Introduction
Hepatocellular carcinoma (HCC) is one of the most
common gastrointestinal malignancies and constitutes the leading
cause of cancer-related mortality in East Asia and South Africa
(1). Currently, the first-line
treatment for HCC is liver transplantation or surgical resection
(2). However, the overall survival
rate after curative therapy is not satisfactory due to the highly
chemoresistant nature of this tumor and the frequent intrahepatic
recurrence. Identification of the genes responsible for the onset
and progression of HCC as well as comprehension of the clinical
significance of these genes are critical for the development of
successful therapies.
The cylindromatosis (CYLD) gene was
originally identified as a tumor suppressor, the mutation of which
predisposes patients to the development of tumors of hair follicles
(cylindromas) (3). It has been
reported that CYLD acts as a negative regulator of the nuclear
factor-κB (NF-κB) signaling pathway by deubiquitinating NF-κB
essential modulator (NEMO), IκB kinase (IKK)-γ, and IKK upstream
regulators, including the tumor necrosis factor (TNF),
receptor-associated factor 2 (TRAF2), TRAF6, TRAF7 and
receptor-interacting protein 1 (RIP1) (4–10).
CYLD also regulates transforming growth factor-β (TGF-β) signaling
via the deubiquitination of Akt in lung fibrosis (11).
Recent studies have demonstrated that CYLD
deficiency may promote the development of several types of cancer
in addition to skin tumors caused by mutations and loss of the
heterozygosity (LOH) of CYLD. LOH of chromosome 16q, which
includes the CYLD gene, has been detected in a large
proportion of multiple myeloma cases and has been associated with
poor overall survival (12–14).
Comparative genomic hybridization (CGH) assays have also suggested
potential genetic abnormalities of CYLD (reduction in copy
number) in HCC, uterine carcinoma and renal cancer (15–17).
Moreover, suppressed CYLD gene expression may contribute to
tumor development in colon cancer, hepatocellular carcinoma and
melanoma (18,19).
The aim of this study was to investigate the
clinical importance of the CYLD gene by analyzing 124
consecutive patients with HCC who were treated with hepatic
resection. Distribution of the CYLD protein expression was also
examined using immunohistochemistry.
Materials and methods
Clinical tissue samples
Between 2005 and 2010, 124 patients (100 men and 24
women) with HCC were registered at the Department of
Gastroenterological Surgery, of the Kumamoto University Hospital
(Kumamoto, Japan). Specimens of primary HCC and adjacent normal
liver tissues were obtained from the patients after written
informed consent was obtained. This study was approved by the Human
Ethics Review Committee of the Graduate School of Medical Sciences,
Kumamoto University (Kumamoto, Japan).
RNA extraction and quantitative reverse
transcription-polymerase chain reaction (qRT-PCR)
Total RNA was obtained from the frozen tissue
samples and cell lines using a mirVana™ miRNA Isolation kit
(Ambion, Austin, TX, USA) according to the manufacturer’s
instructions. Reverse transcription was performed with 1.0
μg of total RNA as previously described (20). qRT-PCR was performed on a
LightCycler 480 II (Roche Diagnostics, Tokyo, Japan) using 2X PCR
Master mix (Roche Diagnostics) and Universal ProbeLibrary (Roche
Diagnostics). Primers were designed using the Roche website and the
Universal ProbeLibrary according to the manufacturer’s
instructions. The primers used were: CYLD, F: 5′-TCTATGG
GGTAATCCGTTGG-3′ and R: 5′-CAGCCTGCACACTCAT CTTC-3′, and universal
probe no. 83; and hypoxanthine phosphoribosyltransferase (HPRT), F:
5′-TGACCTTGATTTA TTTTGCATACC-3′ and R: 5′-CGA GCAAGACGTTCAGT
CCT-3′, and universal probe no. 73. HPRT, 18S ribosomal
RNA (rRNA) and glyceraldehyde 3-phosphate dehydrogenase
(GAPDH) were examined as the internal controls (21). HPRT was proved to be the
most suitable reference gene. For amplification, an initial
denaturation at 95°C for 10 min was followed by 45 cycles for 15
sec at 95°C, annealing 15 sec at 60°C, and extension 13 sec at
72°C. The experiments were performed twice to confirm
reproducibility.
Immunohistochemistry and evaluation of
CYLD
Paraffin-embedded tissue sections were dewaxed with
xylene and rehydrated using graded concentrations of ethanol. The
samples were then stained for CYLD using our previously described
technique (22). Endogenous
peroxidase activity was blocked using 3% hydrogen peroxide. The
sections were incubated in 200X diluted primary rabbit anti-CYLD
antibody (Sigma, Tokyo, Japan) overnight at 4°C. A subsequent
reaction was performed with a biotin-free horseradish peroxidase
enzyme-labeled polymer of the EnVision Plus detection system (Dako
Co., Tokyo, Japan). A positive reaction was visualized with a
3,3′-diaminobenzidine (DAB) solution, followed by counterstaining
with Mayer’s hematoxylin. Each immunohistochemical marker was
independently evaluated by two blinded investigators. CYLD
expression status in HCC cells was quantified as a percentage of
the total number of stained cells detected in ≥5 random high-power
fields (magnification, ×400) in each section. The positivity of
staining cells with 10% was determined as the cut-off value.
Statistical analysis
Statistical analysis was performed using the
JMP® 8.0 software (SAS Institute., Cary, NC, USA).
Values were presented as the mean ± standard deviation (SD).
Differences between groups were calculated using the Wilcoxon test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of CYLD in clinical tissue
specimens and their clinicopathological characteristics
We performed qRT-PCR analysis in the primary HCC
specimens. CYLD expression was quantified by caluculating the ratio
of CYLD to HPRT1 signal. CYLD expression was
detected in the tumor and non-tumor tissues. CYLD expression of
tumor tissue was not markedly different compared to that of
non-tumor liver tissue. For the clinicopathological evaluation,
patients were allocated into two groups based on the median value
of tumor-to-non-tumor (T/N) ratio of CYLD expression.
Patients with a T/N ratio larger than the median T/N ratio of
CYLD expression were allocated to the high expression group,
while the remaining patients comprised the low expression group.
Clinicopathological characteristics associated with the CYLD
expression status of the 124 patients are summarized in Table I. CYLD expression was only
correlated with the serum α-fetoprotein (AFP) value (P=0.0093).
 | Table ICYLD-mRNA expression and
patient clinicopathological characteristics. |
Table I
CYLD-mRNA expression and
patient clinicopathological characteristics.
| | CYLD (T/N ratio)
| |
|---|
| Clinicopathological
characteristics | No. of patients | High | Low | P-value |
|---|
| Agea (years) | | | | |
| <66 | 63 | 30 | 33 | 0.7637 |
| ≥66 | 61 | 32 | 29 | |
| Gender | | | | |
| Male | 100 | 49 | 51 | 0.4103 |
| Female | 24 | 13 | 11 | |
| AFPb (U/ml) | | | | |
| <15.2 | 68 | 41 | 27 | 0.0093 |
| ≥15.2 | 56 | 21 | 35 | |
| PIVKA-IIa (U/ml) | | | | |
| <108 | 61 | 31 | 30 | 0.5000 |
| ≥108 | 69 | 31 | 32 | |
| Tumor
diametera (mm) | | | | |
| <35.5 | 62 | 32 | 30 | 0.4288 |
| ≥35.5 | 62 | 30 | 32 | |
| No. of tumors | | | | |
| Solitary | 94 | 47 | 47 | 0.5829 |
| Multiple | 30 | 15 | 15 | |
|
Differentiation | | | | |
| Well/mod | 103 | 52 | 51 | 0.5000 |
| Poor | 21 | 10 | 11 | |
| Vascular
invasionc | | | | |
| Negative | 66 | 36 | 30 | 0.1345 |
| Positive | 56 | 24 | 32 | |
| HCV-Ab | | | | |
| Negative | 70 | 38 | 32 | 0.1826 |
| Positive | 54 | 24 | 30 | |
| HBs-Ag | | | | |
| Negative | 86 | 45 | 41 | 0.2796 |
| Positive | 38 | 17 | 21 | |
| Liver
cirrhosisd | | | | |
| Negative | 87 | 43 | 44 | 0.8444 |
| Positive | 37 | 19 | 18 | |
Correlation between CYLD expression and
prognosis
The correlation between each clinicopathological
characteristic and prognosis was analyzed by univariate analyses
(Table II). The data indicated
that poor prognosis in HCC patients correlated with tumor a
diameter of >35.5 mm (P<0.0001), multiple tumors (P=0.0048),
positive vascular invasion (P=0.0021), the protein induced by
vitamin K absence or antagonist (PIVKA)-II >108 (P=0.0278), and
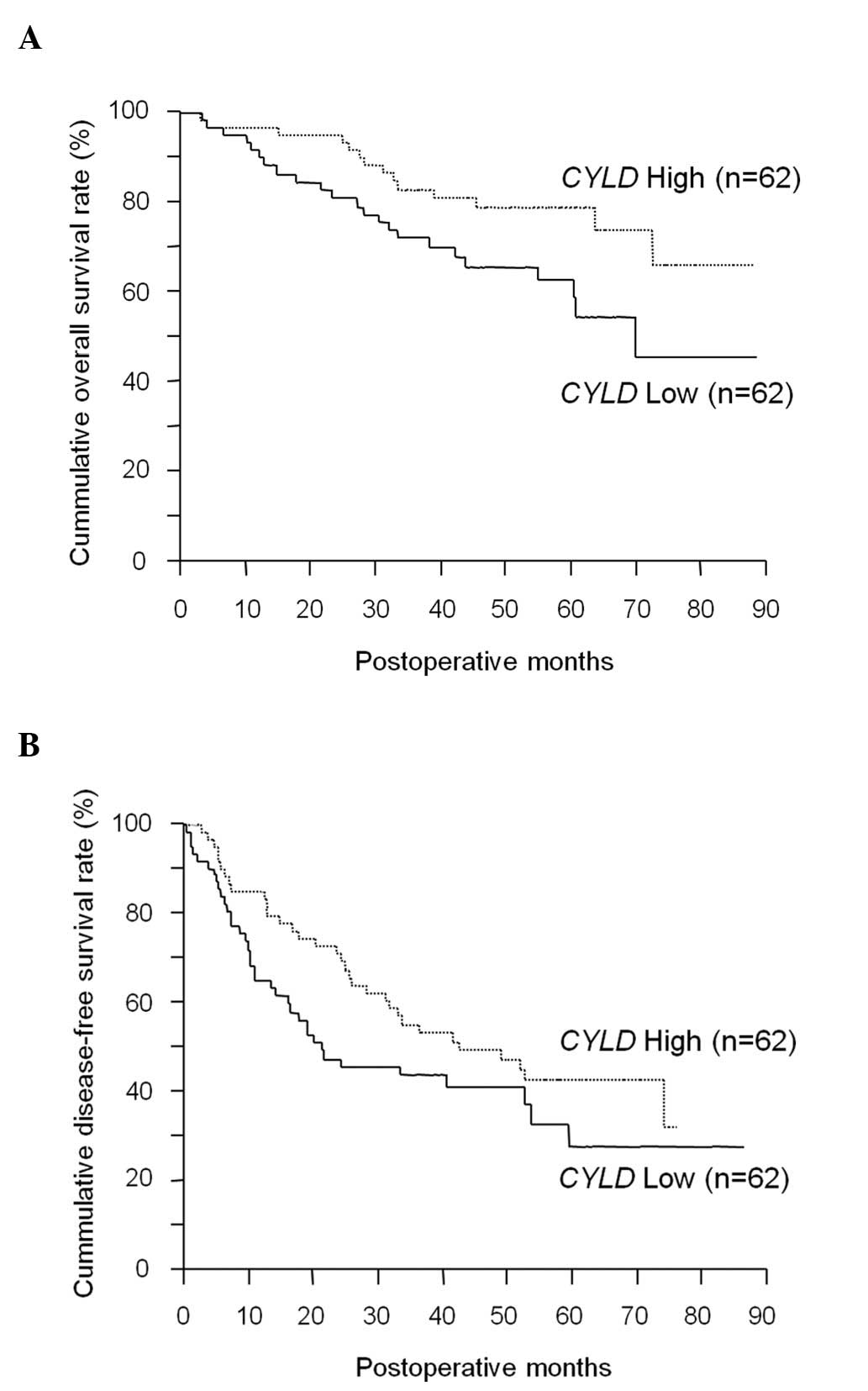
low CYLD expression (P=0.0406) (Fig. 1A). In the multivariate analysis,
CYLD expression was not an independent factor for predicting
poor prognosis (data not shown). Although CYLD expression
was not significantly correlated with disease-free survival
(P=0.1021) (Fig. 1B), the low
CYLD expression group had more patients with early
recurrence within 2 years (30/37 patients) compared to the high
CYLD expression group (17/31 patients; P=0.016).
 | Table IIUnivariate analysis of
clinicopathological characteristics for overall survival of
patients. |
Table II
Univariate analysis of
clinicopathological characteristics for overall survival of
patients.
| Clinicopathological
characteristics | No. of
patients | Median survival
(months) | P-value |
|---|
| Agea (years) | | | |
| <66 | 63 | 36.0 | 0.4168 |
| ≥66 | 61 | 21.6 | |
| Gender | | | |
| Male | 106 | 46.7 | 0.5799 |
| Female | 24 | 41.3 | |
| AFPb | | | |
| <15.2 | 68 | 38.7 | 0.5008 |
| ≥15.2 | 56 | 41.1 | |
| PIVKA-IIa | | | |
| <108 | 61 | 42.2 | 0.0278 |
| ≥108 | 63 | 38.2 | |
| Tumor
diametera (mm) | | | |
| <35.5 | 62 | 45.2 | <0.0001 |
| ≥35.5 | 62 | 33.1 | |
| No. of tumors | | | |
| Solitary | 94 | 41.2 | 0.0048 |
| Multiple | 30 | 36.6 | |
|
Differentiation | | | |
| Well/mod | 103 | 41.7 | 0.129 |
| Poor | 21 | 37.3 | |
| Vascular
invasionc | | | |
| Negative | 66 | 42.7 | 0.0021 |
| Positive | 56 | 38.1 | |
| HCV-Ab | | | |
| Negative | 72 | 42.0 | 0.8255 |
| Positive | 58 | 44.7 | |
| HBs-Ag | | | |
| Negative | 91 | 44.8 | 0.3037 |
| Positive | 39 | 42.0 | |
| Liver
cirrhosisd | | | |
| Negative | 87 | 42.7 | 0.7831 |
| Positive | 37 | 43.9 | |
| CYLD (T/N
ratio) | | | |
| Low | 62 | 41.1 | 0.0406 |
| High | 62 | 37.0 | |
Expression of CYLD protein
Among 70 HCC cases, 53 (75.7%) were positive for
CYLD expression. CYLD expression was heterogeneously distributed in
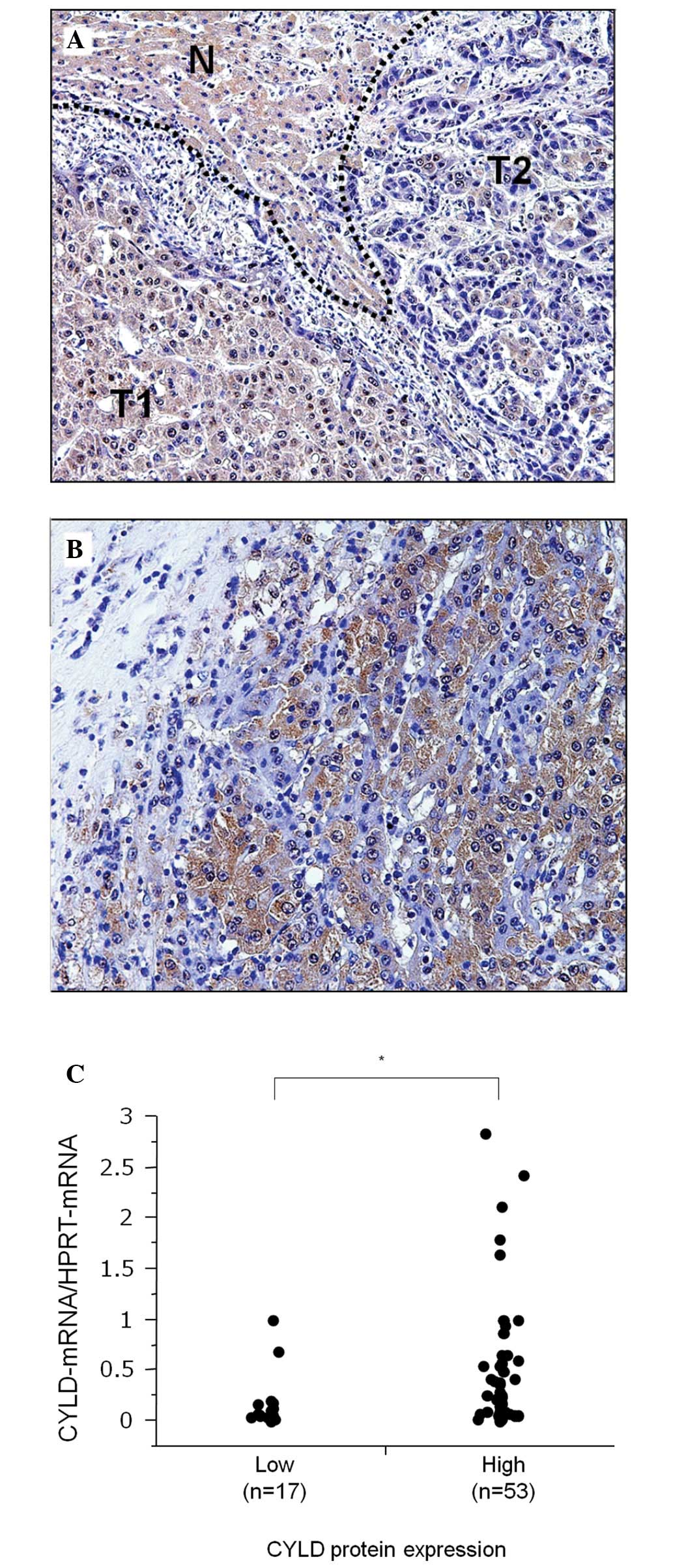
the tumor tissue and downregulated in tumor cells. In Fig. 2A, a representative case of HCC
shows that a number of tumor cells (T1) with a high CYLD expression
are well-differentiated and that they demonstrate a trabecular
pattern. Conversely, other tumor cells (T2) with low CYLD
expression lost their cell polarity and demonstrated dense
chromatin in the nucleus. Another case of HCC comprising tumor
cells with dense chromatin and a small nucleus that lost CYLD
expression, despite being surrounded by CYLD-expressing tumor cells
with more cytoplasm and only faint chromatin in the nucleus
(Fig. 2B). However, CYLD protein
expression was not associated with tumor-related factors, such as
tumor size, tumor diameter, vascular invasion, tumor
differentiation and prognosis (data not shown). To confirm the
correlation of CYLD-mRNA expression with protein expression,
CYLD-mRNA expression normalized by HPRT-mRNA
expression in tumor tissue was compared between the high and
low-CYLD protein expression groups. This finding showed that the
high-CYLD protein expression group demonstrated a markedly higher
CYLD-mRNA expression compared to the low-CYLD protein
expression group (P=0.036) (Fig.
2C).
Discussion
In this study, we showed that reduced
CYLD-mRNA expression is associated with a poor prognosis in
HCC patients, since the incidence of early recurrence (i.e., within
2 years) was higher in the low compared to the high-CYLD
expression group. The pattern of recurrence was similar between the
two groups. Since intrahepatic recurrence within 2 years is
considered an intrahepatic metastasis from the primary tumor, this
outcome suggests that CYLD is associated with metastatic
potential and, thus, a poor prognosis. CYLD-mRNA expression
demonstrated no correlation with tumor-related factors with the
exception of serum AFP. AFP production has been strongly associated
with specific molecular subtypes of HCC, such as hepatoblastoma
(23), while a reduced CYLD
expression may therefore be associated with a specific molecular
phenotype.
A recent in vivo study demonstrated that a
liver-specific conditional knockout of CYLD induced
apoptosis in hepatocytes via the chronic activation of
TGF-β-activated kinase 1 and c-Jun N-terminal kinase (JNK) in the
periportal area. As a result, this promoted progressive fibrosis
and inflammation, resulting in cancer development (24). Although CYLD expression was
expected to be potentially associated with certain types of
carcinogenesis from viral hepatitis or liver cirrhosis due to
chronic inflammation, no correlation was observed between
CYLD expression and non-tumor liver tissue. A previous in
vitro study demonstrated that HCC cells transfected with the
CYLD gene showed an increased NF-κB reporter activity
(18). The present study supports
the clinical and oncological importance of CYLD in HCC
progression.
A limited number of clinical studies have
investigated the protein expression and distribution of CYLD in
solid types of cancer such as HCC. Notably, in this study,
immunohistochemical analysis showed that CYLD expression was
distributed according to tumor cell morphology within the same
tumor, and tumor cells that lost their cell polarity tended to lose
CYLD expression. The mechanism underlying staining pattern remains
unclear, and further investigation is required to better understand
the role of CYLD in dysplastic cell morphology and chromatin
structure.
In conclusion, the present study suggests that CYLD
is associated with tumor development in HCC patients. This is a
preliminary study and, as a result, the functional aspect of CYLD
in HCC patients needs to be further investigated. However, the
present study is considered to be useful in investigating whether
CYLD may be a future molecular target in HCC patients.
Abbreviations:
|
AFP
|
α-fetoprotein;
|
|
CYLD
|
the cylindromatosis gene;
|
|
HCC
|
hepatocellular carcinoma;
|
|
PIVKA-II
|
protein induced by vitamin K absence
or antagonist-II;
|
|
qRT-PCR
|
quantitative reverse
transcription-polymerase chain reaction
|
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics. CA Cancer J Clin. 62:10–29. 2012.
|
|
2
|
Carr BI: Hepatocellular carcinoma: current
management and future trends. Gastroenterology. 127:S218–S224.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bignell GR, Warren W, Seal S, et al:
Identification of the familial cylindromatosis tumour-suppressor
gene. Nat Genet. 25:160–165. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jin W, Chang M, Paul EM, et al:
Deubiquitinating enzyme CYLD negatively regulates RANK signaling
and osteoclastogenesis in mice. J Clin Invest. 118:1858–1866. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reiley WW, Jin W, Lee AJ, et al:
Deubiquitinating enzyme CYLD negatively regulates the
ubiquitin-dependent kinase Tak1 and prevents abnormal T cell
responses. J Exp Med. 204:1475–1485. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wright A, Reiley WW, Chang M, et al:
Regulation of early wave of germ cell apoptosis and spermatogenesis
by deubiquitinating enzyme CYLD. Dev Cell. 13:705–716. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Stirling B, Temmerman ST, et al:
Impaired regulation of NF-kappaB and increased susceptibility to
colitis-associated tumorigenesis in CYLD-deficient mice. J Clin
Invest. 116:3042–3049. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brummelkamp TR, Nijman SM, Dirac AM and
Bernards R: Loss of the cylindromatosis tumour suppressor inhibits
apoptosis by activating NF-kappaB. Nature. 424:797–801. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kovalenko A, Chable-Bessia C, Cantarella
G, et al: The tumour suppressor CYLD negatively regulates NF-kappaB
signalling by deubiquitination. Nature. 424:801–805. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Trompouki E, Tsagaratou A, Kosmidis SK, et
al: Truncation of the catalytic domain of the cylindromatosis tumor
suppressor impairs lung maturation. Neoplasia. 11:469–476.
2009.PubMed/NCBI
|
|
11
|
Lim JH, Jono H, Komatsu K, et al: CYLD
negatively regulates transforming growth factor-β-signalling via
deubiquitinating Akt. Nat Commun. 3:7712012.
|
|
12
|
Jenner MW, Leone PE, Walker BA, et al:
Gene mapping and expression analysis of 16q loss of heterozygosity
identifies WWOX and CYLD as being important in determining clinical
outcome in multiple myeloma. Blood. 110:3291–3300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Annunziata CM, Davis RE, Demchenko Y, et
al: Frequent engagement of the classical and alternative NF-kappaB
pathways by diverse genetic abnormalities in multiple myeloma.
Cancer Cell. 12:115–130. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Keats JJ, Fonseca R, Chesi M, et al:
Promiscuous mutations activate the noncanonical NF-kappaB pathway
in multiple myeloma. Cancer Cell. 12:131–144. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hashimoto K, Mori N, Tamesa T, et al:
Analysis of DNA copy number aberrations in hepatitis C
virus-associated hepatocellular carcinomas by conventional CGH and
array CGH. Mod Pathol. 17:617–622. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hirai Y, Kawamata Y, Takeshima N, et al:
Conventional and array-based comparative genomic hybridization
analyses of novel cell lines harboring HPV18 from glassy cell
carcinoma of the uterine cervix. Int J Oncol. 24:977–986.
2004.PubMed/NCBI
|
|
17
|
Ströbel P, Zettl A, Ren Z, et al:
Spiradenocylindroma of the kidney: clinical and genetic findings
suggesting a role of somatic mutation of the CYLD1 gene in the
oncogenesis of an unusual renal neoplasm. Am J Surg Pathol.
26:119–124. 2002.PubMed/NCBI
|
|
18
|
Hellerbrand C, Bumes E, Bataille F, et al:
Reduced expression of CYLD in human colon and hepatocellular
carcinomas. Carcinogenesis. 28:21–27. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Massoumi R, Kuphal S, Hellerbrand C, et
al: Down-regulation of CYLD expression by Snail promotes tumor
progression in malignant melanoma. J Exp Med. 206:221–232. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Okabe H, Beppu T, Ueda M, et al:
Identification of CXCL5/ENA-78 as a factor involved in the
interaction between cholangiocarcinoma cells and cancer-associated
fibroblasts. Int J Cancer. Feb 15–2012.(Epub ahead of print).
|
|
21
|
Fu LY, Jia HL, Dong QZ, et al: Suitable
reference genes for real-time PCR in human HBV-related
hepatocellular carcinoma with different clinical prognoses. BMC
Cancer. 9:492009. View Article : Google Scholar
|
|
22
|
Okabe H, Beppu T, Hayashi H, et al:
Hepatic stellate cells may relate to progression of intrahepatic
cholangiocarcinoma. Ann Surg Oncol. 16:2555–2564. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee JS and Thorgeirsson SS: Functional and
genomic implications of global gene expression profiles in cell
lines from human hepatocellular cancer. Hepatology. 35:1134–1143.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nikolaou K, Tsagaratou A, Eftychi C, et
al: Inactivation of the deubiquitinase CYLD in hepatocytes causes
apoptosis, inflammation, fibrosis, and cancer. Cancer Cell.
21:738–750. 2012. View Article : Google Scholar : PubMed/NCBI
|