| III | 1 (Ref.) | 0.0005 | 1 (Ref.) | 0.0386 |
| IV | 4.03
(1.82–9.54) | | 9.72
(1.14–59.82) | |
| Interferon-α
treatment | | | | |
| Not received | 1 (Ref.) | 0.5558 | | |
| Received | 1.28
(0.57–3.28) | | | |
| miR-486
expression | | | | |
| Low | 1 (Ref.) | 0.0202 | 1 (Ref.) | 0.0064 |
| High | 3.38
(1.18–14.26) | | 4.33
(1.45–18.71) | |
Survival analysis was performed using an
alternatively defined cut-off point. When low-miR-486
expression was defined on the basis of lower than median levels of
expression of miR-486, the number of high- as well as
low-miR-486 miR-486 cases was 75. Kaplan-Meier analysis
revealed that cancer-specific mortality did not exhibit
statistically significant differences in high- and
low-miR-486 cases (data not shown). When low-miR-486
expression was classified according to the lowest tertile, the
number of high- and low-miR-486 cases was 100 and 50,
respectively. Kaplan-Meier analysis revealed that cancer-specific
mortality did not exhibit statistically significant differences in
high- and low-miR-486 cases (data not shown).
Expression of miR-486 and therapeutic
outcomes
Biomarkers that predict therapeutic outcomes may
provide tools to allow physicians to better stratify patients to
more effective treatments. Therefore, we analyzed the association
between miR-486 expression and therapeutic outcomes in stage
III and IV RCC patients treated with interferon-α (n=32). However,
expression of miR-486 was not significantly associated with
therapeutic outcome (P=0.2931, log-rank test, Fig. 1D).
Expression of OLFM4 in RCC
OLFM4, which encodes the OLFM4 protein, has
been reported as one of the direct targets of miR-486
(14). Although alteration of
OLFM4 expression has been reported in several types of human cancer
(16–18), expression of OLFM4 in RCC has not
been investigated. Therefore, the expression of OLFM4 was analyzed
using immunohistochemistry in 86 RCC cases. In non-neoplastic
kidney, OLFM4 staining was observed in uriniferous tubules but not
in glomeruli (Fig. 2A). By
contrast, OLFM4 staining was not expressed in RCC cells (Fig. 2B). No OLFM4 staining was detectable
in the 86 RCC cases.
Discussion
Several lines of evidence have suggested that miRNAs
are useful as biomarkers as well as therapeutic targets for cancer.
Prediction of the survival of patients with stage III and IV RCC
remains difficult, and these groups would benefit from the
detection of prognostic markers that identify individuals for whom
adjuvant or post-operative treatment would be advantageous. In the
present study, RNA from 150 FFPE RCC tissues was prepared, since
biomarkers developed from FFPE samples may be more readily
translated into clinical application. Expression of miR-486
in patients with stage III and IV RCC was significantly associated
with cancer-specific mortality according to a Kaplan-Meier
analysis. The univariate and multivariate Cox proportional hazards
analyses demonstrated that expression of miR-486 was an
independent prognostic classifier. Furthermore, since treatment
with interferon-α for patients with stage III and IV RCC affects
patient survival, individuals who did not receive interferon-α
treatment were assessed. Although cancer-specific mortality among
patients with high- and low-miR-486 expression showed no
statistically significant difference, cancer-specific mortality of
patients with high-miR-486 expression tended to be worse
compared to that of patients with low-miR-486 expression.
This finding suggests that miR-486 expression is associated
with a more aggressive RCC histology. Taken together, these results
indicate that miR-486 is a promising biomarker to identify
patients with poor prognosis in stage III and IV RCC.
In the present study, we demonstrated that
miR-486 expression was associated with cancer-specific
mortality in patients with stage III and stage IV RCC. In addition,
miR-486 expression was associated with cancer-specific
mortality in stage III and IV RCC patients who were not treated
with interferon-α. Therefore, measurement of miR-486
expression may help identify patients with a high risk of disease
recurrence, while treatment with interferon-α may be indicative for
patients with miR-486-positive RCC. However, it is unclear
whether such patients may benefit from interferon-α treatment. To
address this issue, we examined whether or not miR-486
expression was able to identify patients for whom interferon-α
treatment is beneficial in stage III and IV RCC. However,
expression of miR-486 was not significantly associated with
therapeutic outcome. These results suggest that even when patients
with a high risk of disease recurrence are identified by
miR-486 measurement, those patients may not benefit from
interferon-α treatment. Novel therapeutic methods may be more
effective for these patients.
miR-486 has been reported to have oncogenic
properties. Overexpression of miR-486 has been reported in
several human malignancies. In the present study, although
expression of miR-486 in RCC samples was significantly
higher compared to that in corresponding non-neoplastic kidney
samples, expression of miR-486 was not associated with tumor
stage. Therefore, high-miR-486 expression is likely to be
involved in carcinogenesis, but not in tumor progression. Since
targets of miR-486 are PTEN and FOXO1
(12), expression of PTEN
and FOXO1 may be low in high-miR-486 RCC cases. In
addition to PTEN and FOXO1, one of the targets of
miR-486 is OLFM4 (14). In the present study, although OLFM4
staining was observed in uriniferous tubules, staining of OLFM4 was
not detected in the 86 RCC cases. Previously, we showed that
patients with OLFM4-positive gastric cancer had a better survival
rate compared to patients with OLFM4-negative gastric cancer
(16). Forced expression of OLFM4
has also been reported to decrease cell adhesion and migration
(17,18). Taken together, OLFM4 is likely to
have tumor suppressive properties in RCC. Given that staining of
OLFM4 was not detected in the 86 RCC cases, overexpression of
miR-486 is unlikely to have a major role in the loss of
OLFM4.
In summary, we showed that a high-miR-486
expression is an independent prognostic classifier in stage III and
IV RCC. Therefore, measurement of miR-486 helps identify
high-risk patients. In this study, expression of miR-486
from FFPE samples was assessed. Therefore, measurement of
miR-486 can be readily translated into clinical
applications.
Acknowledgements
This study was supported in part by
grants-in-aid for Cancer Research from the Ministry of Education,
Culture, Science, Sports and Technology of Japan and in part by a
grant-in-aid for the Third Comprehensive 10-Year Strategy for
Cancer Control from the Ministry of Health, Labor and Welfare of
Japan. The authors would like to thank Mr. Shinichi Norimura for
his excellent technical assistance and advice. This study was
conducted in collaboration with the Research Center for Molecular
Medicine of the Faculty of Medicine of Hiroshima University. We
also thank the Analysis Center of Life Science of Hiroshima
University for the use of their facilities.
References
|
1
|
Srinivasan M, Sedmak D and Jewell S:
Effect of fixatives and tissue processing on the content and
integrity of nucleic acids. Am J Pathol. 161:1961–1971. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Specht K, Richter T, Muller U, Walch A,
Werner M and Hofler H: Quantitative gene expression analysis in
microdissected archival formalin-fixed and paraffin-embedded tumor
tissue. Am J Pathol. 158:419–429. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schetter AJ, Heegaard NH and Harris CC:
Inflammation and cancer: interweaving microRNA, free radical,
cytokine and p53 pathways. Carcinogenesis. 31:37–49. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ueda T, Volinia S, Okumura H, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: a microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gottardo F, Liu CG, Ferracin M, et al:
Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
White NM, Bao TT, Grigull J, et al: miRNA
profiling for clear cell renal cell carcinoma: biomarker discovery
and identification of potential controls and consequences of miRNA
dysregulation. J Urol. 186:1077–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Narducci MG, Arcelli D, Picchio MC, et al:
MicroRNA profiling reveals that miR-21, miR486 and miR-214 are
upregulated and involved in cell survival in Sézary syndrome. Cell
Death Dis. 2:e1512011.PubMed/NCBI
|
|
10
|
Hu Z, Chen X, Zhao Y, et al: Serum
microRNA signatures identified in a genome-wide serum microRNA
expression profiling predict survival of non-small-cell lung
cancer. J Clin Oncol. 28:1721–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Konishi H, Ichikawa D, Komatsu S, et al:
Detection of gastric cancer-associated microRNAs on microRNA
microarray comparing pre- and post-operative plasma. Br J Cancer.
106:740–747. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Small EM, O’Rourke JR, Moresi V, et al:
Regulation of PI3-kinase/Akt signaling by muscle-enriched
microRNA-486. Proc Natl Acad Sci USA. 107:4218–4223. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Crackower MA, Oudit GY, Kozieradzki I, et
al: Regulation of myocardial contractility and cell size by
distinct PI3K-PTEN signaling pathways. Cell. 110:737–749. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oh HK, Tan AL, Das K, et al: Genomic loss
of miR-486 regulates tumor progression and the OLFM4 antiapoptotic
factor in gastric cancer. Clin Cancer Res. 17:2657–2667. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Medical Research Council Renal Cancer
Collaborators: Interferon-alpha and survival in metastatic renal
carcinoma: early results of a randomised controlled trial. Medical
Research Council Renal Cancer Collaborators Lancet. 353:14–17.
1999.
|
|
16
|
Oue N, Sentani K, Noguchi T, et al: Serum
olfactomedin 4 (GW112, hGC-1) in combination with Reg IV is a
highly sensitive biomarker for gastric cancer patients. Int J
Cancer. 125:2383–2392. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Koshida S, Kobayashi D, Moriai R, Tsuji N
and Watanabe N: Specific overexpression of OLFM4(GW112/HGC-1) mRNA
in colon, breast and lung cancer tissues detected using
quantitative analysis. Cancer Sci. 98:315–320. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu W, Liu Y, Zhu J, Wright E, Ding I and
Rodgers GP: Reduced hGC-1 protein expression is associated with
malignant progression of colon carcinoma. Clin Cancer Res.
14:1041–1049. 2008. View Article : Google Scholar : PubMed/NCBI
|